Abstract
The inositol phosphatase, MTMR4 (myotubularin-related protein 4), was identified as a novel interactor of the ubiquitin ligase Nedd4 (neural-precursor-cell-expressed developmentally down-regulated 4). hMTMR4 (human MTMR4) and Nedd4 co-immunoprecipitated and co-localized to late endosomes. The PY (Pro-Tyr) motif of hMTMR4 binds to WW (Trp-Trp) domains of hNedd4. MTMR4 expression was decreased in atrophying muscle, whereas Nedd4 expression was increased and hMTMR4 was ubiquitinated by hNedd4, suggesting that this novel interaction may underlie the biological process of muscle breakdown.
Keywords: inositol phosphatase, muscle atrophy, myotubularin, myotubularin-related protein 4 (MTMR4), neural-precursor-cell-expressed developmentally down-regulated 4 (Nedd4), ubiquitin ligase
Abbreviations: BCA, bicinchoninic acid; CCD, charge-coupled device; DMEM, Dulbecco's modified Eagle's medium; DSP, dual-specificity phosphatase; ENaC, epithelial sodium channel; GFP, green fluorescent protein; GST, glutathione transferase; h, human; HA, haemagglutinin; HECT, homologous with E6-associated protein C-terminus; HEK, human embryonic kidney; HPRT, hypoxanthine–guanine phosphoribosyltransferase; LAMP, lysosome-associated membrane protein; MTMR4, myotubularin-related protein 4; Nedd4, neural-precursor-cell-expressed developmentally down-regulated 4; PTP, protein tyrosine phosphatase; PX, phox homology domain; XLMTM, X-linked myotubular myopathy
INTRODUCTION
Cellular ubiquitination is a vital process regulating protein stability, internalization and endosomal trafficking [1–3]. It is a multi-enzyme cascade whereby ubiquitin moieties are selectively transferred on to target molecules, which, in many cases, tags them for degradation by the 26S proteasome. Specificity, in large part, is determined by the HECT (homologous with E6-associated protein C-terminus)- and RING (really interesting new gene)-type ubiquitin ligases that interact with their substrates via specific protein–protein interaction domains. Nedd4 (neural-precursor-cell-expressed developmentally down-regulated 4) is a ubiquitously expressed HECT-domain-containing E3 ubiquitin–protein ligase that has been shown to regulate the stability and/or localization of several membrane proteins through interactions of its WW (Trp-Trp) domain with substrate PY (Pro-Tyr) motifs [4–9].
Nedd4 has been shown previously to increase in models of skeletal muscle atrophy [10–12] and has been shown to target Notch1 in skeletal muscle [11], although the mechanisms by which Nedd4 contributes to the biological process of muscle breakdown are unknown. We sought to identify novel protein substrates of Nedd4 in muscle. Using a bioinformatics approach, we identified a PY-motif-containing protein expressed in skeletal muscle, MTMR4 (myotubularin-related protein 4), that belongs to the family of PTPs (protein tyrosine phosphatases)/DSPs (dual-specificity phosphatases) [13]. Myotubularins are 3′-phosphatases specific for PtdIns3P and PtdIns(3,5)P2. It has been proposed that myotubularins regulate endosome trafficking by dephosphorylation of PtdIns3P [14]. Interestingly, MTMR4 is the only family member that possesses a PY motif [15] and has a unique distribution to endosomes [14], the major site of substrate lipid accumulation. In the present study, we found that MTMR4 and Nedd4 co-immunoprecipitate, interact via a WW domain–PY motif interaction and co-localize in cells to the PtdIns3P-rich late/recycling endosomes. Using a rat model of denervation atrophy, we have confirmed that the increase in Nedd4 expression in atrophying muscle (compared with control muscle) is coincident with a decrease of MTMR4 in atrophying muscle. We demonstrate that Nedd4 ubiquitinates MTMR4 and that mutation of the MTMR4 PY motif inhibits Nedd4 binding and MTMR4 ubiquitination, suggesting that MTMR4 is a bona fide Nedd4 target. We propose that MTMR4 may play an important role in the progression of Nedd4-mediated skeletal muscle atrophy.
EXPERIMENTAL
Cell culture
All cells were cultured at 37 °C under 5% CO2. For biochemical characterization of the Nedd4–MTMR4 interaction, plasmids expressing wild-type hNedd4 (h indicates human) (GenBank® accession number NM_006154), catalytically inactive hNedd4 (Cys→Ser) and human MTMR4 (accession number NM_004687), a gift from Dr Joe Zhang (Vanderbilt-Ingram Cancer Center, Nashville, TN, U.S.A.), either untagged or HA (haemagglutinin)-tagged, wild-type or PY motif mutant (whereby the second invariant proline residue of the XPPXY motif was mutated to alanine using Stratagene QuikChange® mutagenesis kit), were transfected into Phoenix HEK (human embryonic kidney)-293 or HeLa cells grown in DMEM (Dulbecco's modified Eagle's medium), 10% FBS (fetal bovine serum), 100 units/ml penicillin and 100 mg/l streptomycin using standard CaCl2 transfection. At 48 h post-transfection, cells were harvested and lysed in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2 and 1 mM EGTA) vortex-mixed for 10 s and incubated on ice for 10 min. The lysate was then cleared by centrifugation at 12000 g for 10 min, and the protein concentrations in the supernatant were quantified using BCA (bicinchoninic acid) protein assay (Pierce).
