Abstract
Mutations and polymorphisms in complement genes have been linked with numerous rare and prevalent disorders, implicating dysregulation of complement in pathogenesis. The 3 common alleles of factor B (fB) encode Arg (fB32R), Gln (fB32Q), or Trp (fB32W) at position 32 in the Ba domain. The fB32Q allele is protective for age-related macular degeneration, the commonest cause of blindness in developed countries. Factor B variants were purified from plasma of homozygous individuals and were tested in hemolysis assays. The protective variant fB32Q had decreased activity compared with fB32R. Biacore comparison revealed markedly different proenzyme formation; fB32R bound C3b with 4-fold higher affinity, and formation of activated convertase was enhanced. Binding and functional differences were confirmed with recombinant fB32R and fB32Q; an intermediate affinity was revealed for fB32W. To confirm contribution of Ba to binding, affinity of Ba for C3b was determined. Ba-fB32R had 3-fold higher affinity compared with Ba-fB32Q. We demonstrate that the disease-protective effect of fB32Q is consequent on decreased potential to form convertase and amplify complement activation. Knowledge of the functional consequences of polymorphisms in complement activators and regulators will aid disease prediction and inform targeting of diagnostics and therapeutics.
Keywords: alternative pathway, AMD
Dysregulation of the alternative pathway (AP) of complement is associated with numerous pathologies, including age-related macular degeneration (AMD), atypical hemolytic uremic syndrome (aHUS), rheumatoid arthritis, dense deposit disease (DDD), and lupus nephritis (1). The AP “ticks over” continuously in plasma, enabling rapid response to pathogen, and it also amplifies the other complement activation pathways (2). The critical AP activation step is cleavage of C3 to C3b. The C3-cleaving enzyme, or convertase, is formed through the Mg2+-dependent binding of factor B (fB) to C3b, forming the proenzyme, C3bB. Factor B within the complex is then cleaved by a plasma serine protease, fD, releasing an amino terminal fragment, Ba (comprising 3 short consensus repeats; SCR). This fragment takes no further part in the complement cascade. Bb, comprising a serine protease domain and a von Willebrand factor Type A (vWF-A) domain, remains bound to C3b, and is an active serine protease capable of cleaving further C3. Convertase-generated C3b can itself form more C3bBb, providing amplification of activation. As the C3b clusters around the surface-bound convertases a C5-cleaving enzyme is formed (C3bBbC3b), C5b is generated and the lytic pathway proceeds, with formation of the membrane attack complex (MAC) (3).
To protect against the tendency of the AP to rapidly amplify, host cells express an armory of complement regulatory proteins (CReg), which inhibit convertases or prevent MAC formation on their surfaces (4, 5). These CReg are either membrane-associated (CD55, CD46, and CD59) or fluid phase (fH, C4b-binding protein) proteins. Dysregulation of the AP is brought about by loss-of-function/expression mutations in CReg or gain-of-function mutations in components, both scenarios result in uncontrolled complement activation on self-surfaces and subsequent tissue damage and/or inflammation. The best characterized disease of AP dysregulation is aHUS, where loss-of-function mutations in the CReg or gain-of-function mutations in components lead to increased complement activation at cell surfaces, resulting in renal damage (6, 7).
More remarkably, common polymorphisms in AP components and CReg have also been linked to disease. Of particular interest are 3 common polymorphic variants of fB that differ at position 32 in the Ba domain (amino acid 7 in the mature protein; fB32R, fB32Q, and fB32W; rs12614 and rs641153) (8–10); fB32R is the most frequent allele in Caucasians (allele frequency 0.79), and was originally described as fB-S (“slow,” defined by electrophoretic mobility); the fB-F (“fast”) allele was further defined as fB-FA (fB32Q; allele frequency 0.05) and fB-FB (fB32W; allele frequency 0.16) (11). Factor B-S and fB-F have long been associated with various pathologies, including susceptibility to pathogenic infection, for example, with Trypanosoma cruzi, where fB-S is protective in cardiomyopathy associated with Chagas disease (12). Factor B-S is reported to be overrepresented in multiple sclerosis (13), and weakly associated with Type I diabetes mellitus, where it is present in a HLA haplotype strongly linked to disease (14). However, the most striking example of linkage of common variants in fB to disease is in AMD, the commonest cause of irreversible blindness in the elderly in the Western world (15).
The hallmark lesion of AMD is the development of drusen, lipoproteinaceous deposits located between the retinal pigment epithelium (RPE) and Bruch's membrane (16). Deposition of drusen is followed by extensive atrophy of the RPE and overlaying photoreceptor cells (geographic atrophy, GA), or aberrant choroidal neovascularization (CNV). CNV under the macula is the primary cause of blindness. Although the pathogenesis of AMD is still unclear, inflammatory responses are implicated (17, 18). AMD is a multifactorial disease, influenced by age, ethnicity, and environmental and genetic risk factors (19). Two major AMD susceptibility loci (1q31, CFH, and 10q26, LOC387715/HTRA1) that independently contribute to disease risk have been recently identified by candidate region linkage and whole genome association analyses (20–23). Linkage to CFH, the gene encoding fH, was followed by reports describing association of AMD with polymorphic variations in other complement genes, fB, C3, and the fH-related genes CFHR1 and CFHR3 (24–28). Remarkably, although most AMD-associated polymorphisms are linked to increased disease risk, several replication studies have demonstrated that the fB32Q variant confers significant protection from development of AMD (26, 27, 29).
