Abstract
The recycling of the amyloid precursor protein (APP) from the cell surface via the endocytic pathways plays a key role in the generation of amyloid β-peptide (Aβ) in Alzheimer’s Disease (AD). We report here that inherited variants in the SORL1 neuronal sorting receptor are associated with late-onset AD. These variants, which occur in at least two different clusters of intronic sequences may regulate tissue-specific expression of SORL1. We also show that SORL1 directs trafficking of APP into recycling pathways, and that when SORL1 is under-expressed, APP is sorted into Aβ-generating compartments. These data suggest that inherited or acquired changes in SORL1 expression or function are mechanistically involved in causing AD.
INTRODUCTION
The accumulation of Aβ peptide, a neurotoxic proteolytic derivative of the amyloid precursor protein (APP) is a central event in the pathogenesis of Alzheimer’s Disease (AD) 1. Thus, accumulation of Aβ in the brain is associated with disease-causing inherited variants in the amyloid precursor protein (APP) 2, presenilin 1 (PS1) 3 presenilin 2 (PS2) 4 and apolipoprotein E (APOE) genes 5,6. The generation of Aβ occurs in several subcellular compartments, but a principle location is during the re-entry and recycling of APP from the cell surface via the endocytic pathway (Figure 1B) 7–11. We reasoned that inherited variants in these pathways might modulate APP processing, and thereby might affect risk for AD. This concept is supported by prior reports that: 1) the expression of several candidate proteins within these pathways (e.g. SORL112, VPS35 13) are reduced in AD brain tissue; and 2) reductions in the expression of some of these proteins is associated with increased Aβ production 13–15. However, it is unclear whether these changes are causal or are simply reactive to AD.
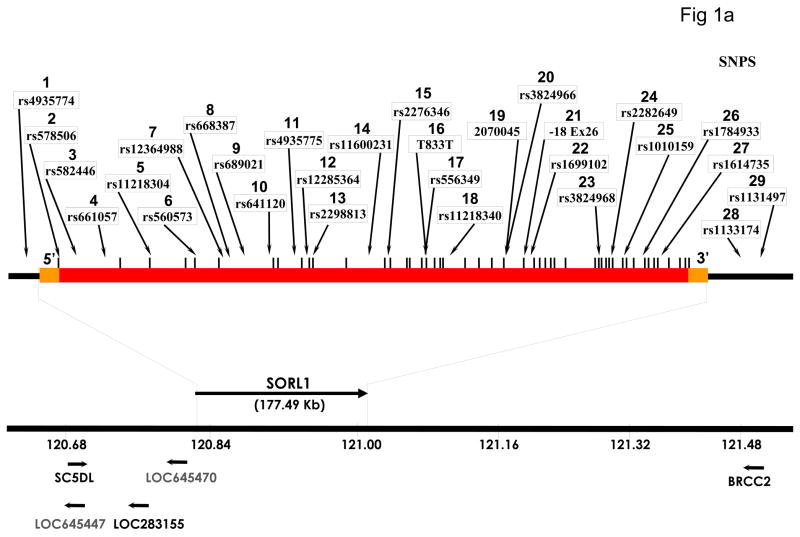
Figure 1.
Genomic map of SORL1 gene showing the location of SNPs genotyped in this study. Orange bars represent the 5′UTR and 3′UTR, red bar represents intragenic regions, vertical bars represent each of the 48 exons. SNPs 1, 28 and 29 are located in extragenic intervals.
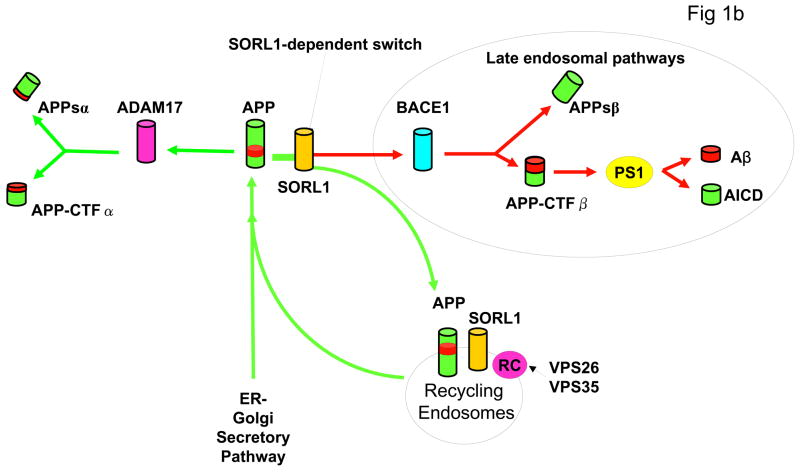
B: Diagram of APP processing pathways. APP holoprotein is synthesized in the endoplasmic reticulum (ER) and Golgi. Proteolytic cleavage through the Aβ peptide domain by ADAM17 and other α-secretase enzymes generates N-terminal soluble APPsα and membrane-bound APP-CTFα fragments. Sequential cleavage by BACE1 (β-secretase) generates N-terminal APPsβ and membrane bound APP-CTFβ fragments. The latter undergoes presenilin-dependent γ-secretase cleavage to generate Aβ and amyloid intracellular domain (AICD). SORL1 binds both APP holoprotein (see Fig. 3) and VPS35 (not shown) and acts as a sorting receptor for APP holoprotein. Absence of SORL1 switches APP holoprotein away from the retromer recycling pathway, and instead directs APP into the β-secretase cleavage pathway, increasing APPsβ production (Fig 2c) and then into the γ-secretase cleavage pathway to generate Aβ (see Fig. 2b). Blockade of the retromer complex (RC) by inhibiting retromer complex proteins such as VPS26 (Fig. 2d) or VPS35 has a similar effect, also increasing APPsβ and Aβ production.
To address this question, we investigated genetic associations between AD and single nucleotide polymorphisms (SNPs) in selected members of the vacuolar protein sorting (VPS) gene family including VPS35 (16q12); VPS26 (10q21); sortilin - SORT1 (1p21-p13); sortilin-related VPS10 containing receptors - SORCS1 (10q23-q25), SORCS2 (4p16), and SORCS3 (10q23-q25); and sortilin-related receptor, low density lipoprotein receptor class A repeats-containing - SORL1 (11q23-q24)]. The inheritance of SNPs from these genes was explored in six independent datasets that have sufficient power to detect modest gene effects (λs = 1.5). These datasets were collected from restricted ethnic origins in order to minimize the confounding effects of allelic heterogeneity 16,17. Indeed, two of these six datasets (Caribbean-Hispanic FAD and Israeli-Arab datasets), were drawn from population isolates with a limited number of founders 18,19.
These six datasets were divided into a “discovery cohort” composed of families with late-onset familial AD and a “replication cohort” composed of discordant sibships and case:control datasets. The FAD pedigrees in the discovery cohort (North European FAD = 124 families 20,21 and Caribbean-Hispanic FAD = 228 families 22 - Table 1) were interrogated with conservative family-based-association (FBAT) methods, which are less sensitive to population stratification. Positive results from the “discovery cohort” were then re-investigated in the “replication cohort” (Table 1). This replication cohort contained: 1) North European case:controls (178 sporadic AD cases, 242 controls of northwest European Caucasian) 20; 2) MIRAGE Caucasian sibships (276 Caucasian sibships from the MIRAGE Study) 23,24; 3) MIRAGE African-American sibships (238 African-American sibships from the MIRAGE Study) 23,24; and 4) Israeli-Arab case:controls (all 111 AD cases and 114 normal controls from the Wadi Ara population study) 19,25.
Table 1.
A: Characteristics of genotyped subjects in six independent datasets of families multiply affected by late-onset familial AD and sporadic case-control dataset. NA = not applicable.
B: Characteristics of the three Mayo datasets used for independent confirmation of the results in Caucasians. Age at diagnosis is shown for the JS and RS series, age at death for the AUT series.
| Sample Characteristics | Number of genotyped subjects in Familial datasets | Number of genotyped subjects in case:control datasets | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Source | North European | Caribbean Hispanic | MIRAGE Caucasian | MIRAGE African American | North European | Israeli Arab | Mayo JS | Mayo RS | Mayo AUT |
| Total number of AD families | 124 | 228 | 276 | 238 | |||||
| Total Subjects | 685 | 1180 | 531 | 371 | |||||
| Patients with AD | 321 | 605 | 279 | 244 | 178 | 111 | 549 | 433 | 423 |
| Controls | 242 | 114 | 477 | 1217 | 430 | ||||
| At-risk Relatives | 342 | 517 | 252 | 127 | |||||
| Nuclear sibships (≥2 siblings genotyped) | 163 | 303 | 186 | 122 | |||||
| Patients per family, mean (range) | 2.6 (1–6) | 2.7 (1–12) | 1.0 (1–2) | 1.0 (1–2) | |||||
| At-risk persons per family, mean (range) | 2.8 (0–18) | 2.3 (0–15) | 0.9 (0–4) | 0.5 (0–3) | |||||
| Mean age at onset (years) | 70 ± 9 | 73 ± 11 | 74 ± 8 | 77 ± 8 | 76 ± 7 | 83 ± 8 | 77±5 | 76±6 | 76±6 |
| Mean age of controls (years) | 73 ± 8 | 76 ± 6 | 75±6 | 77±5 | 74±6 | ||||
| Autopsy Confirmation (one per family or cases/controls) | 58 | 5 | 0 | 0 | NA/NA | NA/NA | 11/0 | 91/42 | 423/430 |
Independent replication was obtained from three large datasets of American Caucasians that were independently ascertained, genotyped and analyzed statistically at the Mayo Clinic.
