Abstract
Comparative genomics has provided evidence for numerous conserved protein domains whose functions remain unknown. We identified a protein harboring “domain of unknown function 860” (DUF860) as a component of group II intron ribonucleoprotein particles in maize chloroplasts. This protein, assigned the name WTF1 (“what's this factor?”), coimmunoprecipitates from chloroplast extract with group II intron RNAs, is required for the splicing of the introns with which it associates, and promotes splicing in the context of a heterodimer with the RNase III-domain protein RNC1. Both WTF1 and its resident DUF860 bind RNA in vitro, demonstrating that DUF860 is a previously unrecognized RNA-binding domain. DUF860 is found only in plants, where it is represented in a protein family comprising 14 orthologous groups in angiosperms. Most members of the DUF860 family are predicted to localize to chloroplasts or mitochondria, suggesting that proteins with this domain have multiple roles in RNA metabolism in both organelles. These findings add to emerging evidence that the coevolution of nuclear and organellar genomes spurred the evolution of diverse noncanonical RNA-binding motifs that perform organelle-specific functions.
Keywords: DUF860, mitochondria, plastid
The evolution of mitochondria and chloroplasts from bacterial endosymbionts was accompanied by large scale transfer of genes to the nucleus (1). Accordingly, many organellar proteins are encoded by nuclear genes of bacterial ancestry that retain their ancestral function. However, during the long coevolution of mitochondria and chloroplasts with their host cell, both organelles acquired features that are not typical of their bacterial ancestors. The origin of the genes that confer such traits is only beginning to be elucidated.
The complex RNA metabolism characteristic of plant mitochondria and chloroplasts provides striking examples of acquired, nonprocaryotic traits. For example, both organellar genomes are rich in introns, RNAs in both organelles are modified by RNA editing, and posttranscriptional events have the predominant role in determining gene product abundance (2, 3–5). Many proteins that participate in such processes were not derived from the endosymbiont, but rather emerged in the context of nuclear-organellar coevolution. For example, the pentatricopeptide repeat (PPR) protein family is found only in eucaryotes, where it has been implicated in RNA-related processes in mitochondria and chloroplasts (6). Current data argue that PPR proteins generally function as RNA-interaction platforms, but they appear to be derived from the tetratricopeptide repeat (TPR) motif, a more ancient motif that binds protein ligands.
Here, we present evidence that a previously uncharacterized protein family defined by “domain of unknown function 860” (DUF860) fits this general paradigm. We show that the DUF860 protein WTF1 (“what's this factor?”) is required for the splicing of group II introns in chloroplasts, that it associates in vivo with its genetically-defined RNA ligands, and that both WTF1 and its resident DUF860 exhibit RNA-binding activity in vitro. Most members of the DUF860 family are predicted to localize to chloroplasts or mitochondria, suggesting that proteins with this domain have multiple roles in gene expression in both organelles. Although DUF860 is found only in land plants, it is distantly related to a class of ubiquitin hydrolases (UBH) found throughout the eucaryotes. However, structural modeling suggests that DUF860 adopts a structure that differs from UBH enzymes, and that has a surface that is reminiscent of helical repeat RNA-binding motifs such as the PPR and PUM-HD motifs.
Results
WTF1 Is Found in Group II Intron Ribonucleoprotein Particles (RNPs).
WTF1 came to our attention during our analysis of the machinery that promotes the splicing of group II introns in chloroplasts. Nine nucleus-encoded chloroplast splicing factors have been identified in land plants, each involved in the splicing of distinct intron subsets (7–14). In an effort to identify additional factors, we used antibodies to the splicing factors CRS1, CAF1, and CAF2 to immunopurify intron RNPs from chloroplast extract, and we identified coimmunoprecipitating proteins by mass spectrometry. Two proteins identified in this manner were shown previously to be authentic components of intron RNPs (7, 15).