The risk allele in LOC387715 causes destabilization of the mRNA and subsequent reduced expression of the encoded protein, a mitochondrial protein normally expressed at high levels in the retinal photoreceptor cells (30). Thus, a functional consequence that increases risk of AMD is plausible. In contrast, no functional explanation of the association of any complement polymorphism with AMD has yet been provided. Here, we demonstrate the functional mechanism underlying the association of fB variants with pathology. Using purified and recombinant (r) fB variants, we demonstrate that the fB32Q variant, identified by genetic association studies to be protective in AMD, is less efficient at forming the amplifying AP convertase. This variant likely protects from development of pathology in AMD, and perhaps other chronic inflammatory diseases, by dampening complement activation, but may predispose to infection due to reduced amplification activity.
Results
Purification of fB Variants and Differential Activity in Hemolysis Assays.
To investigate whether the R32Q polymorphism in fB influenced function, fB was purified from plasma of a donor identified by genotyping to be R32Q heterozygote. The 2 variants were separated by anion exchange chromatography at pH 6.0 [supporting information (SI) Fig. S1A], and individually gel filtered on Superdex 200 equilibrated in CFD. Fractions containing pure, monomeric fB were identified and used immediately for functional analyses. To compare functional activity of the 2 variants, fB was immunodepleted from human serum (NHSΔfB) and used as a source of all other complement components. Rabbit erythrocytes were incubated with NHSΔfB and different concentrations of fB variants, lysis was measured (Fig. S1B). The fB32Q variant was less hemolytically active than fB32R, at least 2-fold more protein was required to achieve equivalent lysis.
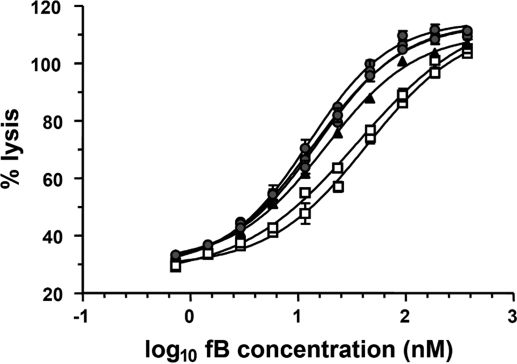
To confirm and extend these data, fB was purified by affinity chromatography, and gel filtration from plasma of 6 individuals identified by genotyping to be homozygote for fB32R (3 individuals), fB32Q (2 individuals), and fB32W (1 individual). The 3 variants showed consistent differences in hemolytic activities, with fB32Q the least lytic and fB32R the most lytic (Fig. 1). Calculated EC50s were 35.6 and 43.5 nM (for the fB32Q individuals); 15.4, 12.6, and 13.9 nM (for the fB32R individuals); and 17.9 nM (for the fB32W individual). The difference in means between fB32R and fB32Q was statistically significant (2-tailed unpaired T test; P < 0.0038).
Fig. 1.
Hemolytic activity of the fB variants. The variant fB proteins were purified from plasma of 6 donors known to be homozygous for fB32R (3 donors, circles), fB32Q (2 donors, squares), or fB32W (1 donor, triangles). Rabbit erythrocytes were incubated with fB-depleted serum and different concentrations of the purified fB variants. Lysis was developed for 45 min and hemoglobin release was used to calculate percentage lysis. Data points represent mean ± SD of 3 determinations. The log10 of fB concentration was plotted against percentage lysis, and curves were fitted by using nonlinear regression analysis to calculate the EC50.
Formation of AP Convertase by fB32R, fB32Q, and fB32W Variants.
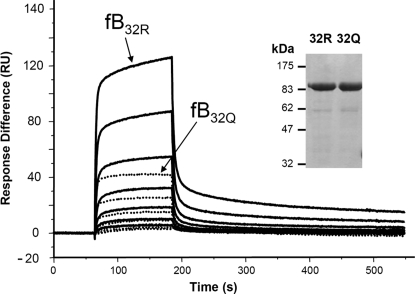
We previously used SPR (Biacore) to monitor proenzyme (C3bB) formation and convertase activation (C3bBb) in real-time. To dissect the mechanisms underlying differential hemolytic activities of fB32R and fB32Q, fB, purified from homozygote donors (see inset in Fig. 2), was gel filtered into Biacore buffer, and analyzed immediately. Proenzyme formation was analyzed by SPR by flowing fB over the C3b-immobilized surface in the presence of Mg2+, without fD. Factor B32R was more efficient in forming proenzyme, with higher levels of C3bB32R formed at identical fB concentrations. Decay was rapid for each variant. We have previously shown that several points of contact and conformational transitions are involved in the interaction between fB and C3b, which is best modeled by using a “2-state transition” model (31). Analysis of sensorgrams (Fig. 2), revealed a 4-fold higher affinity of binding for fB32R over fB32Q; KD calculated as a mean of 3 independent experiments by using “2-state reaction” model: fB32R, 0.17 ± 0.03 μM; fB32Q, 0.74 ± 0.25 μM.