Immunoprecipitation and Western blotting
For co-immunoprecipitation of MTMR4 and Nedd4, equal amounts of lysate from hMTMR4 or HA–hMTMR4 wild-type or PY mutant transfected or untransfected HeLa cells were incubated with 2 μg of anti-MTMR4 or anti-Nedd4 antibodies for 1 h on ice. Then, 50 μl of Protein A–Sepharose beads (50% slurry) was added to the lysate and incubated for 1 h at 4 °C while spinning. Beads were then precipitated by centrifugation at 10000 g for 1 min at 4 °C and washed twice in high-salt HNTG buffer (500 mM NaCl, 20 mM Hepes, pH 7.5, 10% glycerol and 0.1% Triton X-100) and three times in low-salt HNTG buffer (150 mM NaCl, 20 mM Hepes, pH 7.5, 10% glycerol and 0.1% Triton X-100). The beads were mixed with 45 μl of 1× Sample Buffer and boiled at 95 °C for 5 min to elute proteins. Protein eluate was resolved by SDS/8% PAGE and transferred on to Protran 0.2 μm pore-size nitrocellulose membranes (PerkinElmer).
Western blotting was performed using the following primary antibodies: polyclonal anti-Nedd4 WW2 antibodies (Upstate Biotechnology), 1:1000 dilution; polyclonal anti-MTMR4 antibodies (Abgent), 1:200 dilution; monoclonal anti-α-tubulin antibodies (Sigma–Aldrich), 1:10000 dilution; monoclonal anti-HA antibodies (Covance), 1:1000 dilution; monoclonal anti-ubiquitin antibodies (Covance), 1:1000 dilution and polyclonal anti-HPRT (hypoxanthine–guanine phosphoribosyltransferase) antibodies (Abcam), 1:1000 dilution. Protein bands were detected with horseradish-peroxidase-linked goat anti-rabbit or anti-mouse secondary antibodies (Cell Signaling Technology) used at a 1:20000 dilution.
Immunostaining
HeLa cells grown in DMEM were plated on coverslips. After 24 h, cells were transfected as indicated using FuGene™ 6 reagent (Hoffmann-La Roche). After a further 24 h, cells were fixed in 3.7% (w/v) paraformaldehyde, permeabilized in 0.2% Triton X-100 and blocked in 10% (w/v) non-fat dried skimmed milk powder in PBS. Primary antibodies were incubated for 1 h at room temperature (20 °C), and then were washed in PBS. Secondary antibodies conjugated to fluorophores were added for 45 min and washed, and coverslips were mounted on slides in Vectashield mounting medium (Vector Laboratories). Images were acquired using a spinning-disc confocal microscope (Zeiss) and Volocity software (Improvision, PerkinElmer). Primary antibodies used were: mouse anti-GM130 (BD Biosciences), mouse anti-LAMP-1 (lysosome-associated membrane protein 1) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, U.S.A.), mouse anti-(transferrin receptor) (Zymed Laboratories) and mouse anti-HA (Covance). Secondary antibodies used were Cy3 (indocarbocyanine)-conjugated anti-mouse and Cy2 (carbocyanine)-conjugated anti-mouse (Jackson ImmunoResearch Laboratories). In addition to HA-tagged MTMR4, some cells were transfected with either GFP (green fluorescent protein)-tagged Rab5, GFP-tagged Rab7, PX (phox homology domain)–mCherry (gifts from Dr Sergio Grinstein, Hospital for Sick Children, Toronto, Ontario, Canada) and/or GFP–hNedd4. For quantification of co-localization, images were analysed with the Volocity software co-localization feature that calculates measurement statistics including the Manders correlation coefficient.
In vitro binding assays
Human Nedd4-1 WW domains in pQE-30 (Qiagen) were a gift from Daniela Rotin (Hospital for Sick Children, Toronto). The WW domains were cloned with the following boundaries: WW I, 638–760 bp; WW II, 1112–1231 bp; WW III, 1318–1425 bp; WW IV, 1487–1606 bp. His6-tagged proteins were produced in M15 [pREP4] Escherichia coli bacteria and purified following the manufacturer's instructions (Qiagen). GST (glutathione transferase)-fusion proteins of hMTMR4 were produced by PCR of the PY-motif-containing region (2965–3081 bp) and a proline-rich region (1972–2091 bp), TA cloning into pCR2.1-TOPO (Invitrogen), subsequent cloning into pGEX-5X1 and expression in BL21(DE3) pLysS E. coli bacteria according to the manufacturer's protocol (Promega). GST-fusion proteins were bound on glutathione–agarose beads (50% slurry) by incubating for 1 h at 4 °C. Proteins were eluted with 30 mM reduced glutathione by incubating for 1 h at 4 °C. Eluted GST-fusion proteins and His6-bound proteins were quantified, and equal amounts of GST-fusion proteins of regions of hMTMR4 encompassing the PY motif (GST–PY) or a proline-rich region N-terminal to the PY motif (GST–Pro) were incubated with equal amounts of His6-bound proteins for 2 h at 4 °C. The beads and bound proteins were washed three times with high-salt HNTG buffer (as above) and twice with low-salt HNTG buffer. Beads were then mixed with 20 μl of 1× sample buffer and boiled at 95 °C for 5 min to elute proteins. The protein eluate was resolved by SDS/15% PAGE and transferred on to Protran 0.2 μm pore-size nitrocellulose and subjected to Western blotting with anti-His5 (Qiagen) and anti-GST antibodies (Sigma).