RESULTS
SNPs in SORL1 are associated with late-onset AD
At least two SNPs in the intragenic sequences of the SORL1, VPS26, VPS35, SORCS1, SORCS3, SORCS2, and SORT1 genes were initially screened for association with AD in the two independent FAD “discovery datasets”. No allelic associations were observed with VPS26, VPS35, SORCS3, or SORT1 (Supplemental Table 1, 2). However, one SNP in SORCS1 (rs7082289: p = 0.013), one SNP in SORCS2 (rs7694823: p = 0.01) and two SNPs in SORL1 exhibited nominally significant association in at least one of the FAD datasets (rs2298813: p = 0.012; rs2070045: p = 0.031).
To validate these initial results, a second series of SNPs from the SORCS1, SORCS2 and SORL1 genes were investigated in the two FAD discovery datasets. No association was detected with the additional SNPs in SORCS1 (total = 9 SNPs) or in SORCS2 (total = 6 SNPs) (Supplemental Table 1, 2). However, six SNPs clustered in two distinct regions of the SORL1 gene were significantly associated with AD in at least one discovery dataset, and also in at least one replication dataset (Table 3 and Supplemental Table 3) (0.0031 ≤ p ≤ 0.014). Importantly, at five of these SNPs, the alleles associated with AD were identical in both the discovery and replication datasets (Table 3 and Supplemental Table 3). Thus, at the 5′-end of SORL1, AD was associated with the “C”, “G” and “C” alleles at SNPs 8, 9 and 10 respectively in the Caribbean-Hispanic FAD (p = 0.013, 0.017, and 0.021), Israeli-Arab (p = 0.002, 0.007, and 0.005), and North European case:control datasets (p = 0.021, 0.04, and 0.067) (Table 3 and Supplemental Table 3). Similarly, at the 3′-end of SORL1, AD was associated with the “G” and “T” alleles at SNPs 19 and 23 respectively in the North European FAD (p= 0.031; 0.0031) and North European case:control datasets (p = 0.00082, 0.00073) (Table 3 and Supplemental Table 3). Post-hoc statistical adjustment for APOE genotype, age and gender did not alter the conclusions that: 1) there were allelic associations between AD and two clusters of SNPs in distinct regions of SORL1 in different datasets; and 2) that these associations replicated in multiple independent datasets.
Table 3.
Single SNP association results (FBAT p-values) generated for the primary analyses in two independent “discovery datasets” consisting of families with multiple cases of late-onset AD (North European FAD and Caribbean-Hispanic FAD datasets). After adjustment for multiple testing with an FDR level of 0.1, the cutoff p-values for significant association are 0.024 in the North European family dataset and 0.003 in the Hispanic family dataset. MAF = minor allele frequency. Supplemental Table 2A also shows allele and genotype frequencies. Nominally significant p-values are in bold; * = association is significant after correction for multiple testing. The alleles putatively associated with AD are depicted only for SNPs generating nominal p-values of p ≤ 0.10.
Separate confirmatory analyses were performed in four independent “replication cohorts” including two case:control datasets (of North European origin or from an Israeli-Arab inbred population isolate), and two familial datasets (siblings) from the MIRAGE study (Caucasian Americans and the other of African-Americans). Corrections for multiple testing were not applied in these directed replication analyses. Boxes highlight identical alleles that are nominally associated with disease in at least two independent datasets. ND –not determined. NA – not available. The p-values in the case:control cohort are for allelic association. Alleles putatively associated with AD are depicted only for those SNPs generating nominal p-values of p < 0.10. Complete data for all SNPs is contained in Supplemental Table 3.
| SNP | Discovery Datasets | Replication Datasets | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| North European Families | Caribbean Hispanic Families | Israeli Arab case:control | North European case:control | ||||||||||||||
| SNP# | Minor Allele | MAF | Info Families | P | Risk Allele | MAF | Info Families | P | Risk Allele | MAF | P | OR (95%CI) | Risk Allele | MAF | P | OR (95%CI) | Risk Allele |
| 8 | T | 0.429 | 51 | 0.515 | 0.388 | 80 | 0.013 | C | 0.399 | 0.002 | 1.84 (1.25 – 2.71) | C | 0.392 | 0.021 | 46 (1.06 – 2.01) | C | |
| 9 | A | 0.429 | 54 | 0.708 | 0.379 | 78 | 0.017 | G | 0.397 | 0.007 | 1.70 (1.15 – 2.50) | G | 0.398 | 0.040 | 37 (1.01 – 1.86) | G | |
| 10 | T | 0.429 | 55 | 0.565 | 0.374 | 79 | 0.021 | C | 0.385 | 0.0051 | 1.74 (1.18 – 2.56) | C | 0.390 | 0.067 | 32 (0.98 – 1.77) | C | |
| 17 | T | 0.277 | 51 | *0.0057 | T | 0.488 | 91 | 0.170 | 0.384 | 0.042 | 1.49 (1.01 – 2.18) | G | 0.321 | 0.205 | 24 (0.89 – 1.72) | ||
| 19 | G | 0.207 | 42 | 0.031 | G | 0.245 | 84 | 0.617 | 0.236 | 0.499 | 1.16 (0.75 – 1.80) | 0.287 | 0.00082 | 79 (1.27 – 2.53) | G | ||
| 23 | T | 0.264 | 53 | *0.0031 | T | 0.288 | 78 | 0.513 | 0.345 | 0.672 | 1.09 (0.74 – 1.61) | 0.125 | 0.00073 | 16 (1.37 – 3.40) | T | ||
Haplotypic analyses using a sliding window method 26 and window size of three contiguous SNPs confirmed the single SNP analyses, demonstrating replicated haplotypic associations in two regions of SORL1 in different datasets (Table 4 and Supplemental Table 4). Thus, at the 5′ end of SORL1, the “CGC” haplotype at SNPs 8, 9, 10 was associated with AD in the Caribbean-Hispanic FAD (global-p = 0.0098, haplotype-p = 0.0053, haplotype frequency = 0.614 v. 0.583 in controls); Israeli-Arab (global-p = 0.023 haplotype-p = 0.0085, frequency = 0.661 in cases vs.0.539 in controls); and North European case:control datasets (haplotype-p = 0.045, frequency = 0.638 in cases vs. 0.566 in controls) (Table 4, and Supplemental Table 4). In the Israeli-Arab dataset, the overlapping “GCC” haplotype at SNPs 9, 10 and 11 showed even greater evidence for association (global-p = 0.0080; haplotype-p = 0.0047). As might be expected, SNPs 8, 9, and 10 also posses a protective haplotype. The “TAT” haplotype at SNPs 8, 9, 10 was associated with decreased risk of AD in these datasets (Hispanic FAD: haplotype-p = 0.0086; haplotype frequency = 0.353 vs. 0.394 in controls); Israeli-Arab: frequency = 0.301 in cases vs. 0.434 in controls, haplotype-p = 0.0037; North European Caucasian: frequency = 0.351 in cases vs. 0.417 in controls, haplotype-p = 0.068).
Table 4.