Fig. 2.
SPR analysis of proenzyme formation by native fB. The variant fB proteins were purified from the plasma of donors known to be homozygous for fB32R or fB32Q, representative preparation is indicated (reducing SDS/PAGE gel stained with Coomassie Blue R250). FB was flowed over the surface of the C3b-coated chip at concentrations between 460 and 7 nM in Biacore buffer (10 mM Hepes, pH 7.4/50 mM NaCl/1 mM MgCl2/0.005% surfactant P20). Sensorgrams from fB32R are solid lines and fB32Q are dotted lines; identical concentrations are illustrated for the 2 proteins.
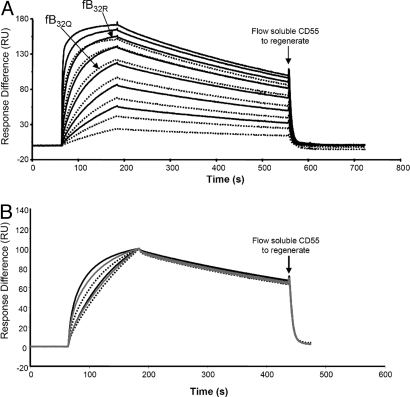
To analyze formation of the convertase C3bBb, fD was included with the fB variants. Although the kinetics of convertase formation by fB32R and fB32Q were similar, because fB32Q bound with lower affinity, lower levels of convertase were formed (Fig. 3A).
Fig. 3.
SPR analysis of activated convertase formation by native fB. (A) The variant fB proteins were purified from the plasma of donors known to be homozygous for fB32R or fB32Q. FB was flowed over the surface of the C3b-coated chip in the presence of 1 μg/mL fD at concentrations between 460 and 7 nM in Biacore buffer (10 mM Hepes, pH 7.4/50 mM NaCl/1 mM MgCl2/0.005% surfactant P20). Sensorgrams from fB32R are solid lines and fB32Q are dotted lines; identical concentrations are illustrated for the 2 variants. Arrows indicate sensorgram generated by either variant at 115 nM for comparison. Kinetic information from 2 independent experiments (different surface and preparation of fB each time) were analyzed by using 2-state reaction model, and data were identical for both experiments. KD: fB32R, 2.9 nM; and fB32Q, 7.1 nM (χ2 in replicate experiments was between 6 and 13). Data were also analyzed by using a 1:1 Langmuir model; although the fit was not as good, kinetic analysis revealed similar affinities (3.2 nM, fB32R; 7.7 nM, fB32Q) (χ2 in replicates was between 7 and 32). (B) All 3 variants were purified from plasma of homozygous donors, and flowed over a C3b surface at 2 concentrations (156, 39 nM). Sensorgrams from fB32R are solid lines, fB32Q are dotted lines, and fB32W are in gray. Data were normalized in the y axis, and sensorgrams overlaid.
To investigate the half-lives of the activated convertase, purified fB variants were flowed over a C3b surface in the presence of fD. Dissociation rates (kd) do not depend on concentration; therefore, decay curves were normalized in the y axis. There was an obvious gradation of enzyme formation (fB32R > fB32W > fB32Q), whereas rates of decay of the 3 enzymes were identical (Fig. 3B), as expected, because Ba, containing the variant amino acid, is released from the convertase, and cannot influence decay of Bb from C3b.
Formation of AP Convertase by Recombinant fB32R, fB32Q, and fB32W Variants.
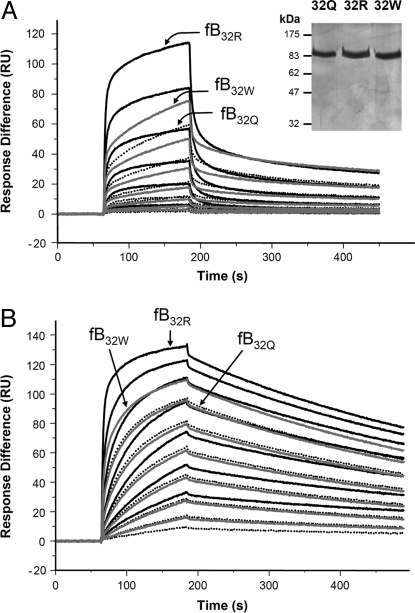
To confirm these kinetic data, we expressed recombinant forms of fB differing only at position 32, rfB32R, rfB32Q, and rfB32W in CHO cells, and purified from supernatant by affinity chromatography and gel filtration (see inset in Fig. 4A). Analysis of proenzyme formation by SPR revealed higher affinity binding of rfB32R compared with rfB32Q, confirming results obtained with the native proteins (Fig. 4A). The third variant, rfB32W, revealed an intermediate efficiency of binding. Kinetic profiles differed slightly when compared with native protein, likely due to different glycosylation; however, measured affinities were similar for native and recombinant. Formation of activated convertase using rfB variants confirmed that rfB32R generated most convertase and rfB32Q the least, with rfB32w intermediate (Fig. 4B); decay rates of Bb were identical for convertases generated by each recombinant protein.