Experimental denervation model and muscle preparation
The model of muscle breakdown/atrophy was the gastrocnemius muscle denervation model described previously [16,17]. Briefly, the right tibial nerve of 18 male Lewis rats (200 g each) was transected under inhalational halothane anaesthesia completely denervating the gastrocnemius muscle. The contralateral leg served as an internal control in each animal. Rats were maintained under conditions of routine care for 30 days, after which they were killed, and the gastrocnemius muscles were harvested from the operated experimental limb and non-operated control limb. All procedures involving animals were carried out in accordance with the Canadian Council on Animal Care guidelines and were approved by the Animal Research Ethics Board of McMaster University. After rapid atraumatic dissection, the muscle was snap-frozen in liquid nitrogen. Half of the muscle was used for total RNA isolation, and the other half was used to extract soluble and insoluble cellular protein. Total protein was extracted by homogenizing (Polytron PT 1200E, Kinematica) the muscle in muscle lysis buffer (5 mM Tris/HCl, pH 8.0, 1 mM EDTA, 1 mM EGTA, 1 mM 2-mercaptoethanol, 1% glycerol, 1 mM PMSF, 10 μg/ml leupeptin and 10 μg/ml aprotinin) three times for 30 s, and homogenates were centrifuged at 1600 g for 10 min at 4 °C. The supernatant (soluble fraction) was centrifuged further at 10000 g for 10 min at 4 °C. The pellet (insoluble fraction) was washed with PBS, resuspended, dissolved by sonication in 200 μl of muscle lysis buffer containing 0.5% SDS and centrifuged further at 10000 g for 10 min at 4 °C, and the supernatant was designated as the insoluble fraction. All fractions were quantified using BCA protein assay and normalized for equal loading. Then, 25 μg of the muscle lysate was separated by SDS/8% PAGE, and Western blotting was performed using anti-MTMR4, anti-Nedd4 and anti-HPRT antibodies. The chemiluminescence signal was acquired using a CCD (charge-coupled device) camera (Bio-Rad Laboratories Fluor-S Max), and the total signal was quantified using Quantity One software (Bio-Rad Laboratories) with volume analysis.
Statistical analyses
Continuous data are reported as a mean±S.D. and were compared using ANOVA, followed by Tukey's post-analysis to compare multiple means if P<0.05 (statistical significance).
RESULTS
MTMR4 is a PY-motif-containing protein expressed in muscle that interacts with Nedd4
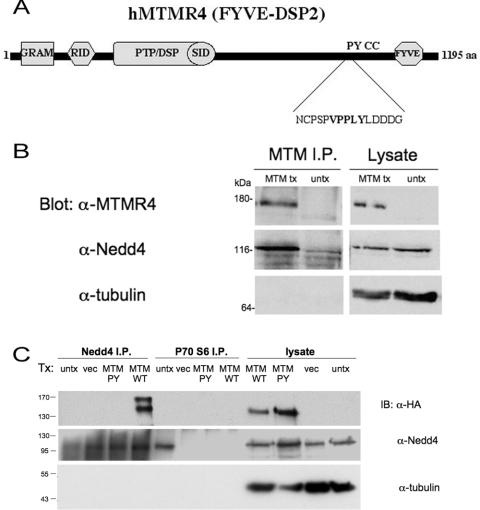
As Nedd4 overexpression was correlated with the progression of atrophic processes in muscle cells, we sought potential substrates in muscle that may be targeted by Nedd4 to elicit its effects. To this end, we took a bioinformatics approach to elucidate PY-motif-containing proteins that were expressed in skeletal muscle. Nedd4 consists of an N-terminal C2 domain, three or four WW domains and the ubiquitin ligase HECT domain and, in many cases, engages in interactions with its substrates via a WW domain–PY motif interaction [4–9]. Searching a SIDNET (Signalling Identification Network) MGC (Mammalian Gene Collection) clone database (http://www.sickkids.ca/Research/SIDNET/SIDNET.html) for PY-motif-containing proteins that were expressed in skeletal muscle limited potential substrates of Nedd4 to several proteins, including MTMR4, which belongs to a family of lipid tyrosine phosphatases with specificity towards PtdIns3P and PtdIns(3,5)P2 [14,15]. MTMR4 possesses several protein–protein and protein–lipid interaction domains (Figure 1A) including the FYVE domain, shown to mediate its localization to endosomes through its interaction with PtdIns3P and PtdIns(3,5)P2 [14] and is the only family member possessing a PY motif. The MTMR4 PY-motif (VPPLY) at the C-terminus (positions 1004–1008) is conserved across mammalian species (results not shown), adhering to the canonical L/XPPXY consensus sequence for WW domain type I binding. To verify that Nedd4 and MTMR4 can interact, cDNA for hMTMR4 was transfected into HEK-293 cells, and lysate from these cells was subjected to immunoprecipitation with anti-MTMR4 antibodies. Immunoprecipitates were separated by SDS/PAGE and probed with anti-MTMR4 and anti-Nedd4 antibodies to determine whether the two proteins interacted, with anti-tubulin antibodies used as a control. Immunoprecipitated transfected hMTMR4 interacted with endogenous Nedd4, but no band was detected in untransfected lysate (Figure 1B). Similarly, cell lysate transfected with HA-tagged hMTMR4 wild-type and PY-motif mutant was immunoprecipitated with anti-Nedd4 antibodies, and subjected to Western blotting with anti-Nedd4, anti-HA and anti-tubulin antibodies. Endogenous Nedd4 was able to immunoprecipitate transfected wild-type hMTMR4, but not the MTMR4 PY mutant, suggesting that the endogenous association of the two proteins is likely to be mediated through the WW domain–PY motif interaction (Figure 1C).
