Abstract
The matrix protein (M) of respiratory syncytial virus (RSV), the prototype viral member of the Pneumovirinae (family Paramyxoviridae, order Mononegavirales), has been crystallized and the structure determined to a resolution of 1.6 Å. The structure comprises 2 compact β-rich domains connected by a relatively unstructured linker region. Due to the high degree of side-chain order in the structure, an extensive contiguous area of positive surface charge covering ≈600 Å2 can be resolved. This unusually large patch of positive surface potential spans both domains and the linker, and provides a mechanism for driving the interaction of the protein with a negatively-charged membrane surface or other virion components such as the nucleocapsid. This patch is complemented by regions of high hydrophobicity and a striking planar arrangement of tyrosine residues encircling the C-terminal domain. Comparison of the RSV M sequence with other members of the Pneumovirinae shows that regions of divergence correspond to surface exposed loops in the M structure, with the majority of viral species-specific differences occurring in the N-terminal domain.
Keywords: CD, crystal structure, respiratory syncytial virus, sequence alignment
Respiratory syncytial virus (RSV) is the prototype member of the Pneumovirinae, a subfamily of the Paramyxoviridae (order Mononegavirales). Morphologically, the extracellular virion consists of a lipid bilayer envelope, within which are embedded 3 glycoproteins, 2 of which (F and G) are important in cell attachment and viral entry into target cells. The third, the SH protein, contributes to pathology in the host (1). Internally, virions contain helical nucleocapsids that consist of N protein tightly bound to the negative-sense nonsegmented genomic RNA. The nucleocapsid in turn is associated with components of the viral RNA-dependent RNA polymerase (L, P, M2-1, and M2-2 proteins), forming the holo-nucleocapsid (2–4). Between the holo-nucleocapsid and the outer envelope there is a layer of matrix protein (M), which is associated peripherally with the membrane (5). The other family members of the Mononegavirales (Rhabdoviridae, Filoviridae, and Bornaviridae) all subscribe to this basic arrangement of the virion, although the overall morphology can vary between the families. For example, Paramyxoviridae virions are pleiomorphic, whereas the Rhabdoviridae have a regular bullet shape structure, and the Filoviridae have a more filamentous shape.
Extracellular RSV virions form by a budding process that occurs at the plasma membrane within specialized lipid domains (5, 6) and M appears to drive the final assembly process, which is the incorporation of the holo-nucleocapsid and initiation of the budding process (7, 8). Before budding, there is a coordinated assembly of viral components; and it is evident that the glycoproteins and M proteins are important determinants of the location on the plasma membrane at which the virus buds (9). It is also possible that the interaction between M and the glycoproteins, possibly mediated with the cytoplasmic tails, are important in the budding process. Genome silencing to prevent transcription and replication by the viral polymerase before incorporation of the holo-nucleocapsid in the nascent virion is a related function of the M. M are also implicated in host cell transcriptional cut-off (10), possibly via a direct interaction with RNA (11).
A number of matrix-like proteins are known to bind membranes or lipid vesicles in vitro, most likely through a combination of hydrophobic and electrostatic interactions (12–14). Expression of certain Ms in eukaryotic cells in the absence of other viral proteins can induce formation of virus-like particles (VLPs). The efficiency of VLP generation can be increased if the M is coexpressed with a viral glycoprotein (15–18). Ms share a tendency to oligomerize, a feature likely to be important in the self-assembly and budding processes (19). In tissue culture, RSV induces formation of long slender projections from the surface of the cell known as viral filaments. It was found that removal of the lipid membrane from viral filaments left an M containing sheath (5); it is possible that M oligomerization and self-assembly is the driving force behind the formation of viral filaments. In this article we report the structure of the full-length RSV M protein solved at a resolution of 1.6 Å, and discuss the implications of this structure for the function of the protein.
Results and Discussion
Structure of the RSV M.