Fig. 4.
SPR analysis of enzyme formation by rfB. The variant rfB proteins were purified from supernatant, representative preparation is indicated (reducing SDS/PAGE gel stained by silver). (A) Recombinant fB was flowed over the C3b-coated chip at concentrations between 233 and 4 nM in Biacore buffer (10 mM Hepes, pH 7.4/50 mM NaCl/1 mM MgCl2/0.005% surfactant P20). Sensorgrams from rfB32R are solid lines, rfB32Q are dotted lines, and rfB32W are in gray; identical concentrations are indicated for the 3 variants. Arrows indicate sensorgram generated by each variant at 233 nM for comparison. KD calculated by using 2-state reaction model: rfB32R, 0.06 μM; rfB32W, 0.07 μM; and rfB32Q, 0.12 μM. (B) Recombinant fB was flowed over the C3b-coated chip at concentrations between 233 and 4 nM in the presence of 1 μg/mL fD. Sensorgrams from rfB32R are solid lines, rfB32Q are dotted lines, and rfB32W are in gray; identical concentrations are indicated for the 3 variants. KD calculated by using 2-state reaction model: rfB32R, 2.7 nM; rfB32W, 6.0 nM; and rfB32Q, 9.5 nM.
Isolation of Ba and Binding to C3b.
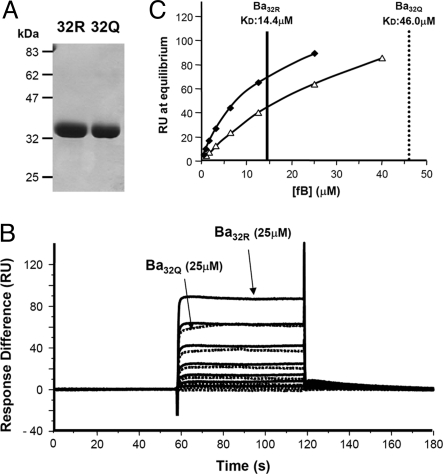
The above data imply that variation in Ba influences proenzyme formation and the amount of enzyme formed. FB binds C3b through a multipoint contact, involving Ba and the vWF-A domain. To confirm differential binding of the variant Ba domains to C3b, Ba fragments were generated from purified fB32R and fB32Q by incubating fB and fD with C3b immobilized on Sepharose. Bb was removed on an anti-Bb affinity column, Ba was “polished” by gel filtration, and used immediately in SPR binding studies with immobilized C3b (Fig. 5A). In contrast to fB, binding was very weak, necessitating high concentrations of Ba to determine binding affinity. Ba purified from fB32R bound C3b with 3-fold higher affinity than Ba generated from fB32Q (Fig. 5 B and C).
Fig. 5.
SPR analysis of Ba binding to C3b. (A) The variant fB proteins were purified from plasma of donors known to be homozygous for fB32R or fB32Q, and Ba was isolated. (B) Ba was flowed over the surface of the C3b-coated chip at concentrations between 25 and 0.4 μM in Biacore buffer (10 mM Hepes, pH 7.4/50 mM NaCl/1 mM MgCl2/0.005% surfactant P20). Sensorgrams from Ba32R are solid lines and Ba32Q are dotted lines; identical concentrations are indicated for the 2 variants. (C) Steady state analysis of the data indicate the affinities for C3b in these buffer conditions are: KD Ba32R, 14.4 μM; and KD Ba32Q, 46.0 μM. For Ba32Q, a sample at 40 μM was included, because the affinity was markedly lower than Ba32R.
Effect of Ionic Strength on Convertase Formation.
To dissect out the differences between the fB variants, enzyme formation and Ba binding were analyzed in low salt concentrations. To confirm that the differential binding occurred at physiological salt, proenzyme and convertase formation by fB32R and fB32Q were analyzed in Hepes-buffered saline (150 mM). The fB32R variant again showed a 3- to 4-fold higher affinity in proenzyme formation compared with fB32Q, with resultant enhanced convertase formation (Fig. S2).