Haplotypes for all three-SNP windows that have a global p-value for association with or against AD of p ≤ 0.05 in at least one dataset. In this table all p-values of p ≤ 0.05 are in bold. ND = not done due to a deviation from HWE in control samples in this dataset. Haplotypes that show increased risk for AD in at least two independent datasets are highlighted in black. Haplotypes showing increased risk in one dataset and reduced risk in a dataset with different ethnic/racial origins are in dark grey. Haplotypes that show reduced risk in at least two independent datasets are highlighted in light grey. Complete data for all SNPs is contained in Supplemental Table 4.
| SNP | Discovery Datasets | Replication Datasets | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP# | HAP | North European Families | Caribbean Hispanic Families | Israeli Arab case:control | North European case:control | MIRAGE African-Americans Sibs | ||||||||||||||||||||||||
| Hap frequency |
Info Families |
Z Score | Hap p-value |
Global sim p |
Hap frequency |
Info Families |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Hap frequency |
Info Families |
Z Score | Hap p-value |
Global sim p |
||||||
| 8 | 9 | 10 | C | G | C | 0.579 | 52 | −0.392 | 0.695 | 0.152 | 0.638 | 75 | 2.786 | 0.0053 | 0.0098 | 0.539 | 0.661 | 2.633 | 0.0085 | 0.023 | 0.566 | 0.638 | 2.001 | 0.045 | 0.154 | 0.473 | 50 | −0.439 | 0.661 | 0.727 |
| 8 | 9 | 10 | T | A | T | 0.395 | 52 | 0.893 | 0.372 | 0.317 | 72 | −2.628 | 0.0086 | 0.434 | 0.301 | −2.901 | 0.0037 | 0.417 | 0.351 | −1.826 | 0.068 | 0.076 | 26 | 0.510 | 0.610 | |||||
| 9 | 10 | 11 | G | C | C | 0.404 | 57 | −1.259 | 0.208 | 0.553 | 0.217 | 62 | −1.202 | 0.229 | 0.022 | 0.282 | 0.407 | 2.828 | 0.0047 | 0.0080 | 0.385 | 0.455 | 1.972 | 0.049 | 0.223 | 0.071 | 23 | −0.550 | 0.582 | 0.682 |
| 9 | 10 | 11 | A | T | A | 0.380 | 57 | 0.860 | 0.390 | 0.319 | 83 | −2.006 | 0.045 | 0.410 | 0.247 | −3.390 | 0.0007 | 0.393 | 0.323 | −1.904 | 0.057 | 0.112 | 23 | 0.280 | 0.780 | |||||
| 9 | 10 | 11 | G | C | A | 0.178 | 42 | 0.465 | 0.642 | 0.428 | 79 | 2.557 | 0.011 | 0.256 | 0.251 | −0.258 | 0.796 | 0.184 | 0.191 | 0.337 | 0.736 | 0.399 | 55 | 0.148 | 0.883 | |||||
| 22 | 23 | 24 | C | T | T | 0.188 | 45 | 2.779 | 0.0054 | 0.018 | 0.205 | 66 | −0.103 | 0.918 | 0.554 | 0.250 | 0.227 | −0.579 | 0.563 | 0.286 | 0.069 | 0.134 | 2.795 | 0.0052 | 0.00065 | 0.035 | 11 | 0.589 | 0.556 | 0.314 |
| 22 | 23 | 24 | C | A | T | 0.003 | 2 | * | * | 0.008 | 3 | * | * | * | * | * | * | 0.170 | 0.082 | -3.096 | 0.0020 | 0.001 | 1 | * | * | |||||
| 22 | 23 | 24 | T | T | T | 0 | 4 | * | * | 0.019 | 7 | * | * | 0.015 | 0.041 | 1.387 | 0.165 | 0.011 | 0.033 | 2.272 | 0.023 | 0.007 | 3 | * | * | |||||
| 23 | 24 | 25 | A | C | T | 0.735 | 58 | −1.742 | 0.082 | 0.041 | 0.551 | 80 | 1.351 | 0.177 | 0.824 | 0.535 | 0.563 | 0.668 | 0.504 | 0.547 | 0.667 | 0.674 | 0.129 | 0.897 | 0.00035 | 0.513 | 44 | 3.029 | 0.0025 | 0.0043 |
| 23 | 24 | 25 | T | T | C | 0.196 | 47 | 1.878 | 0.060 | 0.220 | 70 | −0.811 | 0.417 | 0.265 | 0.259 | −0.135 | 0.892 | 0.085 | 0.167 | 3.268 | 0.0011 | 0.047 | 18 | −0.252 | 0.801 | |||||
| 23 | 24 | 25 | A | C | C | 0.037 | 13 | 0.475 | 0.635 | 0.183 | 51 | −0.608 | 0.543 | 0.103 | 0.103 | −0.057 | 0.955 | 0.066 | 0.046 | −1.080 | 0.280 | 0.403 | 53 | −2.852 | 0.0044 | |||||
| 23 | 24 | 25 | A | T | C | 0.004 | 2 | * | * | 0.008 | 3 | * | * | * | * | * | * | 0.168 | 0.097 | −2.508 | 0.012 | 0.003 | 1 | * | * | |||||
A second cluster of replicated haplotypic associations was observed at the 3′ end of SORL1 in the North European datasets. Thus, the overlapping haplotypes of “CTT” at SNPs 22–24 and “TTC” at SNPs 23–25 were associated with AD in the North European FAD and North European case:control datasets (0.001 < haplotype-p < 0.02; Table 4 and Supplemental Table 4). This region of SORL1 also revealed significant haplotypic associations in the MIRAGE African-American sibships. However, the haplotypic associations at SNPs 23–25 in the MIRAGE African-American sibships were with different haplotypes (global-p = 0.0043; disease-associated “ACT” haplotype-p = 0.0025, frequency = 0.513; protective “ACC” haplotype-p = 0.0044, frequency = 0.403) (Table 4 and Supplemental Table 3). The conclusion that there are at least two distinct regions of SORL1 that are associated with AD in different ethnic groups was supported when shorter or longer haplotypes were examined (Supplemental Tables 5–8).
To provide a completely independent confirmation of the association between AD and SORL1, SNPs 4, 5, 8, 9, 12, 19, and 22–25 were genotyped and analyzed in an independent facility in three series of Caucasian cases and controls ascertained at the Mayo Clinic (n = 1400 late onset AD cases; 2113 controls, Table 1B)27,28. The North European Caucasians and the American Caucasians in the Mayo datasets have slightly different allele frequencies and haplotype structures and may therefore have slightly different ethnic origins. Nevertheless, significant associations were observed at SNPs 4, 12, 19 and 23–25 in the overall Mayo dataset (single SNP: 0.009 ≤ p ≤ 0.046), and two of the three subdatasets individually gave highly significant results (0.003 < p < 0.007) for one or more of these SNPs (Table 5). Importantly, the alleles and haplotypes at SNPs 19, 22–25 that were associated with increased risk for AD in the Mayo datasets (black highlight in Tables 5 and 6) were the same as those associated with increased risk for AD in both the North European FAD dataset and in the North European case:control dataset (black highlight in Tables 3 and 4). Moreover, when all of the Caucasian case:control samples are considered together (n = 1554 AD cases, 2333 controls) the associations remained robust (single SNP: = 0.002 ≤ p ≤,0.04 with three SNPs giving p < 0.008). Intriguingly, both the Mayo dataset and the overall Caucasian case:control also detected association with SNP 4 (p = 0.009 and p = 0.002 respectively), a result not evident in the individual datasets.
Table 5.
A:Independent confirmation of the association of AD with SORL1 in Caucasians was obtained by genotyping 10 SNPs (4, 5, 8, 9, 12, 19, and 22–25) in three additional series of American Caucasians from the Mayo Clinic 27,28. The χ2 test was used to assess allelic associations between AD and SNPs. Corrections for multiple testing were not applied in these directed replication analyses. SNPs that show increased risk for AD are highlighted in black. All six SNPs that showed significant association with AD in the combined Mayo series had ORs >1 in all three series. Meta analysis of each of these six SNPs (Mantel-Haenzsel, fixed or random effects) in the three Mayo series also showed significant association.