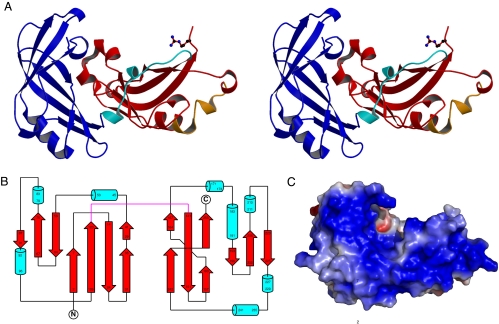
The RSV M was purified by using nickel affinity chromatography. During the cloning process, a methionine to arginine change occurred, and we refer to the resultant form of M as M254R. The crystal structure of M254R was solved by using MIRAS techniques to a resolution of 1.6 Å, representing the first example of an intact M from the Mononegavirales. Despite evidence for higher order oligomers [supporting information (SI) Fig. S1] such as dimers, tetramers, and hexamers, in solution, the crystallized form is monomeric. Crystallographic data are presented in Table S1. The overall fold consists of 2 clear domains connected by a 13-residue linker region. The N-terminal domain comprises residues 1 to 126, whereas the C-terminal domain consists of residues 140 to 255 (Fig. 1A and B). Only residues 99 and 100 could not be assigned a clear location within the electron density. The N-terminal domain consists of a twisted β-sandwich comprised of 2 β-sheets, 1 of 3 and 1 of 4 strands, positioned almost perpendicular to each other. The overall topology of this domain is of a curved horseshoe-like arrangement with β-sheet 1 forming the concave, inner face flanked by loop regions, and β-sheet 2 forming the convex, outer face. The C-terminal domain consists of a flattened β-barrel, comprising 2 3-stranded anti-parallel β-sheets. The regions linking the sheets between strands 2 and 3, and strands 5 and 6, are largely helical in nature (Fig. 1B). There is no evidence for any complexed metal ions or for a potential zinc finger motif. The 254R substitution in the protein lies at the very end of the C-terminal domain in an area largely devoid of secondary structure; therefore, it is unlikely to have a significant effect on the protein structure or function.
Fig. 1.
Three-dimensional structure of the RSV M protein. The crystal structure of M254R (resolution 1.6 Å) shows 2 domains composed largely of β-sheets. Statistical information for the X-ray data are provided in SI Materials and Methods. (A) Divergent (wall eyed) stereoview of M254R colored according to domain with the linker shown in cyan, the N-terminal domain in blue and the C-terminal domain in red. Residue R254 is shown in ball-and-stick representation. (B) A topology diagram of the protein. The linker between the N- and C-terminal domains is shown in magenta. Residues (numbers refer to Met as +1) in β-sheets are represented by broad arrows and helices as cylinders. (C) Electrostatic surface potential (calculated with APBS) for M254R, presented in a color range from red to blue (−5 to +5 kT/e); uncharged residues are uncolored.
The linker region is largely lacking in secondary structure features, with the exception of a short helical region. The presence of this linker is consistent with structures obtained for fragments of the Ebola virus (EBOV) VP40 and vesicular stomatitis virus (VSV) Ms, and from capsid proteins of retroviral origin that fulfil similar in vivo roles (20–24), and its unstructured nature suggests that the N- and C-terminal domains may be able to occupy different orientations, relative to each other, than that observed in the crystal. The 2 domains are only loosely associated, with the major interactions between them being hydrophobic in nature, supported by a small number of water-mediated hydrogen bonds. We have observed that, in common with EBOV VP40, proteolysis of RSV M254R occurs in solution and results in the dissociation of the N- and C-terminal domains, the weak interdomain interactions being insufficient to hold them together. We have mapped the cleavage site of M254R by limited mass spectrometry to the linker region between amino acids Thr-136 and Leu-137 (see Fig. S1). These weak interdomain interactions further suggest that the protein may exist in alternative quaternary structures in solution, and that the interdomain packing observed in the crystal may be metastable, driven mainly by sequestering of the hydrophobic residues, which form the major part of the interface, away from the bulk solvent. Flexibility of this type has been speculated as being important for the M of the influenza virus, as well as for EBOV VP40 and VSV M, and is thought to be necessary to accommodate the different functions of the M throughout the viral life cycle.