Discussion
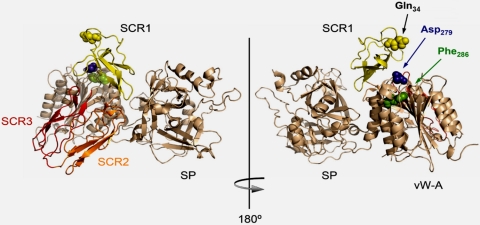
The common allotypes of fB affect risk of disease, the most striking example being AMD, where fB32Q is protective (OR 0.32; 95% CI: 0.21–0.48; see refs. 26, 27). The variant amino acid in fB is just 7 aa from the amino terminus (25-aa leader sequence removed) within the Ba domain; a location that is paradoxical in that Ba is not part of the activated convertase. Therefore, it cannot affect enzyme activity (Fig. 6). The crystal structure of fB demonstrated that Ba was folded back onto the Bb domain, with SCR2 and SCR3 of Ba packed tightly into an antiparallel dimer capped by SCR1, this amino-terminal SCR likely hindering access of C3b to the vWF-A domain in Bb (32). It was suggested that initial binding of the 3 SCRs to C3b displaced them from the vWF-A and serine protease domains, allowing access of C3b, and triggering structural rearrangements that enabled proteolytic cleavage of fB by fD. The amino terminus of fB is unstructured, and the crystal structure commences at Q34 (amino acid 9 in the mature protein; see ref. 32); Q34 (and, by inference, the variant amino acid R32/Q32) is in close proximity to 2 residues in the vWF-A domain (D279 and F286), known to affect proenzyme formation (Fig. 6; see ref. 7). The location of the variant amino acid in fB, and data indicating a binding interaction between Ba and C3b (33), led us to hypothesize that the polymorphism would affect proenzyme formation.
Fig. 6.
Structural analysis of the fB R32Q amino acid substitution. (Left) The crystal structure of human fB shows the SCRs of Ba folded back across the VWF-A domain [Milder et al. (32), Protein Data Bank accession number 2OK5]. (Right) Amino acid Q34 is illustrated in the structure, this amino acid is 2 residues downstream of Q32, and lies in close proximity to D279 and F286, alterations in which are known to affect proenzyme formation (C3bB). The molecular graphic was generated by using PyMOL Molecular Graphics System (DeLano, 2002, available at www.pymol.org).
To test the hypothesis, fB variants were isolated from plasma. In hemolysis assays using rabbit erythrocytes as target, they showed differential activities, with fB32Q being the least lytic (Fig. 1). The underlying mechanism was dissected by using SPR. When proenzyme formation was analyzed, fB32Q bound C3b with a 4-fold lower affinity compared with fB32R; the fB32W variant had an intermediate affinity (Fig. 2). These data were confirmed by using rfB variants (Fig. 4). The influence of the polymorphism in the Ba domain on C3b binding was confirmed by isolating the variant Ba, and directly comparing binding to C3b (Fig. 5). Although the affinity of Ba for C3b is weak, avidity effects from simultaneous multisite binding in the Ba and Bb domains likely explains the striking effect of variation in Ba on apparent affinity of fB. When effects of fB variants on formation of the activated convertase, C3bBb were studied, fB32R generated 2.5-fold more enzyme than fB32Q under the experimental conditions (Fig. 3). These data show that fB32Q is less efficient at maintaining amplification in the AP, and provide a mechanism for the observation that fB32Q protects from pathology in AMD.
Previous investigations into functional differences between the fB variants have produced variable results. Early studies using hemolytic overlay assays indicated that fB32Q had either decreased or equivalent functional activity compared with fB32R (34–36). These data were complicated by variable plasma concentrations of fB and differences in expression levels of the allele products, making quantitative comparisons difficult. A later study by Horiuchi et al. (37) analyzed the functional activity of recombinant forms of fB32R and fB32Q, and concluded that there was no functional difference. In this study, convertase was assembled on the surface of C3b-coated sheep erythrocytes by using native properdin and fD, and rfB (quantified by ELISA in cell culture supernatant). We have shown that the R32Q polymorphism influences the efficiency of proenzyme formation, rather than the activity of the convertase itself; therefore, the functional difference would only be revealed if the assay reported by Horiuchi et al. (37) was developed at a point where formation of activated convertase was incomplete, and not all of the fB was cleaved to Bb and stabilized by properdin. In our studies, all proteins were highly purified and polished by gel filtration into assay buffer immediately before each assay, the resultant sensitive assays have enabled us to establish beyond doubt functional differences between the variants.
The fB32Q variant is apparently only protective in AMD and not in other diseases of complement dysregulation (e.g., DDD and aHUS). The reasons for disease specificity are unclear. Several studies have demonstrated elevated plasma levels of complement activation products (including C3a, C3d, Ba, and SC5b-9) in AMD compared with controls, indicating systemic dysregulation of complement, and implying that AMD is a systemic disease with local manifestation in the retina (38, 39). This systemic component may amplify the effects of the fB polymorphism, explaining the disease specificity. Comparative expression levels from the fB alleles in controls and AMD are not yet available; early studies using crossed immunoelectrophoresis and radial immunodiffusion assays demonstrated a trend for higher expression from the fB32Q allele in normal individuals (38, 39) although, as with many complement components, the range around the mean was large. Decreased plasma levels of the fB32R allele product may indicate decreased expression, or increased catabolism due to AP tickover or chronic inflammatory processes.