A predicted protein containing DUF860 (IPR008578) was identified in both the CAF1 and CAF2 coimmunoprecipitates [supporting information (SI) Table S1 and Fig. S1]. As no functional information was available for this or any DUF860 protein, we named it WTF1. The rice and Arabidopsis proteomes include 17 and 15 DUF860 proteins, respectively, which comprise 14 orthologous groups (Fig. S1 and Fig. S2). WTF1 and its orthologs are predicted to localize to chloroplasts (Fig. S2), consistent with the isolation of WTF1 from a chloroplast preparation.
Recovery of wtf1 Insertion Mutants.
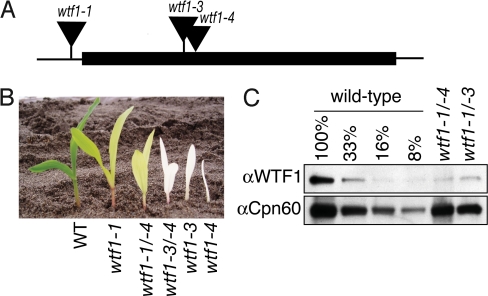
To gain insight into the function of WTF1, we screened our collection of transposon-induced nonphotosynthetic maize mutants for insertions in the wtf1 gene. The mutant alleles used for subsequent experiments are shown in Fig. 1. The wtf1-1 insertion cosegregates with a recessive mutation conferring a pale green phenotype, whereas the wtf1-3 and wtf1-4 insertions cosegregate with recessive mutations conferring an albino phenotype (Fig. 1B). The insertions in wtf1-3 and wtf1-4 disrupt the ORF (Fig. 1A), and are anticipated to be null alleles; the wtf1-1 insertion is upstream of the ORF, consistent with the weaker phenotype observed. The F1 progeny of crosses between plants heterozygous for each allele segregated chlorophyll-deficient, seedling lethal mutants, demonstrating that these phenotypes result from the disruption of wtf1. The chlorophyll deficiency of wtf1-1/wtf1-4 plants is intermediate between that conditioned by the parental alleles (Fig. 1B).
Fig. 1.
Mutant alleles of wtf1. (A) Mu transposon insertions in the wtf1 gene. The wtf1 ORF lacks introns and is indicated by a rectangle. (B) Phenotypes of wtf1 mutants. Plants indicated by 2 alleles are the heteroallelic progeny of complementation crosses. (C) Loss of WTF1 protein in wtf1 mutant chloroplasts. Chloroplasts purified from seedlings of the indicated genotypes were analyzed on immunoblots with anti-WTF1 antiserum. Cpn60 was used as a loading control. Strong wtf1 alleles could not be analyzed in this manner because plastids cannot be purified in sufficient quantity from albino plants.
Polyclonal antibodies were raised to a segment of WTF1 that lacks strong similarity to nonorthologous proteins. These antibodies detect a protein of the size expected for WTF1 in wild-type chloroplasts (Fig. S3A). This protein accumulates to reduced levels in chloroplasts from hypomorphic wtf1 mutants (Fig. 1C), confirming that it is WTF1.
WTF1 Is Found in Complexes Containing Known Chloroplast Splicing Factors.
To confirm and refine the intracellular localization of WTF1, immunoblots of subcellular fractions were probed with the WTF1 antibody (Fig. 2A). WTF1 is enriched in isolated chloroplasts with respect to its concentration in leaf, and was detected solely in the stromal fraction. Antibodies to WTF1 coimmunoprecipitated both CAF1 and CAF2 from stroma (Fig. 2B), confirming that these “bait” proteins, with which WTF1 was initially identified, are associated with WTF1. Furthermore, WTF1 coimmunoprecipitated with several other chloroplast splicing factors (Fig. 2B). Coimmunoprecipitation with RNC1 was particularly strong, whereas weak but reciprocal coimmunoprecipitation was detected with CFM3. WTF1 antibody also coimmunoprecipitated CRS1. CAF1, CAF2, CFM3, RNC1, and CRS1 reside in stromal particles that include group II intron RNAs (7, 10, 12). When stroma was fractionated by sedimentation through sucrose gradients, WTF1 was found in particles spanning the same size range as these group II intron RNPs (≈600–800 kDa; see Fig. 2C).