Structure Comparison.
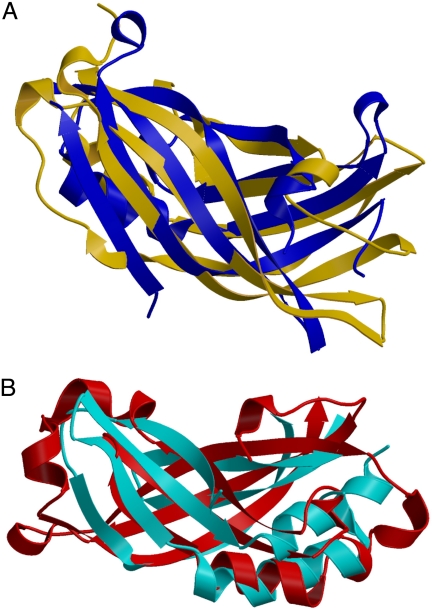
The tertiary structure of M254R is globally similar to that of the EBOV VP40 protein (24), showing the same overall fold with a Z score of 6.6 and a rmsd of 3.7 Å. Searches of the PDB for structurally homologous proteins to M254R using the programs DALI, VAST, and SSM (25–27) yield no further significantly similar structures. Performing the same searches using the N- and C-terminal domains independently demonstrates that, in addition to being closely related to each other, the N-terminal domain shows slight similarities to parts of 2 DNA topoisomerases from Escherichia coli. These similarities are restricted to the shape of the β-sheets in domain 2 of the DNA binding proteins, and are unlikely to be relevant to the function of the N-terminal domain.
Structural information on Ms is scarce partly due to the intrinsic difficulty of working with these hydrophobic proteins that are prone to self-aggregation. Only 2 Ms from the Mononegavirales have been subjected to high-resolution structure determination. For both published structures, a proteolytically resistant core was crystallized as opposed to the full-length structure. Separate overlay of the N- and C-terminal domains of M254R with those of EBOV VP40 (PDB code 1ES6, see Fig. 2 and Fig. S2), allows the difference in angle between the domains in the 2 proteins to be accounted for, and shows the similarities in the protein core of each domain. Despite the clear close relation between the folds of the 2 proteins, the topological arrangement of the 2 differs. The N-terminal domain of RSV M254R contains a mixed 4-stranded and 1 anti-parallel 3-stranded β-sheet as opposed to the 2 3-stranded sheets found in VP40. A similar comparison of M254R and VSV M could not be performed, emphasising the structural, but not functional, diversity of the 2 Ms.
Fig. 2.
Comparison of the RSV M protein topology with that of EBOV VP40. The diagrams show an overlay of the β-sheet arrangements of M254R with EBOV VP40 (PDB code 1ES6). (A) RSV M protein N-terminal domain in blue and VP40 in yellow; and (B) M254R protein C-terminal domain in red and VP40 in cyan. The same images are presented in Fig. S2, in stereoscopic views.
Electrostatic Surface of the RSV M.