The triggers for drusen accumulation and retinal pathology in AMD are unknown; however, autoimmune mechanisms have been suggested. Lipid moieties in the rod photoreceptor membranes become photo-oxidized over time, forming omega-(2-carboxyethyl)pyrrole (CEP) adducts that may elicit an immune response, generating autoantibodies that trigger local inflammatory processes, including complement activation (40, 41). Damaged tissue and accumulation of extracellular insoluble debris is in itself inflammatory, likely driving complement activation and immune responses in AMD, as in other chronic pathologies such as Alzheimer's disease, where damaged neurons and deposits of amyloid β peptide drive complement activation (42).
We here explain the mechanism by which a common polymorphism in a complement component protects an individual from AMD. The R32Q polymorphism in fB (rs12614 and rs641153) affects risk of AMD by altering AP activity. Variations in C3 (R102G) and CFH (V62I and Y402H) are also linked to altered risk of AMD, although the functional consequences of these changes have not been characterized; early reports that fH402H bound less well to CRP in drusen have not been substantiated (43). Because fB32Q is less efficient at amplifying the AP, it protects from AMD and perhaps other chronic inflammatory pathologies; however, it may have the opposite effect in infection, where fB32Q may predispose to disease due to suboptimal AP activation on pathogens. Indeed, the association of variants of multiple complement components or CReg with disease (e.g., C3, fB, and fH in AMD; fH, fI, fB, C3, and CD46 in aHUS) suggest that particular combinations of variants, or “complotypes,” may combine to influence systemic complement activity in an individual; thus, affecting risk of chronic inflammatory damage and susceptibility to infection. The multilocus risk model of AMD suggests that the effects of common variants are additive, and the model can identify individuals whose lifetime risk of AMD ranges from <1 to >50% (20–28). Thus, specific variant sets of complement components and CReg can dramatically influence outcome, an observation that is increasingly important as the population ages, likely impacting not only in AMD, but also in other chronic inflammatory diseases such as Alzheimer's disease and atheroma. Understanding of the mechanisms underlying these associations is an essential prerequisite to the design of therapies that target complement activation in these diseases. For example, our data suggest that anti-Ba antibodies may have potential in the clinic, particularly for those individuals who carry high risk combinations of complement components and CReg.
Materials and Methods
Genotyping.
Healthy volunteers were screened for mutations/polymorphisms in the CFB gene by DNA sequencing of PCR amplified fragments. Genomic DNA was prepared from peripheral blood cells according to standard procedures. Each exon of the CFB gene was amplified from genomic DNA by using specific primers derived from the 5′ and 3′ intronic sequences as described (7). Sequencing was performed in an ABI 3730 sequencer by using a dye terminator cycle sequencing kit (Applied Biosystems).
Preparation of Complement Components and Activation Fragments.
C3 and factor B were purified from normal human serum by classical chromatography (C3) or affinity chromatography (fB); details are given in SI Methods. Ba was purified as a by-product of AP activation as detailed in SI Methods. All proteins used for Biacore studies were gel-filtered into the appropriate Biacore buffer before experimentation.
Production of Recombinant fB.
A cDNA encoding full-length fB32R was introduced in the eukaryote expression vector pCI-Neo (Promega), and R32Q-aa and R32W-aa substitutions introduced by using QuikChange site-directed mutagenesis kit (Stratagen) and appropriate primers. All fB cDNA clones were fully sequenced to confirm fidelity. CHO cells, maintained in Ham-F12 medium (GIBCO-BRL) supplemented with 10% FCS, L-glutamine (2 mM final concentration), penicillin, and streptomycin (10 U/mL and 100 μg/mL), were plated in p60 plates at 5 × 105 cells per well. Transfections were carried out 1 day later with 10 μg of the pCI-Neo constructs and 24 μL of Lipofectine (Invitrogen) in a total volume of 1 mL of medium per well. Transfected cells were selected in G418 sulfate (Geneticin; GIBCO-BRL) at 500 μg/mL, cloned by limiting dilution, and clones producing the highest levels of rfB as assessed by ELISA (7) were expanded for production; rfB was purified from tissue culture supernatant by affinity chromatography on anti-Bb as described above. Eluate was concentrated, and rfB was polished by gel filtration on Superdex200 in Biacore buffer.
Hemolysis Assays.