Figure 1. MTMR4 is a PY-motif-containing protein expressed in muscle that interacts with Nedd4.
(A) Schematic diagram of human MTMR4, based on [33]. Protein domains predicted with SMART (http://smart.embl-heidelberg.de/) and PFSCAN (http://hits.isb-sib.ch/cgi-bin/PFSCAN) include GRAM (glucosyltransferase, Rab-like GTPase activator of myotubularins) domain, RID (Rac-induced recruitment domain), PTP/DSP domain, SID (SET-interacting domain), PY motif (XPPXY), CC (coiled-coil) domain and FYVE domain. (B) Western blot of hMTMR4, Nedd4 and tubulin from transfected (MTM tx) or untransfected (untx) HEK-293 lysate immunoprecipitated with anti-MTMR4 antibodies (MTM I.P.). Equal amounts of whole lysate were run as a control. (C) Lysates (lysate) and immunoprecipitates using anti-Nedd4 antibodies (Nedd4 I.P.) or anti-(p70 S6 kinase) antibodies (p70 S6 I.P.) as negative control, of HeLa cells transfected with HA–hMTMR4 wild-type (MTM WT), HA–hMTMR4 PY mutant (MTM PY), pcDNA 3.1 vector alone (vec) or untransfected (untx). Immunoblotting (IB) was performed with anti-HA (monoclonal) antibodies followed by anti-Nedd4 (polyclonal) antibodies and blots were stripped and re-probed with anti-tubulin antibodies as control. Molecular masses are indicated in kDa.
Nedd4 and MTMR4 co-localize in cells
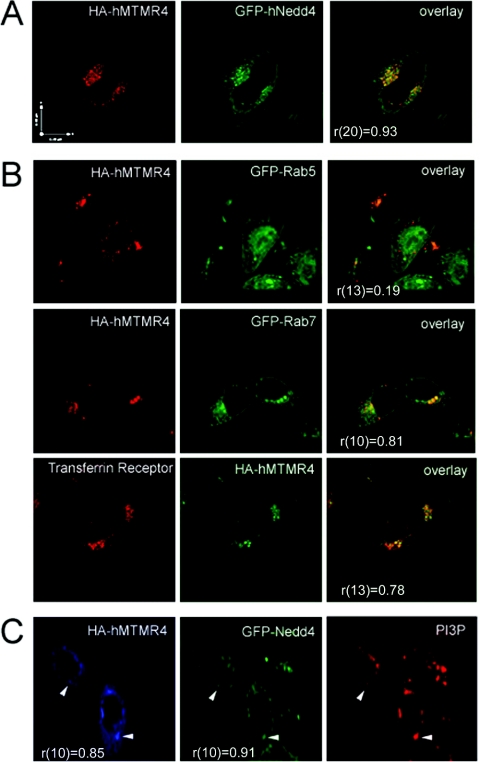
Previous studies have found that MTMR4 has unique distribution to endocytic compartments [14]. We examined co-localization of transfected hMTMR4 and hNedd4 in HeLa cells. In a representative image, the two proteins show a striking overlap with a punctate distribution (Figure 2A). Examination and counting of cells in 14 fields of view at 63× magnification showed that Nedd4 and MTMR4 co-localized in 72% of cells that were coexpressing both transfected proteins. Quantitative analysis of co-localization yielded high Mander's correlation coefficients, indicative of significant overlap of the Nedd4 and MTMR4 signal [r(20)=0.93, with r(n) being Mander's co-localization coefficient for n degrees of freedom, P<0.05]. To verify that these punctae are endosomal in nature, we examined the co-localization of hMTMR4 in HeLa cells with several markers of early, late and recycling endosomes, as well as Golgi and lysosomal markers (Figure 2B). No co-localization was observed of hMTMR4 and the early endosomal marker Rab5 [r(13)=0.19], nor was there significant overlap in MTMR4 signal with the lysosomal marker LAMP [r(9)=0.16] or the Golgi marker GM130 (cis-Golgi matrix protein of 130 kDa) [r(8)=0.09] (results not shown). There was significant overlap between MTMR4 and Rab7/transferrin (Figure 2B), indicative of localization to late and recycling endosomes, an impression that was validated quantitatively [r(10)=0.81, P<0.05 and r(13)=0.78, P<0.05 respectively]. Since PtdIns3P is a major lipid substrate of MTMR4, we co-transfected hMTMR4, GFP–hNedd4 and PX–mCherry (the PX domain of NADPH oxidase binds to PtdIns3P with high specificity [18]) to determine localization. hMTMR4 and hNedd4 co-localized specifically to areas of PX (PtdIns3P) staining (Figure 2C) consistent with endosomal localization [r(10)=0.85, P<0.05 and r(8)=0.91, P<0.05 respectively].