To fulfil its structural role, RSV M must be able to form protein–protein and protein–lipid interactions. Consequently, one would expect to observe surface areas with significant hydrophobic patches, as well as positively charged regions, that would favor protein–membrane association. Examination of the surface of M reveals an extensive positively charged area of ≈600 Å2, extending across both N- and C-terminal domains encompassing, and including, a significant contribution from the linker (Fig. 1 C). Because the binding of M to cell membranes is thought to be mediated largely by electrostatic contacts (12, 28), this region provides a mechanism by which the protein is able to associate with negatively charged host membranes. Comparison of the electrostatic surface of M254 with VP40 is hampered by the fact that in the latter the linker region is not modeled. However, it does appear that a significant positively-charged patch is a feature present on all Ms for which there is structural information available, including retroviral homologues. Looking at the rest of M254R, there is a distinctive negatively charged lobe on the N-terminal domain and a positively charged pocket on the C-terminal domain, which are potential sites for directing interactions with binding partners. Work on the isolated N-terminal domain of EBOV M, VP40 (29), indicates that this domain has the capacity to oligomerize in the presence of nucleic acids, pointing to a more prominent role for this domain in RNP association, with the C-terminal domain interacting predominantly with the membrane. The precise role of each domain in the full-length proteins (RSV M and EBOV VP40) has yet to be determined, although the extent of the positively-charged surface of RSV M indicates that functional surfaces have the potential to extend across the domain boundary and the linker to some degree.
Comparison of Structures in Solution and in Crystal.
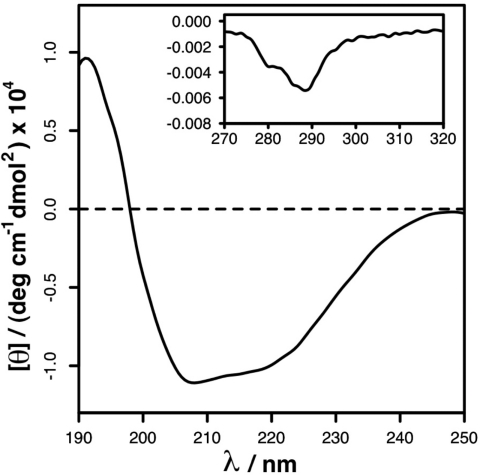
To determine whether the crystal structure was also that of the protein in solution, we performed a CD analysis of the protein. Data collected included wavelengths down to 190 nm, which significantly increases confidence in the interpretation of CD derived structural information for comparison with crystallographic data (29). Evaluation of CD (Fig. 3 and Table 1; for more details of CD spectrum deconvolution, see Figs. S3 and S4 and Tables S2 and S3) and X-crystallographic data indicate that the secondary structure content of M254R in solution is lower than that observed in the crystalline state. This observation implies that assembly of the protein is associated with significant unfavorable entropic contribution from protein folding, in common with the binding of disordered proteins to their counterparts (30–33). Such proteins achieve a favorable binding free energy through large enthalpic contributions from intramolecular electrostatic interactions to offset negative entropic contributions from protein folding. In order for this to be achieved, the number of contacts between the protein and its binding partners are numerous. In the case of M, analogy with disordered proteins would suggest that the contact area for protein–protein and protein–membrane interactions is similarly large. These requirements are consistent with the observed distribution of aromatic and positively charged residues in the protein, particularly with regard to the large patch of positive electrostatic potential described above. Intrinsic disorder has been proposed as a mechanism by which viral proteins are able to form multiple binding interactions with different partners; thus, expanding protein functionality without a concomitant increase in the size of the genome. RSV M fits this description having multiple interactions, such as with itself, the nucleocapsid, the viral glycoproteins, such as the F protein, via their cytoplasmic tails and with the host cell and viral membranes. All of these interactions are essential for the assembly and budding of a virion particle, and could contribute to stabilizing the structure.
Fig. 3.
CD spectrum of RSV M. Datasets of the far-UV (main box, 190–250 nm) and near- UV (inset box, 270–320 nm, axes are the same as the main plot) spectra were collected as indicated in Experimental Procedures. The data were analyzed for secondary structure information by using the CDSSTR, SELCON3, and CONTIN/LL programs. A more comprehensive dataset on M254R structure in solution is provided in Fig. S3, Fig. S4, Table S2, and Table S3; also, see Table 1. A comparison with X-ray data with calculated CD spectra indicates that the crystal is more ordered than the solution structure.
Table 1.