Fig. 2.
WTF1 is associated with splicing factors in chloroplast stroma. (A) Chloroplasts (Cp) and chloroplast subfractions (Thy, thylakoid; Env, envelope) are from the fractionated chloroplast preparation described previously (30); material in each lane is derived from the same quantity of chloroplasts. The blot was reprobed to detect mitochondrial MDH and the thylakoid protein D1. The Ponceau S-stained blot below illustrates the distribution of the stromal protein RbcL and the loading of the leaf and mitochondrial (Mito) samples. (B) Coimmunoprecipitation of WTF1 with chloroplast splicing factors. Stromal extract was used for immunoprecipitations with the antibodies listed at top. Immunoprecipitates were analyzed by immunoblot analysis with the antibodies listed at left. (C) Cosedimentation of WTF1 with intron RNPs. Stromal extract was fractionated by sucrose gradient sedimentation. An equal proportion of each fraction and of the pelleted material (P) was analyzed by probing immunoblots with the antibodies indicated at left. The ribosomal protein Rpl2 marks the position of ribosomes. RbcL marks the position of Rubisco (≈550 kDa). The WTF1, RNC1, CAF1, and CAF2 peaks coincide with those of CRS1, CFM3, and group II intron RNAs in analogous assays (9, 10, 12, 13).
WTF1 Is Associated with Group II Intron RNAs in Chloroplast Extract.
The coimmunoprecipitation of WTF1 with group II intron splicing factors and its cosedimentation with intron RNPs suggested that WTF1 is itself associated with group II introns. To identify RNAs that associate with WTF1, we used a genome-wide RNA coimmunoprecipitation assay (“RIP-chip”) as an initial screen. WTF1 was immunoprecipitated from stromal extract; RNAs purified separately from the pellet and supernatant were labeled with red- or green-fluorescing dye, respectively, combined and hybridized to a tiling microarray of the maize chloroplast genome. Two replicate experiments were performed with WTF1 antiserum; the results were compared with those obtained by using an antibody to OE16, which does not bind RNA. The data are summarized in Table S2 and Fig. S3B. All of the array elements demonstrating highly significant differential enrichment in the experimental versus control assays were from loci containing group II introns (trnK, trnG, atpF, trnV, petB, petD, rpl2, ndhB, rps12-2, trnI, and trnA) or from loci that are cotranscribed with genes containing group II introns (e.g., psbH, ndhJ, and rpl23).
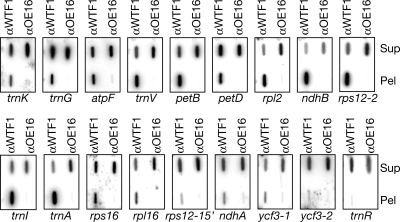
To validate the RIP-chip data, RNAs obtained from replicate immunoprecipitations were analyzed by slot-blot hybridization (Fig. 3). All of the highly-significant RIP-chip peaks were confirmed by the slot-blot data. In addition, the rps16 and rpl16 introns, which were detected as small peaks of marginal significance by RIP-chip, were validated in the slot-blot assay. Several RNAs that did not emerge as peaks in the RIP-chip assay likewise showed little or no enrichment in the slot-blot assay (ycf3-2, rps12-1, and trnR). The ndhA and ycf3-1 introns proved to be weakly enriched when assayed by slot-blot hybridization. The slight enrichment of the ycf3-2 and rps12-1 introns can be accounted for by their presence on the same RNA molecules as the ycf3-1 and rps12-2 introns, respectively.
Fig. 3.
WTF1 is associated with intron RNAs in chloroplast extract. RNA purified from the pellets and supernatants of immunoprecipitations with antisera to WTF1 or OE16 was applied to slot blots and hybridized with the indicated probes. All probes were intron-specific, except that for trnR, which lacks introns. One tenth of the RNA from each immunoprecipitation supernatant (Sup) and one fifth of the RNA from the corresponding pellet (Pel) was analyzed with each probe.