| SNP | JS series | RS series | AUT series | Combined Mayo series | Combined Caucasian datasets | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP# | Minor Allele | MAF | P | OR (95%CI) | Risk Allele | MAF | P | OR (95%CI) | Risk Allele | MAF | P | OR (95%CI) | Risk Allele | MAF | P | OR (95%CI) | Risk Allele | MAF | P | OR (95%CI) | Risk Allele |
| 4 | C | 0.418 | 0.306 | 1.10 (0.92–1.31) | 0.455 | 0.504 | 1.05 (0.90–1.23) | 0.424 | 0.076 | 1.19 (0.98–1.45) | 0.437 | 0.009 | 1.14 (1.03–1.25) | T | 0.431 | 0.002 | 1.16 (1.05–1.27) | T | |||
| 5 | C | 0.413 | 0.876 | 0.99 (0.83–1.18) | 0.397 | 0.373 | 1.08 (0.92–1.26) | 0.397 | 0.301 | 1.11 (0.91–1.35) | 0.402 | 0.227 | 1.06 (0.96–1.17) | 0.403 | 0.081 | 1.09 (0.99–1.19) | |||||
| 8 | C | 0.431 | 0.187 | 1.13 (0.94–1.34) | 0.440 | 0.692 | 0.97 (0.83–1.13) | 0.431 | 0.071 | 1.20 (0.98–1.45) | 0.436 | 0.113 | 1.08 (0.98–1.19) | 0.432 | 0.027 | 1.11 (1.01–1.22) | C | ||||
| 9 | A | 0.436 | 0.202 | 1.12 (0.94–1.34) | 0.443 | 0.640 | 0.96 (0.82–1.13) | 0.448 | 0.313 | 1.10 (0.97–1.34) | 0.442 | 0.322 | 1.05 (0.95–1.16) | 0.438 | 0.109 | 1.08 (0.98–1.18) | |||||
| 12 | T | 0.044 | 0.548 | 1.14 (0.74–1.75) | 0.050 | 0.606 | 1.10 (0.77–1.55) | 0.052 | 0.003 | 1.98 (1.26–3.12) | T | 0.049 | 0.046 | 1.25 (1.00–1.56) | T | 0.049 | 0.087 | 1.20 (0.97–1.48) | |||
| 19 | G | 0.224 | 0.210 | 1.14 (0.93–1.41) | 0.242 | 0.055 | 1.19 (1.00–1.42) | 0.230 | 0.225 | 1.15 (0.92–1.44) | 0.234 | 0.038 | 1.13 (1.01–1.26) | G | 0.238 | 0.0023 | 1.18 (1.06–1.31) | G | |||
| 22 | C | 0.321 | 0.052 | 1.20 (1.00–1.45) | 0.349 | 0.413 | 1.07 (0.91–1.26) | 0.331 | 0.227 | 1.13 (0.92–1.39) | 0.336 | 0.108 | 1.09 (0.98–1.20) | 0.334 | 0.119 | 1.08 (0.98–1.19) | |||||
| 23 | T | 0.296 | 0.006 | 1.31 (1.08–1.59) | T | 0.321 | 0.287 | 1.09 (0.93–1.29) | 0.302 | 0.294 | 1.12 (0.91–1.37) | 0.309 | 0.031 | 1.12 (1.01–1.24) | T | 0.292 | 0.0075 | 1.15 (1.04–1.27) | T | ||
| 24 | T | 0.278 | 0.007 | 1.31 (1.08–1.59) | T | 0.299 | 0.513 | 1.06 (0.89–1.25) | 0.281 | 0.199 | 1.15 (0.93–1.42) | 0.289 | 0.042 | 1.12 (1.00,1.24) | T | 0.286 | 0.040 | 1.11 (1.00–1.23) | T | ||
| 25 | C | 0.333 | 0.001 | 1.35 (1.12–1.63) | C | 0.355 | 0.712 | 1.03 (0.88–1.21) | 0.340 | 0.163 | 1.15 (0.94–1.41) | 0.345 | 0.026 | 1.12 (1.01–1.24) | C | 0.343 | 0.033 | 1.11 (1.01–1.22) | C | ||
Table 6.
Independent confirmation of the association of haplotypes formed by SNPs 22–25 in three additional series from the Mayo Clinic 27,28. The North European Caucasians and the American Caucasians of mixed ancestry in the Mayo datasets have slightly different allele frequencies and haplotype structures. Nevertheless, the CTT haplotype at SNP 23–25, and the overlapping TTC haplotype at SNPs 23–25 (highlighted in black) show increased risk for AD, and are the same haplotypes that show increased risk for AD in North European Caucasians (highlighted in black in Table 4). Haplotypes associated with reduced risk of AD are highlighted in light grey, these are different from the protective haplotypes in the North European datasets, suggesting the potential existence of several protective alleles in this region.
| SNP | JS series | RS series | AUT series | Combined Mayo series | Combined Caucasian datasets | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP# | HAP | Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
Control frequency |
Cases frequency |
Z Score | Hap p-value |
Global sim p |
||||
| 22 | 23 | 24 | C | T | T | 0.235 | 0.285 | 2.641 | 0.0083 | 0.015 | 0.279 | 0.296 | 0.926 | 0.354 | 0.220 | 0.248 | 0.271 | 1.051 | 0.293 | 0.435 | 0.263 | 0.284 | 1.964 | 0.050 | 0.332 | 0.658 | 0.635 | −2.101 | 0.036 | 0.0051 |
| 22 | 23 | 24 | T | A | C | 0.690 | 0.644 | −2.248 | 0.025 | 0.641 | 0.624 | −0.920 | 0.358 | 0.666 | 0.628 | −1.566 | 0.117 | 0.657 | 0.633 | −2.063 | 0.039 | 0.247 | 0.270 | 2.258 | 0.024 | |||||
| 22 | 23 | 24 | C | A | C | 0.039 | 0.034 | −0.732 | 0.464 | 0.041 | 0.036 | −0.644 | 0.520 | 0.038 | 0.050 | 1.146 | 0.252 | 0.040 | 0.039 | −0.178 | 0.859 | 0.042 | 0.043 | 0.005 | 0.996 | |||||
| 22 | 23 | 24 | C | T | C | 0.021 | 0.019 | −0.390 | 0.697 | 0.022 | 0.026 | 0.745 | 0.456 | 0.028 | 0.022 | −0.731 | 0.465 | 0.023 | 0.022 | −0.171 | 0.864 | 0.022 | 0.022 | 0.004 | 0.997 | |||||
| 22 | 23 | 24 | T | T | T | 0.008 | 0.017 | 1.708 | 0.088 | 0.014 | 0.010 | −0.926 | 0.354 | 0.015 | 0.018 | 0.627 | 0.530 | 0.013 | 0.015 | 0.846 | 0.397 | 0.013 | 0.017 | 1.538 | 0.124 | |||||
| 22 | 23 | 24 | C | A | T | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 0.017 | 0.010 | −2.315 | 0.0206 | |||||
| 23 | 24 | 25 | T | T | C | 0.242 | 0.300 | 2.959 | 0.0031 | 0.0070 | 0.293 | 0.305 | 0.681 | 0.496 | 0.386 | 0.263 | 0.288 | 1.115 | 0.265 | 0.432 | 0.275 | 0.298 | 2.039 | 0.041 | 0.099 | 0.663 | 0.636 | −2.443 | 0.015 | 0.017 |
| 23 | 24 | 25 | A | C | T | 0.700 | 0.631 | −3.261 | 0.0011 | 0.646 | 0.629 | −0.913 | 0.362 | 0.668 | 0.633 | −1.445 | 0.148 | 0.662 | 0.631 | −2.670 | 0.0076 | 0.259 | 0.285 | 2.504 | 0.0123 | |||||
| 23 | 24 | 25 | A | C | C | 0.029 | 0.046 | 1.911 | 0.056 | 0.036 | 0.031 | −0.752 | 0.452 | 0.036 | 0.046 | 0.936 | 0.349 | 0.035 | 0.041 | 1.361 | 0.174 | 0.037 | 0.042 | 0.873 | 0.383 | |||||
| 23 | 24 | 25 | T | C | C | 0.022 | 0.017 | −0.767 | 0.443 | 0.019 | 0.026 | 1.121 | 0.262 | 0.022 | 0.021 | −0.208 | 0.835 | 0.020 | 0.021 | 0.118 | 0.906 | 0.019 | 0.020 | 0.341 | 0.733 | |||||
| 23 | 24 | 25 | A | T | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 0.017 | 0.011 | −2.208 | 0.027 | |||||
Cell Biology of SORL1
The SNPs and haplotypes identified here are unlikely to be the actual AD-causing variants. Sequencing of the exons and immediate intron-exon boundaries in carriers of the disease-associated haplotypes at SNPs 8–10 or SNPs 22–24, and investigation of SORL1 splice forms recovered by RT-PCR failed to identify any pathogenic sequence variants that were enriched in AD cases over controls (Supplemental Table 9). It is therefore likely that the associations with SNPs reflect the presence of pathogenic variants within the intronic sequences near SNPs 8–10 and 22–24. The possibility that the observed associations with SNPs inside the SORL1 gene might reflect pathogenic variants outside SORL1 can be excluded because the SNPs flanking the 5′ and 3′ ends of SORL1 all showed no association with AD. We speculate that these putative intronic variants might modulate cell-type-specific transcription or translation of SORL1 in neurons of carriers of the AD-associated haplotypes.
Direct exploration of this hypothesis is difficult. First, the variations in SORL1 expression in AD brain have been cell-type specific, SORL1 expression being depressed in neurons but not glia 12. Second, there are only limited numbers of brain tissue samples from individuals where SORL1 SNP marker phase, and thus haplotypes are known. Nevertheless, tentative support for the hypothesis that AD-associated haplotypes in SORL1 may be associated with reduced SORL1 transcription is provided by quantitative real-time PCR studies of SORL1 expression in lymphoblasts from carriers of the “CTT” AD-haplotype at SNPs 22–24. (Insufficient numbers of samples were available to test the effects of SNPs 8–10). These experiments revealed that SORL1 was expressed in AD-haplotype carriers at less than half the levels that were observed in obligate carriers of non-AD haplotypes (10324 ± 8215 arbitrary units in carriers versus 23650 ± 17999 in non-carriers; mean ± SD; normalized to β-actin mRNA; p < 0.05, two tailed Mann-Whitney U-test; n = 8 independent samples; n = 3 replications). However, it is also of note that univariate regression analyses revealed that SORL1 haplotype status accounted for only ~14% of this variance (p = 0.08). This latter result implies that other genetic and non-genetic factors can also modulate SORL1 expression, and perhaps therefore risk for AD.