Comparison of the solution and crystallographic structure of the the RSV M protein
| α-Helix | 310-Helix | Strand | Turn | Coil | Bridge | |
|---|---|---|---|---|---|---|
| % of total, CD* | 12 | 12 | 23 | 22 | 31 | — |
| % of total, X-ray† | 12 | 8 | 47 | 21 | 12 | 1 |
Summary of the output resulting from analysis of the M254R structure using the Stride program. Comparison of the CD and X-ray data. The output from the Contin/LL program is presented as this gave the best fit between experimental and computed structures. For more details of the various programs used to fit the CD data to known structures, see SI Materials and Methods. The CD spectrum shows that the solution structure is more disordered that that of the crystallized form, compare ″strand″ (23 vs. 47%) and ″coil″ (31 vs. 12%).
*Output from the CONTIN/LL program.
†Output from Stride analysis.
Comparison with Other Pneumovirinae Ms.
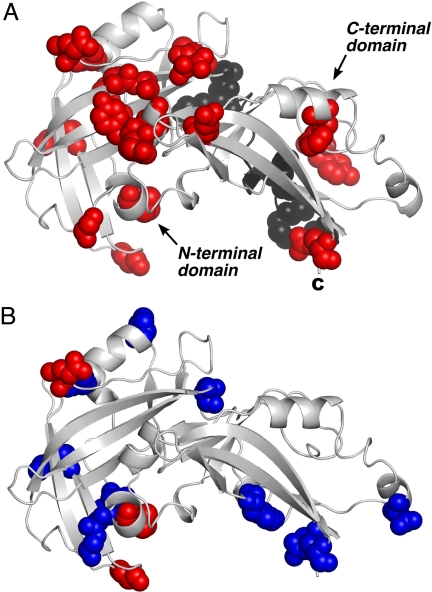
Analysis of sequence alignments of a number of pneumovirus and metapneumovirus M proteins with M254R reveals that the majority of significant amino acid sequence diversity (Q-scores of 20 and above) can be mapped to external loop and edge regions of the β-sheets and to the linker region (Fig. 4A and B; for alignments, see Fig. S5). An alignment of the more closely related bovine and ovine RSV M proteins with human RSV M proteins demonstrates that the major variations in amino acid sequence are mostly found in regions at each end of the horseshoe structure, with the remainder occurring in the linker and the ends of the helix that lies on the outer surface of the N-terminal domain. Inclusion of the more distantly related metapneumoviral Ms in the alignment, such as those from human and avian metapneumoviruses, produces a similar pattern for the C-terminal domain; however, the differences are much more pronounced in the N-terminal domain, with significant sequence diversity occurring in the linker and in the regions adjacent to the linker. We hypothesize that these differences correlate with surface residues that mediate species-specific interactions, such as M with its cognitive nucleocapsid or viral glycoproteins. Given that membrane interactions are unlikely to be sequence-specific, and that protein activity is not transferred between species outside experimental conditions, this hypothesis is consistent with the involvement of loop regions in protein–protein and/or protein–RNA interactions, particularly for the N-terminal domain, where sequence diversity is greatest.
Fig. 4.
Distribution of residues over the surface of RSV M that display significant sequence diversity from related proteins. (A) Residues with a Q score ≤20 after alignment of all 8 pneumovirus proteins to M254R are displayed in red (nonlinker residues) or black (linker residues). The C terminus is indicated by C. (B) Residues with a Q score ≤20 or ≤50 after alignment of all RSV proteins are shown in red and blue, respectively. The orientation of the structure is the same as in A. Alignments are presented in Fig. S5.
Model for Membrane Binding.
The large positively-charged area on the surface of RSV M, which spans both domains, is consistent with the role of this protein in membrane association. We hypothesize that it is likely that this patch will be the driving force for association with the negatively-charged lung membrane (34, 35). This deduction would be consistent with biochemical observations of other matrix-like proteins, where electrostatic charge is the major component of the interaction between protein and membrane, at least in vitro. Association of the positively charged patch to the membrane, particularly at N- and C-terminal domain orientations other than those observed in the crystal structure, would leave a significant hydrophobic area on the protein exposed that may drive interactions with other viral components, or become buried at the protein–membrane interface.