Figure 2. MTMR4 and Nedd4 co-localize to late endosomes/recycling endosomes in living cells.
(A) Immunofluorescence of HeLa cells transfected with HA-tagged hMTMR4 (red) and GFP–hNedd4 (green). Overlay shows merging of the two images. (B) Co-localization of HA–hMTMR4 with markers of late endosomes (Rab7) and recycling endosomes (transferrin receptor). HA–hMTMR4 does not co-localize with markers of early endosomes (Rab5). (C) Triple labelling of HeLa cells transfected with HA–hMTMR4 (blue), GFP–Nedd4 (green) and PX–mCherry (red) to represent PtdIns3P localization. Arrowheads point to examples of co-localization. Mander's co-localization coefficients, r (degrees of freedom), are shown for each set of co-staining. Scale bars, 11 μm.
Nedd4 and MTMR4 interact via a WW domain-PY motif interaction
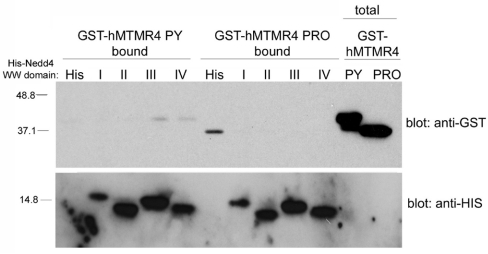
To verify that the interaction between the two proteins was mediated by a WW domain–PY motif interaction, we expressed His6-tagged versions of the four WW domains of human Nedd4 and incubated equal amounts of the fusion proteins (on Ni2+ beads) with GST–PY and GST–Pro as a control. We found that GST–PY consistently bound to WW III and IV of hNedd4 (n=3), but GST–Pro did not bind to any of the His6–WW fusion proteins (Figure 3), verifying that the interaction between hMTMR4 and hNedd4 is mediated through a PY motif–WW domain interaction. This finding with MTMR4 is consistent with other substrates of Nedd4, as hNedd4 WW IV corresponds to the high-affinity WW III in rat, which was found to bind to the PY motif of a subunit of the epithelial sodium channel, ENaC [19]. Mutation or deletion of the ENaC PY motif obliterates Nedd4 binding, resulting in Liddle's syndrome [19].
Figure 3. MTMR4 and Nedd4 binding is mediated through a PY motif–WW domain interaction.
GST-fusion proteins of the area encompassing the PY motif of hMTMR4 (GST-hMTMR4 PY) and of a non-PY-containing proline-rich region of hMTMR4 (GST-hMTMR4 PRO) as control were generated in bacteria as described in the Experimental section. Samples of 0.3 μg of soluble and GST–PY and GST–Pro were incubated with 0.5 μg of His6-tagged hNedd4–WWI, –WWII, –WWIII or –WWIV immobilized on Ni2+-nitrilotriacetate beads (His-hNedd4 WW domain I–IV) or His6–beads alone (His). Following thorough washes of the beads, bound proteins were eluted with sample buffer, separated by SDS/PAGE, and Western blotting was performed with anti-GST and anti-His antibodies. Molecular masses are indicated in kDa.
Nedd4 protein is increased in muscle atrophy
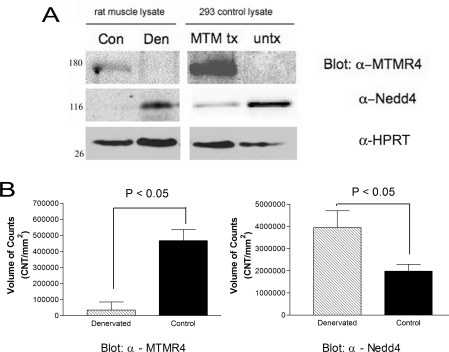
As we, and others, have shown that Nedd4 expression increases upon denervation induced atrophy [10,11], we hypothesized that any potential targets of Nedd4-mediated ubiquitination (and subsequent degradation) would be decreased. To determine whether MTMR4 protein levels are affected by denervation-induced atrophy, we performed Western blot analysis of protein lysate from experimental (denervated atrophied muscle) and control (contralateral innervated rat muscle) with anti-MTMR4 antibodies. We found that the antibody recognized several bands of different size (results not shown); many were differentially expressed in experimental compared with control limbs. The HEK-293 cell lysate, transfected with hMTMR4 cDNA, was run alongside the control and experimental insoluble muscle lysates to confirm the identity of MTMR4 in these fractions. A faint band (approx. 160 kDa) was recognized by the anti-MTMR4 antibodies in the control lysate, corresponding to what has been reported in previous studies with hMTMR4 [15] (Figure 4A). The intensity of the band decreased in the experimental (denervated) lysate compared with the control, whereas Nedd4 expression increased in the denervated muscle lysates compared with the control. The expression of HPRT remained unchanged. Quantification of the blots with CCD acquisition of the chemiluminescence signal shows that MTMR4 decreases approx. 90% with a 2-fold increase in Nedd4 expression (Figure 4B). These data suggest that decreased levels of MTMR4 protein correlate with increased levels of Nedd4 protein, supporting the notion of MTMR4 being a target of Nedd4-mediated degradation.