The observation that specific genetic variants in SORL1 are associated with AD, and these same variants may be accompanied by reduced SORL1 expression are critical on several levels. First, these observations lead to the conclusion that the previously reported reductions in SORL1 expression in neurons in sporadic AD are likely to be causal rather than simply reactive changes. This notion is supported by that fact that SORL1 expression is not altered in other types of AD with known etiology (e.g. PS1-mutant FAD)12,29. Second, these observations raise the question of how changes in SORL1 expression or function might affect risk for AD. To explore this question, we undertook the following cell biological experiments, which demonstrate that SORL1 directly binds APP and differentially regulates its sorting into endocytic or recycling pathways.
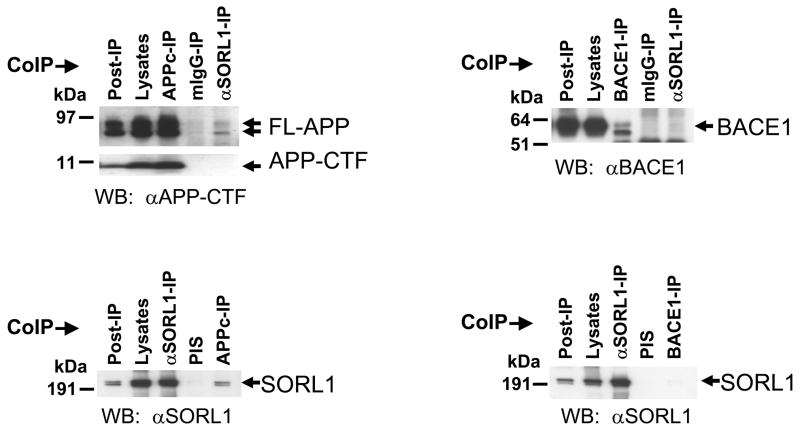
Co-immunoprecipitation experiments in native HEK cells demonstrated that endogenous SORL1 physically interacts with the endogenous APP holoprotein (Figure 2) and with VPS35 (which drives cargo selection in the retromer via VPS10-containing proteins like SORL1 30 - data not shown). SORL1, however, does not bind to APP carboxyl-terminal fragments produced by α-, β- or γ-secretase cleavage (Figure 2). These protein-protein interactions are specific because SORL1 does not bind to other Type 1 membrane proteins (e.g. BACE1 31, Figure 3) or to VPS26 (which links VPS35 to the other structural elements of the retromer30 - data not shown).
Figure 2.
A: Small quantities of endogenous APP holoprotein but not APP C-terminal fragments (APP-CTFs, generated by α- or β-secretase) can be co-immunoprecipitated with endogenous SORL1 (Top panel). Conversely small quantities of endogenous SORL1 can be co-precipitated with endogenous APP holoprotein (Bottom panel).
B: SORL1 does not interact with BACE1 (β-secretase). Co-immunoprecipitations with antibodies to over-expressed BACE1-V5 fail to capture SORL1 (Bottom panel). Conversely, SORL1-directed antibodies do not co-immunoprecipitate BACE1 (Top panel) even though BACE1 also traffics through the endosome to Golgi pathway.
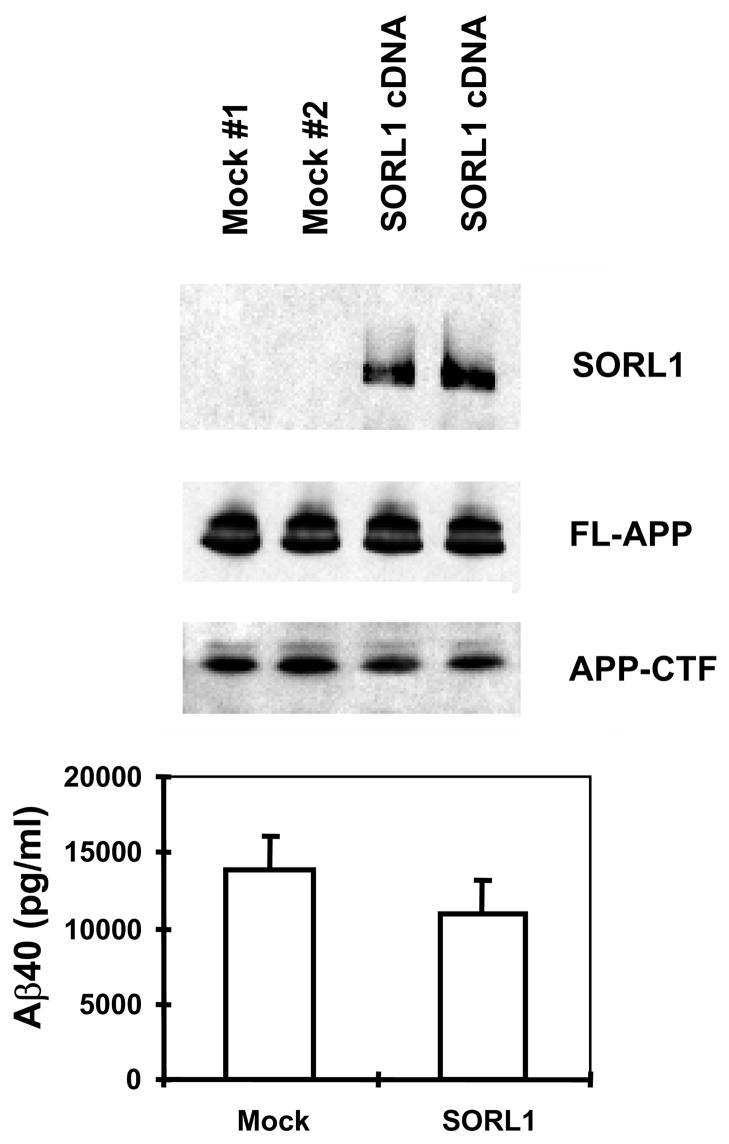
Figure 3.
A: Over-expression of SORL1 reduces Aβ40 (and Aβ42 not shown) secretion (p < 0.05). Upper panel: Representative data of Western blot for SORL1 and APP in HEK293 cells stably expressing APPSwe, and transiently transfected with empty vector (mock) or SORL1 (n = 2 independent transfections). Lower panel: Bar charts of ELISA assays of secreted Aβ40 (and Aβ42 not shown) following SORL1 over-expression. Error bar: SD; *p<0.05 compared to Control (2-tailed t-test); n = 2 replications.
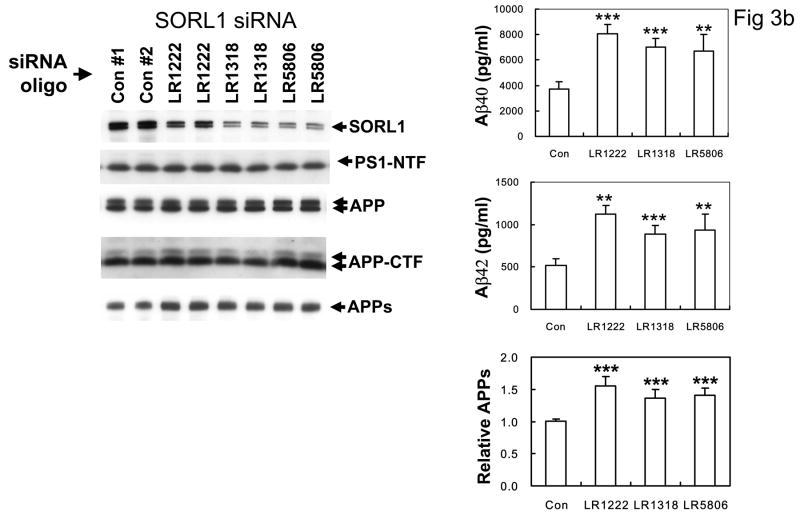
B: Left panel: Suppression of SORL1 expression with three independent siRNA primers (LR1222, LR1318, and LR5806) did not alter the expression levels or maturation of APP, APP-C83 C-terminal fragments or PS1, but (Right panel) significantly increased Aβ40 and Aβ42 secretion and APPs secretion (*p <0.005, ** p < 0.001 2-tailed t-test compared to controls, n = 5 replications, 3 siRNAi oligomers).