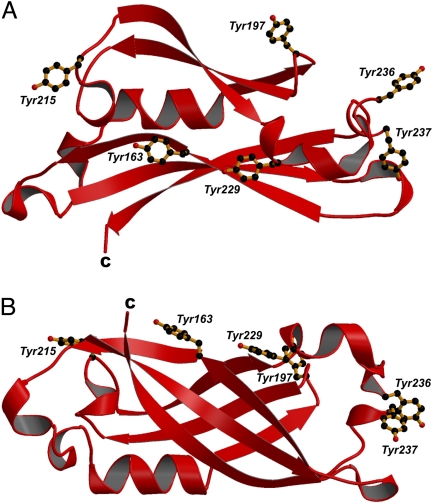
Considering the C-terminal domain alone, although a significant proportion of the surface residues are hydrophobic (36), it is pertinent that this domain has a number of surface exposed arginine and lysine residues that are able to contribute to a favorable interaction with negatively-charged membranes. Membrane binding by the C-terminal domain, driven largely by electrostatic interactions, with a contribution from hydrophobic residues, would leave the N terminus free to perform more species-specific functions, such as protein–protein interactions needed for virion assembly. This interpretation would be consistent with observations on EBOV VP40 (30). We also note that the C-terminal domain has a striking arrangement of tyrosine residues (Y163, Y197, Y215, Y229, Y236, and Y237) forming a planar distribution that encircles the majority of the C-terminal domain (Fig. 5A and B) with a degree of overlap with the positively-charged patch. Tyrosine, among other aromatic amino acids, has been demonstrated to occur with high probability in the interfacial region of membrane proteins (37–39), and in model systems has been shown to associate strongly with phospholipids (40). The functionality of the tyrosine residues has yet to be investigated, but the motif is conserved within all RSV Ms. The metapneumoviral Ms do not have this motif, although, in these proteins, functionally equivalent residues replace the tyrosines. Mutagenesis studies based on the structure presented within this article will further our understanding of the contribution of particular residues or features to the biological functionality of this protein.
Fig. 5.
Distribution of tyrosine residues in the C-terminal domain of the RSV M. The N-terminal domain of M254R has been omitted for clarity. The planar distribution of the residues (shown as ball-and-stick representations) on the surface of the M protein is readily apparent. The C-terminal residue is indicated by C; A and B show orthogonal views of this domain.
Conclusion
The structure of the M of RSV has been solved by X-ray crystallography to a resolution of 1.6 Å. The high resolution and high degree of crystallographic order observed in the structure allow us to throw light on the mode of membrane binding and the mechanism by which this protein performs its varied and critical roles. Central to these roles, there is a significant area (≈600 Å2) of positive electrostatic potential that forms an extended surface for interaction with the membrane, which carries a complementary negative potential. Because this area spans both domains and the linker, the geometry of surface contacts made by the protein will depend on the relative positioning of the domains, should those contacts involve contributions from both domains.
Experimental Procedures
Protein Expression Purification and Crystallization.
A histidine tagged version of M254R was expressed in E. coli strain BL21 (CodonPlus). Cells were lyzed by sonication and M254R purified by nickel-affinity chromatography. After removal of insoluble material, the protein was subjected to crystallization trials. Successful crystallization was obtained in 70% Tacsimate, pH 7.0; later, the conditions were optimized to 55–65% Tacsimate, pH 7.0.
X-Ray Data Collection.
Data from native, seleno-methionine derivatized, and mercury soaked crystals were collected at the European Synchrotron Radiation (Grenoble, France) (native crystal) and Stanford Synchrotron Radiation Lightsource (derivatized forms) facilities. The crystal statistics are presented in the SI Materials and Methods. The 3D figures presented were generated by using Molscript (Fig. 1A), PyMol (Fig. 1B), and the topology diagram rendered in TopDraw (see SI Materials and Methods).