Figure 4. MTMR4 and Nedd4 are differentially regulated in atrophied muscle.
(A) Representative Western blot of MTMR4, Nedd4 and HPRT (as loading control) from gastrocnemius muscle insoluble protein fractions from denervated (Den) and contralateral control (Con) limbs. MTM tx is positive control lysate from MTMR4 transfected or untransfected (untx) HEK-293 cells. Molecular masses are indicated in kDa. (B) Quantification of the chemiluminescent signal from Western blots performed using Bio-Rad Fluor-S Max acquisition system and Quantity One software. Each bar represents the results from eight muscles (mean+S.E.M.). P<0.05 denotes statistically significant differences between mean values. CNT, counts.
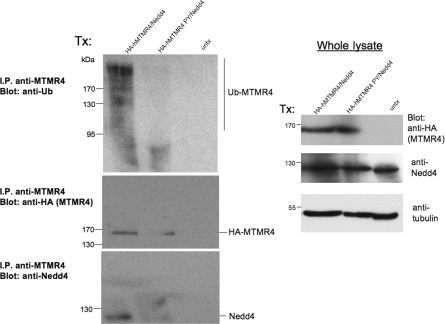
MTMR4 ubiquitination is dependent on an intact PY motif
MTMR4 co-precipitates with Nedd4 and has a differential expression pattern in muscle compared with that of Nedd4, so we hypothesized that it is a potential substrate for ubiquitination and degradation by Nedd4. Conjugation of ubiquitin moieties tags proteins for degradation by the proteasome, so we rationalized that, if MTMR4 was a bona fide target for degradation by Nedd4, then Nedd4 would ubiquitinate MTMR4. Mutation of the MTMR4 PY motif prevents Nedd4 binding and thus would spare the mutant MTMR4 from ubiquitination and subsequent degradation. Cellular lysates from HeLa cells transfected with HA-tagged hMTMR4, HA-tagged PY mutant hMTMR4 (both transfected together with hNedd4) or untransfected lysate were subjected to immunoprecipitation with anti-MTMR4 antibodies (Figure 5). Immunoprecipitates were separated by SDS/PAGE and probed with anti-ubiquitin, anti-HA and anti-Nedd4 antibodies to detect the expression of the proteins. Ubiquitinated MTMR4 (seen as a high-molecular-mass smear) was detected in immunoprecipitates only when wild-type MTMR4 was transfected, but not in immunoprecipitates from the PY-mutant-transfected or untransfected lysate (Figure 5). These data suggest that the MTMR4 PY motif is necessary for Nedd4 interaction and subsequent ubiquitination [20].
Figure 5. MTMR4 ubiquitination is dependent on an intact PY motif.
(A) HeLa cell lysate transfected with HA-tagged hMTMR4 or HA-tagged hMTMR4 PY motif mutant, together with hNedd4 or untransfected lysate (untx) were subjected to immunoprecipitation (I.P.) with anti-MTMR4 antibodies. Beads were washed and separated by SDS/PAGE with total lysate and then subjected to Western blotting with anti-ubiquitin (Ub) antibodies to detect ubiquitinated MTMR4. Blots were re-probed with anti-HA antibodies to detect transfected hMTMR4 and then stripped and re-probed with anti-Nedd4 antibodies to detect transfected Nedd4. Aliquots of whole-cell lysate were subjected to immunoblotting with anti-HA and anti-Nedd4 antibodies. Blots were stripped and re-probed with anti-α-tubulin antibodies as a loading control. Ubiquitinated MTMR4 is seen as a high-molecular-mass smear (top-left-hand panel) in the hMTMR4 transfected lysate only. Molecular masses are indicated in kDa.
DISCUSSION
In the present paper, we describe a novel interaction between the ubiquitin ligase Nedd4 and the DSP MTMR4, a regulator of phosphoinositide metabolism and thus of endosomal function. Although this interaction has not been previously identified, it is not unexpected, as previous studies have shown an association of Nedd4 with phosphoinositides and an involvement in endocytosis. The N-terminal C2 lipid-binding domain of Nedd4 has been shown to mediate Ca2+-dependent phospholipid binding and membrane association [21], in addition to mediating its own protein–protein interactions that serve to localize it to apical rafts [22]. The Nedd4 C2 domain displayed no preferences towards lipid composition in an In vitro binding assay, but was shown to bind phosphoinositides, and this binding was partially augmented in the presence of Ca2+. In addition, the yeast Nedd4 homologue, Rsp5, has a well-defined role in endocytosis; the C2 domain of Rsp5 localizes it to the plasma membrane, where Rsp5 binds membrane phosphoinositides directly and directs ubiquitination of endosomal cargo [23–25].