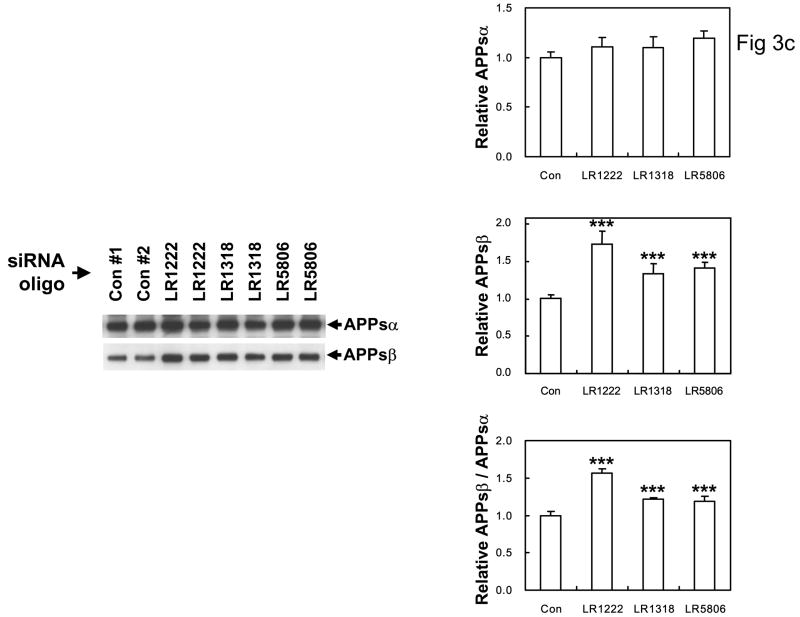
C: anti-SORL1 siRNA treatment results in significant increases in APPsβ secreted into the media, but no significant change in APPsα levels. Left panel: Western blots of conditioned media from cells treated with nonsense siRNA oligo-nucleotides (Controls #1 and #2) or with anti-SORL1 siRNA oligonucleotides investigated with the 2H3 antibody to APPsα or with SW192 antibody to APPsβ (n = 5 replications). Right panel: quantitation normalized to the control. ** p < 0.0001 2-tailed t-test compared to controls, n = 5 replications.
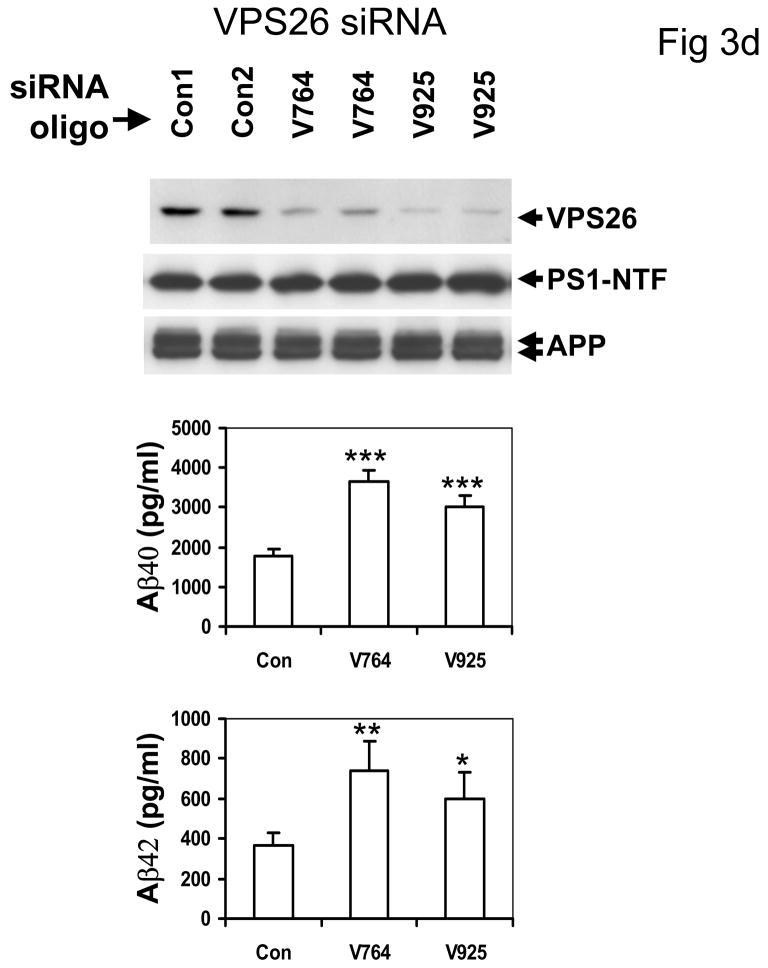
D: Top panel: suppression of VPS26, another member of the VPS10 family involved in the retromer pathways also did not alter APP or PS1 maturation, but (Middle and Bottom panels) did increase both Aβ40 and Aβ42 secretion (*p <0.005, ** p < 0.001 2-tailed t-test compared to controls, n = 5 replications, 2 siRNA oligomers). The control primer had no such effect.
The interaction between SORL1, VPS35 and APP holoprotein provides a mechanism by which SORL1 can regulate differential sorting of APP into the retromer recycling pathway or into the late endosomal pathway (where APP undergoes β- and γ-secretase cleavage to generate Aβ). In agreement with this hypothesis, over-expression of SORL1, which would be predicted to divert APP holoprotein into the retromer recycling pathway, results in decreased Aβ production (82% of control value, p < 0.05, n = 5 replications; Figure 3A). Conversely, siRNA suppression of SORL1 expression, which we speculate mimics the effects of AD-associated variants in SORL1, results in deflection of APP holoprotein away from the recycling retromer pathway and into the late endosome-lysosome pathway. As would be predicted, siRNA suppression of SORL1 causes: 1) overproduction of the APPsβ ectodomain generated by BACE1 cleavage of APP holoprotein (mean ± SEM normalized to control = 149.45% ± 9.66, p < 0.0001, n = 5 replications; Figure 3C); and 2) over-production of Aβ by the subsequent γ-secretase cleavage of the APP C-terminal stub generated by BACE1 (Aβ40 = 189% of control; Aβ42 = 202% of control, p < 0.001; three independent siRNA oligonucleotides with five replications each, Figure 3B). The conclusion that SORL1 regulates sorting of APP into the retromer-recycling pathway is supported by the observation of identical effects following suppression of the retromer proteins VPS26 (Aβ40 = 186% of control value; Aβ42 = 183% of control value, p < 0.001, n = 5 replications; Figure 3D) or VPS35 13. These results are in good agreement with independent reports that appeared during preparation of this manuscript 14,15.
DISCUSSION
Taken together, our results suggest that genetic and possibly environmentally-specified changes in SORL1 expression or function are causally linked to the pathogenesis of AD and have a modest effect on risk for this disease. The precise identity of the genetic effectors in SORL1 remains to be determined. However, the results described here imply that: i) there are several different allelic variants in distinct genomic regions of the SORL1 gene in different populations; ii) that these variants are likely to be in intronic regulatory sequences that might govern cell-type or tissue-specific expression of SORL1; and iii) these variants affect this risk by altering the physiological role of SORL1 in the processing of APP holoprotein.
The observations that: 1) no single SORL1 SNP or haplotype is associated with increased risk for AD in all six datasets; and 2) that some datasets fail to show any association, contrasts sharply with APOE (where APOE ε4 is associated with AD in most datasets 32). However, four points mitigate concerns that the association between SORL1 and AD is spurious. First, the association was initially identified using conservative family-based association tests, which are less sensitive to confounding due to population stratification 33. Second, at each set of SNP clusters, the same alleles and haplotypes were associated with increased risk for AD in at least three unrelated datasets drawn from ethnically different origins. Third, the association of disease with a single allele in all datasets (i.e. an APOE ε4-like association) is not a universal observation for either complex or monogenic diseases 17. Thus, the occurrence of pathogenic mutations across multiple domains of disease genes (i.e. allelic heterogeneity), and the absence of these variants in some datasets (i.e. locus heterogeneity) are not unusual in either monogenic or complex traits 34,35. Fourth, the absence of significant associations in two datasets (MIRAGE Caucasian sibships and Mayo-RS) does not negate the findings from the other datasets. There are several potential explanations for the failure to detect a significant association in these two datasets, including: 1) insufficient power to reliably detect the association in all series; 2) locus heterogeneity (i.e. non-SORL1-causes are over-represented and SORL1-associated causes are under-represented in these datasets); and 3) allelic heterogeneity (i.e. the association was obscured because in these datasets the biologically active SORL1-alleles occur on a different SNP background or on multiple SNP backgrounds). Indeed, the probable existence of population-specific alleles (ie allelic heterogeneity) has important implications for replication studies. Such studies will need to assess a battery of SNPs focused on datasets with as homogeneous a genetic background as possible.