Circular Dichroism Spectroscopy.
Protein samples were dialysed against 5 mM phosphate buffer overnight at 4 °C. Far-UV CD spectra and the corresponding blanks were recorded between 190 and 250 nm in a cuvette of path length of 0.2 cm by using a Jasco J-810 Spectropolarimeter by averaging 8 accumulations recorded at a rate of 10 nm/min, with a pitch of 0.5 nm, a bandwidth of 1 nm, and a response time of 2 s. Near-UV spectra were recorded by using a 1-cm cell, with a pitch 0.2 nm and a response time of 1 s. After subtraction of the appropriate blank, binominal smoothing was carried out within the Jasco Spectra Analysis program. Smoothed data were analyzed for protein secondary structure by using the CDSSTR, SELCON3, and CONTIN/LL programs (41), accessed either via the Dichroweb service (42, 43), or the CDPro package. Both a general protein (SP43/dataset 4) and a membrane protein (SMP56/dataset 10) reference set were used (41, 44).
Full methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Professor E. N. Baker (University of Auckland) for support and advice on the preparation of this manuscript; the European Synchrotron Radiation Facility (Grenoble, France) and Stanford Synchrotron Radiation Lightsource for X-ray data collection facilities; and Dr. G. Sharples and Dr. E. Pohl (Durham University, Durham, United Kingdom) for critical reading of the manuscript. V.A.M. was supported by a European Molecular Biology Organization long-term fellowship. This work was also supported in part by contributions from OneNorthEast and from the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for the RSV M protein have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 2vqp (coordinates) and r2vqpsf (structure factors)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0805740106/DCSupplemental.
References
- 1.Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol. 2007;81:8361–8366. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins PL, Chanock RM, Murphy BR. In: Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Vol 1. Philadelphia: Lippincott-Raven; 2001. pp. 1443–1486. [Google Scholar]
- 3.Murphy LB, et al. Investigations into the amino-terminal domain of the respiratory syncytial virus nucleocapsid protein reveal elements important for nucleocapsid formation and interaction with the phosphoprotein. Virology. 2003;307:143–153. doi: 10.1016/s0042-6822(02)00063-6. [DOI] [PubMed] [Google Scholar]
- 4.Murray J, Loney C, Murphy LB, Graham S, Yeo RP. Characterization of monoclonal antibodies raised against recombinant respiratory syncytial virus nucleocapsid (N) protein: Identification of a region in the carboxy terminus of N involved in the interaction with P protein. Virology. 2001;289:252–261. doi: 10.1006/viro.2001.1150. [DOI] [PubMed] [Google Scholar]
- 5.Henderson G, Murray J, Yeo RP. Sorting of the respiratory syncytial virus matrix protein into detergent-resistant structures is dependent on cell-surface expression of the glycoproteins. Virology. 2002;300:244–254. doi: 10.1006/viro.2002.1540. [DOI] [PubMed] [Google Scholar]
- 6.Brown G, et al. Analysis of the interaction between respiratory syncytial virus and lipid-rafts in Hep2 cells during infection. Virology. 2004;327:175–185. doi: 10.1016/j.virol.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 7.Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Peebles ME. The Paramyxoviruses. New York: Plenum; 1991. pp. 427–456. [Google Scholar]
- 9.Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol R. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghildyal R, Baulch-Brown C, Mills J, Meanger J. The matrix protein of Human respiratory syncytial virus localises to the nucleus of infected cells and inhibits transcription. Arch Virol. 2003;148:1419–1429. doi: 10.1007/s00705-003-0112-y. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez L, Cuesta I, Asenjo A, Villanueva N. Human respiratory syncytial virus matrix protein is an RNA-binding protein: Binding properties, location and identity of the RNA contact residues. J Gen Virol. 2004;85:709–719. doi: 10.1099/vir.0.19707-0. [DOI] [PubMed] [Google Scholar]