NHS was depleted of fB by flowing over the JC1 anti-Bb affinity column in CFD. Run-through was collected, and undiluted fractions pooled and stored at −80 °C. Use of this standard fB-depleted serum eliminated any variation in hemolysis due to polymorphisms or concentration differences in other complement components and regulators. Rabbit erythrocytes (Erb), washed in CFD (Complement Fixation Diluent, Oxoid) and resuspended to 2% (vol/vol), were mixed with fB depleted NHS (NHSΔfB; 1/38 final dilution; 50 μL of cells, 25 μL of diluted NHSΔfB) and 50 μL of a dilution of purified, gel-filtered fB. CFD permits activation of all pathways of complement activation; therefore, lysis resulted from direct activation of the AP on the surface of Erb or from AP-mediated amplification of the other pathways. After 45 min, cells were pelleted by centrifugation, and hemoglobin release was measured by absorbance at 415 nm. Control incubations included 0% lysis (buffer only) and 100% lysis (0.1% Nonidet-P40). Percentage lysis = 100*(A415 test sample-A415 0% control)/(A415 100% control-A415 0% control). The log10 of fB concentration (final concentration in the incubation) was plotted on the x axis, and percentage lysis on the y axis. Nonlinear regression was used to fit the curves (GraphPad Prism), and EC50 was calculated as the concentration of fB giving a response half way between background (no fB) and maximum.
Biosensor Analysis.
All analyses were carried out on a Biacore T100 (GE Healthcare). C3b was amine coupled to the sensor chip as instructed by the manufacturers (NHS/EDC coupling kit). In the example illustrated in Figs. 2 and 3, 880RU C3b are immobilized, although replicate experiments used differing amounts. In Fig. 4, 458RU C3b are immobilized. For kinetic analyses a CM5 (carboxymethylated dextran) chip was used, and data collected at 25 °C at a flow rate of 30 μL/min; data from a reference cell was subtracted to control for bulk refractive index changes. Samples were injected by using the KINJECT command to ensure accurate association kinetics. To analyze proenzyme formation, fB variants were flowed across the C3b surface at different concentrations; the surface was regenerated with EDTA containing buffer as previously described (31). To analyze formation of the activated convertase, fD was included at 1 μg/mL, and the surface was regenerated by using soluble rCD55 (gift of Susan Lea, Oxford, United Kingdom; see ref. 31). In the case of Ba, no regeneration step was required. Due to the low affinity of the Ba interaction, high levels of C3b (4058RU) were immobilized, and steady state analysis was used to quantitate affinity. Data were evaluated by using Biaevaluation T100 evaluation software (Version 1.1), global fitting was used to determine kinetic parameters that fitted all curves (differing concentrations of fB) within an experiment. Concentration of analytes was assessed by using absorbance at A280, molarities were calculated by using the following extinction coefficients (molecular masses and coefficients obtained by using Protean software, DNAStar): fB, 1.43; and Ba, 1.74.
Supplementary Material
Acknowledgments.
We thank the blood donors for their invaluable contribution to the project. This work was supported by Wellcome Trust University Award 068823 (to C.L.H.), Medical Research Council Project Grant 84908 (to C.L.H. and B.P.M.), Ministerio de Ciencia e Innovación Grant SAF 2008-00226 (to S.R.d.C.), and the Centro de Investigación Biomédica en Red de Enfermedades Raras and Fundación Renal Iñigo Álvarez de Toledo (to S.R.d.C.). B.P.M. was funded by a Wellcome Trust Program Grant (06850).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812584106/DCSupplemental.
References
- 1.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 2.Lachmann PJ, Hughes-Jones NC. Initiation of complement activation. Springer Semin Immun. 1984;7:143–162. doi: 10.1007/BF01893018. [DOI] [PubMed] [Google Scholar]
- 3.Morgan BP. The complement system: An overview. Methods Mol Biol. 2000;150:1–13. doi: 10.1385/1-59259-056-X:1. [DOI] [PubMed] [Google Scholar]
- 4.Morgan BP, Meri S. Membrane proteins that protect against complement lysis. Springer Semin Immun. 1994;15:369–396. doi: 10.1007/BF01837366. [DOI] [PubMed] [Google Scholar]
- 5.Morgan BP, Harris CL. Complement Regulatory Proteins. London: Academic; 1999. [Google Scholar]
- 6.Atkinson JP, Liszewski MK, Richards A, Kavanagh D, Moulton EA. Hemolytic uremic syndrome: An example of insufficient complement regulation on self-tissue. Ann N Y Acad Sci. 2005;1056:144–152. doi: 10.1196/annals.1352.032. [DOI] [PubMed] [Google Scholar]
- 7.Goicoechea de Jorge E, et al. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA. 2007;104:240–245. doi: 10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alper CA, Boenisch T, Watson L. Genetic polymorphism in human glycine-rich beta-glycoprotein. J Exp Med. 1972;135:68–80. doi: 10.1084/jem.135.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jahn I, et al. Genomic analysis of the F subtypes of human complement factor B. Eur J Immunogenet. 1994;21:415–423. doi: 10.1111/j.1744-313x.1994.tb00214.x. [DOI] [PubMed] [Google Scholar]