Our results also resolve the conundrum concerning the significance of previous reports of reduced expression in AD-affected brain tissue of several genes potentially involved in APP trafficking. The data reported here argue that the reduction in SORL1 expression is likely to be a primary and pathogenic event, whereas the reduction in VPS35 is likely to be a secondary event.
Finally, our data demonstrate that SORL1 plays a key physiological role in the differential sorting of APP holoprotein. In the presence of SORL1, APP holoprotein is recovered via the retromer. In the absence of SORL1, APP is released into late endosomal pathways where it is subjected to β- and subsequently γ-secretase cleavage that generate Aβ (Figure 1b).
METHODS
Subjects
Informed consent was obtained from all participants. The clinical diagnosis of “probable” or “possible” AD was defined according to the NINCDS-ADRDA diagnosis criteria at clinics specializing in memory disorders. Clinical characteristics of the North European, MIRAGE, Caribbean-Hispanic FAD, Israeli-Arab and Mayo American Caucasian datasets are summarized in Table 1 19,20,22–25,27,28. The North European case:control set is drawn from the same populations as the North European FAD dataset 20,22. The three Mayo datasets were drawn from Caucasian subjects and controls assessed in clinical series at the Rochester (RS) and Jacksonville (JS) Mayo clinics, or from Caucasian brains in which the presence or absence of AD was determined neuropathologically (AUT).
Genetic Analyses
Genotyping was performed using the GenomeLab SNPstream System and primer sets were as in Supplemental Table 2B (Beckman Coulter Inc., CA). 100 DNA samples were genotyped twice for every SNP marker (concordance rate was >99%). APOE was genotyped as described 5. Genotyping of the Mayo samples was performed on an ABI 7900 instrument using TaqMan chemistry with primers and probes designed by Applied Biosystems Inc. The entire open reading frame of the SORL1 gene was sequenced in twelve AD cases, twelve familial AD cases and two normal controls selected from the North European and Caribbean-Hispanic datasets (Supplemental Tables 2C and 9).
Alternatively spliced transcripts were sought by conventional RT-PCR in eight overlapping fragments using total RNA isolated from frontal cortex (Canadian Brain Tissue Bank and the New York Brain Bank) (16 normal controls and 17 sporadic AD cases). (Supplemental Table 2D, Supplemental Figure 2).
Statistical Analyses
SNP marker data were assessed for deviations from Hardy-Weinberg equilibrium (Pedstats program) and for Mendelian inheritance errors (Pedcheck software). Single point family-based association was assessed with FBAT v1.5.5 36, using an additive genetic model with the null hypothesis of no linkage and no association. Allele frequencies were estimated by FBAT using the EM algorithm. APOE ε4 carrier status was included in the analyses using PBAT v2.6 37–40. The χ2 test (or the Fisher’s exact test) was used to assess genotypic and allelic associations between AD. Multivariate logistic regression analysis was performed to adjust for APOE ε4, sex, and age-at-onset/age-at-examination.
Statistical significance and multiple testing corrections
The Benjamini-corrected false discovery rate (FDR) 41 was used with a cut-off level of 0.1 to correct for multiple testing. The p-values presented are nominal p-values. The cut off p-values for significance in each dataset are displayed in the table legends.
Linkage Disequilibrium
LD structure was examined using Haploview (http://www.broad.mit.edu/mpg/haploview/index.php). Haplotype blocks were defined using confidence-intervals algorithm. The default settings were used in these analyses, which create 95% confidence bounds on D′ to define SNP pairs in strong LD.
Haplotype Analyses
Haplotype analyses were carried out with a sliding window of three contiguous SNPs using FBAT for family data and Haplo. stats v1.1.1 for case:control data 16,26,42,43 44. The analyses were repeated using sliding windows of 2, 4, 5 and 6 SNPs.
Expression plasmids and cDNA constructs for human SORL1
The cDNA clones for APP K670N/M671L Swedish mutation (APPSwe) and BACE1 V5-tagged at the C-terminus were described previously 45,46.
Cell culture and transfection
HEK293 cell line stably expressing APPSwe was as described 47. Transient transfection of BACE1 cDNA was performed using LipofectAMINE 2000 (Invitrogen, CA).
RNA Interference
Small interfering RNA (siRNA) oligonucleotides were designed using the online siRNA Design Tool (Dharmacon Research, CO). The siRNAs for SORL1 are: 5′-AAACAACCGCACCAAUUUAUA-3′ (termed LR1222), 5′-AAGUGACACCUUGGUGAGGUA-3′ (termed LR1318), and 5′-AAAGACGGUCAUUGUCAGUAA-3′ (termed LR5806). The siRNAs for VPS26 are: 5′-AAACAAUCGCCAAAUAUGAAA-3′ (termed V764) and 5′-GAAGACCGGAGGUACUUCAAA-3′ (termed V925). The siCONTROL Non-Targeting siRNA #1 (Dharmacon Research, Lafayette, CO) was used as a negative control.
Transfections were performed using LipofectAMINE 2000 according to the manufacturer’s recommendations. In case of consecutive transfections, cells were split after 24 hours and then retransfected 24 hours later. After culturing for an additional 24 hours, the conditioned medium was collected for Aβ assay, and the cells were harvested for Western blotting.
Antibodies, Immunoprecipitation and Western blotting
Antibodies were: mouse monoclonal anti-human LR11/SORL1gp250 (BD Transduction Laboratories) and 5-4-30-19-2; rabbit anti-SORL1 C-terminal antibody (from W. Hampe) rabbit polyclonal antibody to PS1-NTF (Ab14, from S. Gandy); mouse monoclonal anti-myc (Invitrogen); rabbit polyclonal antibody to APP C-terminal (Sigma); anti-BACE1 (EE-17, Sigma). Proteins were immunoprecipitated in 1% digitonin 48 Western blotted, and visualized by ECL (Amersham).
Aβ, APPsα and APPsβ assays
Aβ40 and Aβ42 peptide levels were measured by sandwich ELISA 49. APPs, APPsα and APPsβ were measured by Western blotting using antibodies 22C11 (Chemicon), 2H3 and SW192 (Elan Pharmaceuticals, CA) respectively. Differences were assessed by two-tailed students’ t-test.
Quantitative RT-PCR
PCR primer pairs targeting SORL1 exon 23:5′-ctgcagcaacgggaactgtat (forward) and 5′-tgtctccacagtcgttgtcaaag (reverse). Total RNA (5ug) was reverse-transcribed using a random hexamer. Real-time PCR was performed in a 384-well format using a Prism 7900HT instrument (ABI) and the Sybr Green detection method. Samples were analyzed in triplicate and mean expression levels corresponding to SORL1 mRNA expression were normalized to β-actin mRNA levels.
Supplementary Material
Table 2.
Single Nucleotide Polymorphisms (SNPs) used in this study. Marker intervals are calculated on the basis of NCBI locations (National Center for Biotechnology Information Web site). SNPs are referred to in this paper by sequential numbers (marker number) reflecting their relative physical map positions. Strand orientation information was obtained from NCBI: “fwd/T” refers to forward or top strand; “rev/B” refers to reverse or bottom strand.