Little is known of the regulation and physiological role of MTMR4, but studies show a unique distribution to endosomal structures, the major site of substrate lipid (PtdIns3P) accumulation [14]. In the present study, we have shown that MTMR4 co-localizes with the late endosomal marker Rab7 in addition to the transferrin receptor, a marker for recycling endosomes. In addition, MTMR4 and Nedd4 both localize to areas of PtdIns3P staining, all suggesting that MTMR4 and Nedd4 are co-localized to late endosomes. Phosphatase-inactive forms of MTMR4 or overexpression of wild-type MTMR4 were shown to inhibit EGFR (epidermal growth factor receptor) degradation [14]. This effect was abrogated if the MTMR4 FYVE domain was mutated, and was recapitulated by the expression of the FYVE domain alone at very high levels, promoting the notion that this family member mediates endosomal trafficking by engaging in specific interactions between its FYVE domain and endosomal phosphoinositides, thus affecting the balance of PtdIns3P synthesis and degradation.
Mutations in other MTM family members are associated with muscle dysfunction. This is not unexpected, since muscle is the predominant location of MTM expression. In humans, MTM1 mutations are associated with XLMTM (X-linked myotubular myopathy), which is a congenital myopathy associated with major hypotonia at birth [26,27]. The histopathology of skeletal muscle reveals small rounded fibres with central nuclei. Mtm1-knockout mice recapitulate the histological signs of XLMTM and show a progressive myopathy starting at a few weeks after birth, whereas muscle histology appears normal at birth [26]. Adenoviral-mediated replacement of MTM1 reverses the myopathy completely, restoring muscle mass and contractility [28]. This suggests that the defect in the muscle fibres is due to a defect in structural maintenance rather than an impairment of myogenesis, as was hypothesized previously. Furthermore, knockdown of Caenorhabditis elegans MTMR3, the closest relative of MTMR4 and the only other family member to contain a FYVE domain, results in severe impairment of body movement, revealing a critical role for MTMR3 in maintaining muscle function [29]. In the present study, the finding that increased levels of Nedd4 in atrophied muscle correlate with a decrease in the levels of MTMR4 supports the notion of MTM family regulation of muscle structure and function.
The role of the Nedd4–MTMR4 interaction in the physiological progression of muscle atrophy can be speculated upon. Phosphoinositides are important membrane lipids that regulate cell growth and survival, cell division and membrane trafficking through recruiting and activating effector proteins and enzymes that contain phosphoinositide-binding domains, to specific membrane microdomains [30]. The MTM substrate PtdIns3P, which is highly enriched in early endosomes and internal vesicles of multivesicular endosomes late in the endosome pathway, plays a major role in endocytic trafficking [30,31]. Depletion of PtdIns3P results in inefficient sorting and delayed trafficking of a number of proteins (such as internalized growth factor receptors) through the endocytic pathways to the late endosome and lysosome. A main function of the other MTM substrate, PtdIns(3,5)P2, is to catalyse the budding of vesicles from the late endosome as yeast mutations in Fab1p/PIKfyve kinase [the enzyme that phosphorylates PtdIns3P to PtdIns(3,5)P2] results in enlarged poorly acidified vacuoles and late endosomes [32]. Thus decreased levels of MTM expression, or alternatively MTM loss-of-function mutants, probably result in cellular dysfunction because of inadequate dephosphorylation of PtdIns3P and PtdIns(3,5)P2 and dysregulation of the lipid signaling within the endocytic pathway. Experimental models examining the status of endosomal trafficking and receptor recycling in the absence of MTMR4 and/or mutants that are defective in its ubiquitination can address the contribution of Nedd4 to these processes and the participation of endosomal trafficking to the progression of muscle atrophy.
In conclusion, we have identified a novel Nedd4 target in muscle, the inositol phosphatase MTMR4, which implicates further a role for Nedd4 in regulating endosomal signalling and distinguishes a candidate for future studies examining the physiological role of Nedd4 action in atrophying muscle.
Acknowledgments
We gratefully acknowledge Lucy Osborne (University of Toronto), Chris Fladd (Hospital for Sick Children, SIDNET) and Brent Steer (University of Toronto) for their technical expertise and assistance.
FUNDING
This work was supported by the Canadian Institutes for Health Research. J. Batt was a recipient of a Canadian Institutes for Health Research Open Operating Grant [grant number FRN 74656] and Neuromuscular Research Partnership (an initiative of Amyotrophic Lateral Sclerosis Society of Canada, Muscular Dystrophy Canada and Canadian Institutes for Health Research) [grant number JNM-90959], P. J. P. was a recipient of a Canadian Institutes for Health Research postdoctoral fellowship, and N. G. is a recipient of a Canadian Institutes for Health Research M.D./Ph.D. studentship.