- 10.Abbal M, et al. Two subtypes of BfF by isoelectrofocusing: Differential linkage to other HLA markers. Hum Genet. 1985;69:181–183. doi: 10.1007/BF00293294. [DOI] [PubMed] [Google Scholar]
- 11.Davrinche C, Abbal M, Clerc A. Molecular characterization of human complement factor B subtypes. Immunogenetics. 1990;32:309–312. doi: 10.1007/BF00211644. [DOI] [PubMed] [Google Scholar]
- 12.Messias-Reason IJ, Urbanetz L, Pereira da Cunha C. Complement C3 F and BF S allotypes are risk factors for Chagas disease cardiomyopathy. Tissue Antigens. 2003;62:308–312. doi: 10.1034/j.1399-0039.2003.00101.x. [DOI] [PubMed] [Google Scholar]
- 13.Papiha SS, Duggan-Keen M, Roberts DF. Factor B (BF) allotypes and multiple sclerosis in north-east England. Hum Hered. 1991;41:397–402. doi: 10.1159/000154033. [DOI] [PubMed] [Google Scholar]
- 14.Hagglof B, et al. Studies of HLA, factor B (Bf), complement C2 and C4 haplotypes in type 1 diabetic and control families from northern Sweden. Hum Hered. 1986;36:201–212. doi: 10.1159/000153627. [DOI] [PubMed] [Google Scholar]
- 15.Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration–emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelsalam A, Del Priore L, Zarbin MA. Drusen in age-related macular degeneration: Pathogenesis, natural course, and laser photocoagulation-induced regression. Surv Ophthalmol. 1999;44:1–29. doi: 10.1016/s0039-6257(99)00072-7. [DOI] [PubMed] [Google Scholar]
- 17.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 18.Donoso LA, Kim D, Frost A, Callahan A, Hageman GS. The role of inflammation in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2006;51:137–152. doi: 10.1016/j.survophthal.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 20.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 21.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 23.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maller J, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 25.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 26.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer KL, et al. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum Mol Genet. 2007;16:1986–1992. doi: 10.1093/hmg/ddm146. [DOI] [PubMed] [Google Scholar]
- 28.Spencer KL, et al. Deletion of CFHR3 and CFHR1 genes in age-related macular degeneration. Hum Mol Genet. 2008;17:971–977. doi: 10.1093/hmg/ddm369. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsdottir J, Conley YP, Weeks DE, Ferrell RE, Gorin MB. C2 and CFB genes in age-related maculopathy and joint action with CFH and LOC387715 genes. PLoS ONE. 2008;3:e2199. doi: 10.1371/journal.pone.0002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritsche LG, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 31.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55) J Biol Chem. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 32.Milder FJ, et al. Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol. 2007;14:224–228. doi: 10.1038/nsmb1210. [DOI] [PubMed] [Google Scholar]
- 33.Pryzdial EL, Isenman DE. Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J Biol Chem. 1987;262:1519–1525. [PubMed] [Google Scholar]
- 34.Mauff G, Adam R, Wachauf B, Hitzeroth HW, Hiller C. Serum concentration and functional efficiency of factor B alleles. Immunobiology. 1980;158:86–90. doi: 10.1016/S0171-2985(80)80045-3. [DOI] [PubMed] [Google Scholar]
- 35.Mortensen JP, Lamm LU. Quantitative differences between complement factor-B phenotypes. Immunology. 1981;42:505–511. [PMC free article] [PubMed] [Google Scholar]
- 36.Lokki ML, Koskimies SA. Allelic differences in hemolytic activity and protein concentration of BF molecules are found in association with particular HLA haplotypes. Immunogenetics. 1991;34:242–246. doi: 10.1007/BF00215259. [DOI] [PubMed] [Google Scholar]
- 37.Horiuchi T, et al. Human complement factor B: cDNA cloning, nucleotide sequencing, phenotypic conversion by site-directed mutagenesis and expression. Mol Immunol. 1993;30:1587–1592. doi: 10.1016/0161-5890(93)90450-p. [DOI] [PubMed] [Google Scholar]
- 38.Sivaprasad S, et al. Estimation of systemic complement C3 activity in age-related macular degeneration. Arch Ophthalmol. 2007;125:515–519. doi: 10.1001/archopht.125.4.515. [DOI] [PubMed] [Google Scholar]
- 39.Scholl HP, et al. Systemic complement activation in age-related macular degeneration. PLoS ONE. 2008;3:e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu X, et al. Carboxyethylpyrrole protein adducts and autoantibodies, biomarkers for age-related macular degeneration. J Biol Chem. 2003;278:42027–42035. doi: 10.1074/jbc.M305460200. [DOI] [PubMed] [Google Scholar]
- 41.Hollyfield JG, et al. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14:194–198. doi: 10.1038/nm1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeer PL, McGeer EG. Inflammation and the degenerative diseases of aging. Ann N Y Acad Sci. 2004;1035:104–116. doi: 10.1196/annals.1332.007. [DOI] [PubMed] [Google Scholar]
- 43.Hakobyan S, et al. Complement factor H binds to denatured rather than to native pentameric C-reactive protein. J Biol Chem. 2008;45:30451–30460. doi: 10.1074/jbc.M803648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.