These experiments showed that the trnK, trnG, atpF, trnV, petB, petD, rpl2, ndhB, rps12-2, trnI, and trnA introns are associated with WTF1 in chloroplast extract. This intron set includes known ligands of CAF1 (petD and trnG), CAF2 (petB and ndhB), CFM3 (trnG, petB, petD, and ndhB), and CRS1 (atpF) (10, 11, 16), consistent with the coimmunoprecipitation of WTF1 with these proteins. In addition, the slot-blot data provided evidence that WTF1 associates with the rps16 and rpl16 introns, suggested weak associations with the ndhA and ycf3-1 introns, and argue against an association with the ycf3-2 or rps12-1 intron. However, most striking is the overlap between the intron set that coimmunoprecipitates with WTF1 and that reported previously for RNC1 (7). This similarity suggested that the functions of RNC1 and WTF1 might be coupled, a possibility that was confirmed in subsequent experiments.
WTF1 Is Required for the Splicing of Chloroplast Introns.
To determine whether WTF1 promotes splicing in vivo, the splicing of all chloroplast group II introns was assayed in wtf1 mutants. Noncomplementing progeny of crosses between different wtf1 alleles were used for these experiments to ensure that defects observed result from the disruption of wtf1. Mutations in wtf1 cause a reduction in plastid ribosome content, as revealed by a loss of plastid rRNAs and of all photosynthetic enzyme complexes that include plastid-encoded subunits (Fig. S4). Severe plastid ribosome deficiencies cause pleiotropic effects on plastid RNA metabolism, including the failure to splice introns in subgroup IIA (16, 17). Therefore, we analyzed splicing in wtf1-4/-1 mutants, whose moderate ribosome loss is not expected to disrupt splicing, and in wtf1-4/-3 mutants, whose severe plastid ribosome deficiency is anticipated to disrupt subgroup IIA splicing. Results were compared with those obtained with control mutants having plastid ribosome deficiencies of a similar magnitude (Fig. S4B): wtf1-4/-1 mutants were compared with hcf7 and wtf1-4/-3 mutants were compared with iojap.
Many splicing defects were detected in wtf1 mutants, with the results correlating well with the RNA coimmunoprecipitation data. Poisoned-primer extension assays revealed a reduced ratio of spliced to unspliced RNA from the petB, petD, ndhB, rpl2, and rps12-2 loci (Fig. 4A and Fig. S5B). RNA gel blots demonstrated a reduced ratio of spliced to unspliced RNA from all tRNA loci containing group II introns (trnG, trnV, trnI, trnA, and trnK) (Fig. 4B and Fig. S5A). The loss of spliced tRNAs and of the excised rps12-2 intron was accompanied by an increase in unspliced precursors (Fig. 4B and Fig. S5), indicating a defect in splicing rather than in stabilization of the spliced products. Ribonuclease protection assays demonstrated a defect in rps16 splicing (Fig. 4C), and showed a reduction in rpl16 splicing in strong mutant alleles (Fig. S5C). In contrast, the splicing of the rps12-1 and ycf3-2 introns, which did not coimmunoprecipitate with WTF1, was unaffected in wtf1 mutants (Fig. S5 B and C). Subtle splicing defects were detected for the ycf3-1 and ndhA introns (Fig. S5C), which weakly coimmunoprecipitated with WTF1. The only substantive discord between the RNA coimmunoprecipitation and genetic data concerned the atpF intron: this intron strongly coimmunoprecipitated with WTF1, but its splicing was only subtly disrupted in hypomorphic wtf1-4/-1 mutants (Fig. S5 A and B). Because the atpF intron is in subgroup IIA and fails to splice in mutants lacking plastid ribosomes, analysis of wtf1-4/-3 mutants is uninformative; therefore, the possibility that WTF1 is required for atpF splicing cannot be eliminated.
Fig. 4.