References
- 1.Staub O., Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol. Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 2.Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 4.Bouamr F., Melillo J. A., Wang M. Q., Nagashima K., de Los Santos M., Rein A., Goff S. P. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 2003;77:11882–11895. doi: 10.1128/JVI.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry P. C., Kanelis V., O'Brien M. C., Kim B., Gautschi I., Forman-Kay J., Schild L., Rotin D. Affinity and specificity of interactions between Nedd4 isoforms and the epithelial Na+ channel. J. Biol. Chem. 2003;278:20019–20028. doi: 10.1074/jbc.M211153200. [DOI] [PubMed] [Google Scholar]
- 6.Kanelis V., Bruce M. C., Skrynnikov N. R., Rotin D., Forman-Kay J. D. Structural determinants for high-affinity binding in a Nedd4 WW3* domain-Comm PY motif complex. Structure. 2006;14:543–553. doi: 10.1016/j.str.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 7.Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J. Cell Biol. 2006;175:631–645. doi: 10.1083/jcb.200603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 9.Leykauf K., Salek M., Bomke J., Frech M., Lehmann W. D., Durst M., Alonso A. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J. Cell Sci. 2006;119:3634–3642. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- 10.Batt J., Bain J., Goncalves J., Michalski B., Plant P., Fahnestock M., Woodgett J. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006;20:115–117. doi: 10.1096/fj.04-3640fje. [DOI] [PubMed] [Google Scholar]
- 11.Koncarevic A., Jackman R. W., Kandarian S. C. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J. 2007;21:427–437. doi: 10.1096/fj.06-6665com. [DOI] [PubMed] [Google Scholar]
- 12.Stevenson E. J., Koncarevic A., Giresi P. G., Jackman R. W., Kandarian S. C. Transcriptional profile of a myotube starvation model of atrophy. J. Appl. Physiol. 2005;98:1396–1406. doi: 10.1152/japplphysiol.01055.2004. [DOI] [PubMed] [Google Scholar]
- 13.Nandurkar H. H., Huysmans R. The myotubularin family: novel phosphoinositide regulators. IUBMB Life. 2002;53:37–43. doi: 10.1080/15216540210812. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzo O., Urbe S., Clague M. J. Systematic analysis of myotubularins: heteromeric interactions, subcellular localisation and endosome related functions. J. Cell Sci. 2006;119:2953–2959. doi: 10.1242/jcs.03040. [DOI] [PubMed] [Google Scholar]
- 15.Zhao R., Qi Y., Chen J., Zhao Z. J. FYVE-DSP2, a FYVE domain-containing dual specificity protein phosphatase that dephosphorylates phosphotidylinositol 3-phosphate. Exp. Cell Res. 2001;265:329–338. doi: 10.1006/excr.2001.5185. [DOI] [PubMed] [Google Scholar]
- 16.Bain J. R., Veltri K. L., Chamberlain D., Fahnestock M. Improved functional recovery of denervated skeletal muscle after temporary sensory nerve innervation. Neuroscience. 2001;103:503–510. doi: 10.1016/s0306-4522(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 17.Hynes N. M., Bain J. R., Thoma A., Veltri K., Maguire J. A. Preservation of denervated muscle by sensory protection in rats. J. Reconstr. Microsurg. 1997;13:337–343. doi: 10.1055/s-2007-1006413. [DOI] [PubMed] [Google Scholar]
- 18.Kanai F., Liu H., Field S. J., Akbary H., Matsuo T., Brown G. E., Cantley L. C., Yaffe M. B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 19.Kanelis V., Rotin D., Forman-Kay J. D. Solution structure of a Nedd4 WW domain–ENaC peptide complex. Nat. Struct. Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 20.Patterson C., Cyr D. M., editors. Berlin: Springer; 2005. Ubiquitin–Proteasome Protocols (Methods in Molecular Biology, vol. 301) [Google Scholar]
- 21.Plant P. J., Yeger H., Staub O., Howard P., Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 22.Plant P. J., Lafont F., Lecat S., Verkade P., Simons K., Rotin D. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J. Cell Biol. 2000;149:1473–1484. doi: 10.1083/jcb.149.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn R., Hicke L. Domains of the Rsp5 ubiquitin–protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell. 2001;12:421–435. doi: 10.1091/mbc.12.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn R., Hicke L. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 2001;276:25974–25981. doi: 10.1074/jbc.M104113200. [DOI] [PubMed] [Google Scholar]
- 25.Dunn R., Klos D. A., Adler A. S., Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 2004;165:135–144. doi: 10.1083/jcb.200309026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buj-Bello A., Biancalana V., Moutou C., Laporte J., Mandel J. L. Identification of novel mutations in the MTM1 gene causing severe and mild forms of X-linked myotubular myopathy. Hum. Mutat. 1999;14:320–325. doi: 10.1002/(SICI)1098-1004(199910)14:4<320::AID-HUMU7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Laporte J., Biancalana V., Tanner S. M., Kress W., Schneider V., Wallgren-Pettersson C., Herger F., Buj-Bello A., Blondeau F., Liechti-Gallati S., Mandel J. L. MTM1 mutations in X-linked myotubular myopathy. Hum. Mutat. 2000;15:393–409. doi: 10.1002/(SICI)1098-1004(200005)15:5<393::AID-HUMU1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 28.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C. R., Durand M., Kretz C., Danos O., Douar A. M., et al. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 2008;17:2132–2143. doi: 10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J., Zeng F., Ho W. T., Teng L., Li Q., Fu X., Zhao Z. J. Characterization and functional studies of a FYVE domain-containing phosphatase in C. elegans. J. Cell. Biochem. 2008;104:1843–1852. doi: 10.1002/jcb.21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson F. L., Dixon J. E. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 32.Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 33.Laporte J., Bedez F., Bolino A., Mandel J. L. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum. Mol. Genet. 2003;12:R285–R292. doi: 10.1093/hmg/ddg273. [DOI] [PubMed] [Google Scholar]