| Marker Number | dbSNP rs Number | Alleles | Strand Orientation | Physical Map Location (bp) | Distance (bp) from Previous Marker | SNP Type |
|---|---|---|---|---|---|---|
| 1 | rs4935774 | A/G | rev/B | 120826964 | --- | Upstream of 5′ UTR |
| 2 | rs578506 | C/G | fwd/T | 120828687 | 1,723 | intron |
| 3 | rs582446 | A/G | rev/B | 120833069 | 4,382 | intron |
| 4 | rs661057 | C/T | fwd/T | 120834164 | 1,095 | intron |
| 5 | rs11218304 | C/T | rev/B | 120854321 | 20,157 | intron |
| 6 | rs560573 | A/T | fwd/T | 120866094 | 11,773 | intron |
| 7 | rs12364988 | A/G | rev/B | 120872836 | 6,742 | H269H |
| 8 | rs668387 | T/C | rev/B | 120873131 | 295 | intron |
| 9 | rs689021 | A/G | rev/B | 120876330 | 3,199 | intron |
| 10 | rs641120 | T/C | fwd/T | 120886175 | 9,845 | intron |
| 11 | rs4935775 | C/A | rev/B | 120894712 | 8,537 | intron |
| 12 | rs12285364 | T/C | fwd/T | 120898436 | 3,724 | intron |
| 13 | rs2298813 | A/G | fwd/T | 120898894 | 458 | T528A |
| 14 | rs11600231 | C/T | fwd/T | 120911918 | 13,024 | intron |
| 15 | rs2276346 | T/G | fwd/T | 120919686 | 7,768 | intron |
| 16 | SORL1-T833T | T/A | fwd/T | 120931165 | 11,479 | T833T |
| 17 | rs556349 | G/T | rev/B | 120931417 | 252 | intron |
| 18 | rs11218340 | T/A | fwd/T | 120936564 | 5,147 | intron |
| 19 | rs2070045 | G/T | fwd/T | 120953300 | 16,736 | S1187S |
| 20 | rs3824966 | G/C | fwd/T | 120953393 | 93 | intron |
| 21 | SORL1-ex26 | G/C | fwd/T | 120959359 | 5,966 | (−18) 5′ of exon 26 |
| 22 | rs1699102 | C/T | fwd/T | 120962172 | 2,813 | N1246N |
| 23 | rs3824968 | T/A | fwd/T | 120981132 | 18,960 | A1584A |
| 24 | rs2282649 | C/T | fwd/T | 120984168 | 3,036 | intron |
| 25 | rs1010159 | C/T | rev/B | 120988611 | 4,443 | intron |
| 26 | rs1784933 | G/A | fwd/T | 120994626 | 6,015 | intron |
| 27 | rs1614735 | C/A | rev/B | 120998211 | 3,585 | intron |
| 28 | rs1133174 | A/G | fwd/T | 121006965 | 8,754 | downstream of 3′UTR |
| 29 | rs1131497 | G/C | fwd/T | 121007955 | 990 | downstream of 3′UTR |
Acknowledgments
Supported by the Canadian Institutes of Health Research, Howard Hughes Medical Institute, Canadian Institutes of Health Research-Japan Science and Technology Trust, Alzheimer Society of Ontario, Canada Foundation for Innovation, Ontario Research and Development Challenge Fund, Ontario Mental Health Foundation, Genome Canada, National Institute of Health and the National Institute on Aging: R37-AG15473 and P01-AG07232 (RM), R01-AG09029 (LAF), RO1-HG/AG02213 (RCG), P30-AG13846 (LAF, RCG), R01AG017173 (RPF, LAF), P60-AG16574 (RCP, SGY, NGR), U01-AG06786 (RCP), Alzheimer Association, Alzheimer Society of Canada, Blanchett Hooker Rockefeller Foundation, Charles S. Robertson Gift (RM), Fonds de la Recherche en Santé (YM), Assessorato Regionale alla Sanità-Regione Calabria, Finalized Project of the Ministry of Health (2003–2005) (ACB), Fondation pour la Recherche Médical, Robert and Clarice Smith and Abigail Van Buren, ADRC (RP, SY). The authors thank Dr. Jurg Ott for helpful advice on the statistical analysis. The authors acknowledge the work of Drs. Ben Tycko, Martin Medarano (UTESA), Rafael Lantigua, Yaakov Stern, Abimbola Akomolafe, Jeffrey Browndyke, Helena Chui, Rodney Go, Alexander Kurz, Helen Petrovitch, Norman Relkin, Dessa Sadovnick, Porat Erlich, Shamil Sunyaev, Li Ma, Johann Lok, and Samuel Younkin. The authors’ roles are described in Supplemental Table 10.
Footnotes
The authors’ roles are described in Supplemental Table 10.
COMPETING INTERESTS
The authors have no competing financial interests.
References
- 1.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–9. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goate AM, et al. Segregation of a missense mutation in the amyloid precursor protein gene with Familial Alzheimer Disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 3.Sherrington R, et al. Cloning of a gene bearing missense mutations in early onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 4.Rogaev EI, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a novel gene on chromosome 1 related to the Alzheimer’s Disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 5.Saunders A, et al. Association of Apoliprotein E allele e4 with the late-onset familial and sporadic Alzheimer Disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 6.Bales KR, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nature Genet. 1997;17:254–256. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 7.Golde TE, Estus S, Younkin LH, Selkoe DJ, Younkin SG. Processing of the amyloid protein precursor to potentially amyloidogenic derivatives. Science. 1992;255:728–730. doi: 10.1126/science.1738847. [DOI] [PubMed] [Google Scholar]
- 8.Haass C, Selkoe DJ. Cellular processing of beta-amyloid precursor protein and the genesis of amyloid beta-peptide. Cell. 1993;75:1039–42. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 9.Bayer TA, et al. Key factors in Alzheimer’s disease: beta-amyloid precursor protein processing, metabolism and intraneuronal transport. Brain Pathol. 2001;11:1–11. doi: 10.1111/j.1750-3639.2001.tb00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita A, et al. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–46. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 11.Vetrivel KS, et al. Spatial segregation of gamma-secretase and substrates in distinct membrane domains. J Biol Chem. 2005;280:25892–25900. doi: 10.1074/jbc.M503570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherzer CR, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–5. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 13.Small SA, et al. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58:909–19. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 14.Andersen OM, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Offe K, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–37. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–23. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 18.Athan ES, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. Jama. 2001;286:2257–63. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 19.Bowirrat A, Treves TA, Friedland RP, Korczyn AD. Prevalence of Alzheimer’s type dementia in an elderly Arab population. Eur J Neurol. 2001;8:119–23. doi: 10.1046/j.1468-1331.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- 20.Rogaeva E, et al. Re-analysis of the association of Alpha-2-macroglobulin and Alzheimer’s Disease. Nature Genet. 1999;22:19–22. [Google Scholar]
- 21.Rogaeva E, et al. Evidence for an Alzheimer Disease susceptibility locus on chr 12, and for further locus heterogeneity. JAMA. 1998;280:614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, et al. Fine mapping of 10q and 18q for familial Alzheimer’s disease in Caribbean Hispanics. Mol Psychiatry. 2004;9:1042–1051. doi: 10.1038/sj.mp.4001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graff-Radford NR, et al. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol. 2002;59:594–600. doi: 10.1001/archneur.59.4.594. [DOI] [PubMed] [Google Scholar]
- 24.Green RC, et al. Risk of dementia among white and African American relatives of patients with Alzheimer disease. J Am Med Assoc. 2002;287:329–36. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- 25.Farrer LA, et al. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–22. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 26.Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–8. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 27.Ertekin-Taner N, et al. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer’s disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum Mol Genet. 2005;14:447–60. doi: 10.1093/hmg/ddi041. [DOI] [PubMed] [Google Scholar]
- 28.Ertekin-Taner N, et al. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum Mutat. 2004;23:334–42. doi: 10.1002/humu.20016. [DOI] [PubMed] [Google Scholar]
- 29.Dodson SE, et al. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–72. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaman MN. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 31.He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- 32.Farrer LA, et al. Effects of age, sex, and ethnicity on the asscoiation between apolipoprotein E genotype and Alzheimer’s Disease. A meta-analysis. APOE and Alzheimer’s Disease Meta Analysis consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 33.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–23. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 34.Vermeire S, Rutgeerts P. Current status of genetics research in inflammatory bowel disease. Genes Immun. 2005;6:637–45. doi: 10.1038/sj.gene.6364257. [DOI] [PubMed] [Google Scholar]
- 35.Owen MJ, Craddock N, O’Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21:518–25. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Horvath S, et al. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 37.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–9. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- 39.Lange C, Laird NM. Power calculations for a general class of family-based association tests: dichotomous traits. Am J Hum Genet. 2002;71:575–84. doi: 10.1086/342406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Steen K, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet. 2005;37:683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statistic Soc. 1995;B57:289–300. [Google Scholar]
- 42.Jannot AS, Essioux L, Reese MG, Clerget-Darpoux F. Improved use of SNP information to detect the role of genes. Genet Epidemiol. 2003;25:158–67. doi: 10.1002/gepi.10256. [DOI] [PubMed] [Google Scholar]
- 43.Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol. 1998;9:578–94. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- 44.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F, et al. Nicastrin binds to membrane-tethered Notch. Nature Cell Biology. 2001;3:751–754. doi: 10.1038/35087069. [DOI] [PubMed] [Google Scholar]
- 46.Benjannet S, et al. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J Biol Chem. 2001;276:10879–87. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 47.Yu G, et al. A Novel Protein (Nicastrin) Modulates Presenilin-Mediated Notch/Glp1 And betaAPP Processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 48.Gu Y, et al. The presenilin proteins are components of multiple membrane-bound complexes which have different biological activities. J Biol Chem. 2004;279:31329–31336. doi: 10.1074/jbc.M401548200. [DOI] [PubMed] [Google Scholar]
- 49.Hasegawa H, et al. Both the sequence and length of the C terminus of PEN-2 are critical for intermolecular interactions and function of presenilin complexes. J Biol Chem. 2004;279:46455–63. doi: 10.1074/jbc.M406289200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.