- 12.Ruigrok RW, et al. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J Mol Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 13.Scianimanico S, et al. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 2000;19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timmins J, Ruigrok RW, Weissenhorn W. Structural studies on the Ebola virus matrix protein VP40 indicate that matrix proteins of enveloped RNA viruses are analogues but not homologues. FEMS Microbiol Lett. 2004;233:179–186. doi: 10.1111/j.1574-6968.2004.tb09480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z, et al. Biochemical and functional characterization of the Ebola virus VP24 protein: Implications for a role in virus assembly and budding. J Virol. 2003;77:1793–1800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayakar HR, Jeetendra E, Whitt MA. Rhabdovirus assembly and budding. Virus Res. 2004;106:117–132. doi: 10.1016/j.virusres.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt AP, Leser GP, Waning DL, Lamb RA. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol. 2002;76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugahara F, et al. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology. 2004;325:1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Gaudin Y, Barge A, Ebel C, Ruigrok RW. Aggregation of VSV M protein is reversible and mediated by nucleation sites: Implications for viral assembly. Virology. 1995;206:28–37. doi: 10.1016/s0042-6822(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 20.Riffel N, et al. Atomic Resolution Structure of Moloney Murine Leukemia Virus Matrix Protein and Its Relationship to Other Retroviral Matrix Proteins. Structure. 2002;10:1627–1636. doi: 10.1016/s0969-2126(02)00896-1. [DOI] [PubMed] [Google Scholar]
- 21.Hatanaka H, et al. Structure of equine infectious anemia virus matrix protein. J Virol. 2002;76:1876–1883. doi: 10.1128/JVI.76.4.1876-1883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudier M, Gaudin Y, Knossow M. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 2002;21:2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dessen A, et al. Crystallization and preliminary X-ray analysis of the matrix protein from Ebola virus. Acta Crystallogr D. 2000;56:758–760. doi: 10.1107/s0907444900004388. [DOI] [PubMed] [Google Scholar]
- 24.Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 26.Gibrat J, Made T, Bryant S. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 28.Faaberg KS, Peeples ME. Association of soluble matrix protein of Newcastle disease virus with liposomes is independent of ionic conditions. Virology. 1988;166:123–132. doi: 10.1016/0042-6822(88)90153-5. [DOI] [PubMed] [Google Scholar]
- 29.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nature Protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoenen T, et al. VP40 Octamers Are Essential for Ebola Virus Replication. J Virol. 2005;79:1898–1905. doi: 10.1128/JVI.79.3.1898-1905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 32.Radivojac P, et al. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sickmeier M, et al. DisProt: The Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dombrowsky H, Clark GT, Rau GA, Bernhard W, Postle AD. Molecular species compositions of lung and pancreas phospholipids in the cftr (tm1HGU/tm1HGU) cystic fibrosis mouse. Pediatr Res. 2003;53:447–454. doi: 10.1203/01.PDR.0000049937.30305.8A. [DOI] [PubMed] [Google Scholar]
- 35.Palestini P, et al. Composition, biophysical properties, and morphometry of plasma membranes in pulmonary interstitial edema. Am J Physiol. 2002;282:L1382–L1390. doi: 10.1152/ajplung.00447.2001. [DOI] [PubMed] [Google Scholar]
- 36.Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M. Calculated using the ASC program. J Comp Chem. 1995;16:273–284. [Google Scholar]
- 37.Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 38.Sanderson JM. Peptide-lipid interactions: Insights and perspectives. Org Biomol Chem. 2005;3:201–212. doi: 10.1039/b415499a. [DOI] [PubMed] [Google Scholar]
- 39.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 40.Sanderson JM, Whelan EJ. Characterisation of the interactions of aromatic amino acids with diacetyl phosphatidylcholiney. Phys Chem Chem Phys. 2004;6:1012–1017. [Google Scholar]
- 41.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 42.Lobley A, Whitmore L, Wallace BA. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 43.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004;13:100–112. doi: 10.1110/ps.03258404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.