Splicing defects in wtf1 mutants. Assays used seedling leaf RNA from plants of the indicated genotypes. Introns are designated as subgroup IIA or IIB, according to ref. 35. (A) Poisoned-primer extension assays. Oligonucleotides complementary to exon sequences near the 3′-splice junction of the indicated introns were used to prime reverse transcription in the presence of a dideoxynucleotide that terminates cDNA synthesis after different distances on spliced and unspliced templates. The asterisk marks a product terminating at the branchpoint adenosine formed during the first splicing step. (B) RNA gel blots probed with exon sequences from the tRNA gene indicated at bottom. Asterisks identify unspliced precursors. (C) Ribonuclease-protection assay. The probe was body-labeled and spanned the splice junction diagrammed below. The lengths in nucleotides of probe segments corresponding to intron and exon are indicated in the diagram. S, spliced; U, unspliced; pg, pale green seedlings; iv, ivory seedlings.
These results show that WTF1 promotes the splicing of most or all of the introns with which it associates, with some introns more sensitive to the partial loss of WTF1 than others. The role of WTF1 in the splicing of tRNAs and ribosomal protein mRNAs can account for the loss of plastid ribosomes in wtf1 mutants, as the failure to express any of these genes would prevent chloroplast translation and, thus, block ribosome synthesis itself.
WTF1 and DUF860 Bind RNA.
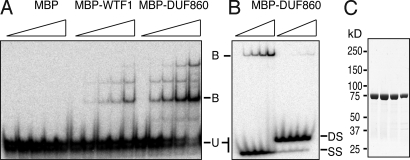
The functional association of WTF1 with RNA in vivo suggested that both WTF1 and its DUF860, which makes up the bulk of the protein (Fig. S1), might have RNA-binding activity. To test this possibility, WTF1 and its DUF860 alone were expressed in Escherichia coli as fusions to maltose-binding protein (MBP), purified, and tested for RNA-binding activity in vitro (Fig. 5A). Both MBP-WTF1 and MBP-DUF860 bound RNA, whereas MBP alone did not. MBP-DUF860 binds with much higher affinity to single-stranded RNA than to the same RNA sequence in double-stranded form (Fig. 5B). These results demonstrate that DUF860 is an RNA-binding domain that binds preferentially to single-stranded RNA. The sequence-specificity of WTF1 was not explored, because it is likely that its specificity in vivo requires its collaboration with RNC1 (see next section).
Fig. 5.
Gel-mobility shift assays demonstrating RNA-binding activity of recombinant WTF1 and DUF860. The proteins indicated at top were incubated with (A) a 141-nt RNA from the petB intron, or (B) a 31-nt RNA from the petA gene. The latter RNA was either heated and snap-cooled to maintain it in single-stranded form (at left, SS), or annealed to its complement (at right, DS). Proteins were used in serial 2-fold dilutions (maximum concentrations of 1 μM for MBP and MBP-WTF1, and 2 μM for MBP-DUF860). RNAs were present at 20 pM. Bound (B) and unbound (U) RNAs were resolved on native polyacrylamide gels. (C) Purity of MBP-DUF860 used for RNA-binding assays. Consecutive fractions from the gel filtration column used as the final purification were analyzed by SDS/PAGE and staining with Coomassie Blue. The purity of MBP-WTF1 is shown in Fig. 6.
WTF1 Functions in the Context of a Heterodimer with RNC1.
The RNAs that coimmunoprecipitate with WTF1 and the splicing defects in wtf1 mutants are strikingly similar to the analogous data for the chloroplast splicing factor RNC1 (7). This observation, together with the fact that WTF1 and RNC1 robustly coimmunoprecipite from stroma (Fig. 2B), suggested that RNC1 and WTF1 might interact directly. To explore this possibility, we assayed the ability of recombinant RNC1 and WTF1 to interact in vitro. RNC1 was retained on amylose-affinity resin after incubation with an MBP-WTF1 fusion protein (Fig. 6A). Also, whereas RNC1 and WTF1 individually eluted from a gel filtration column as expected for their monomeric molecular mass (≈50 kDa), incubation of RNC1 with MBP-WTF1 before fractionation resulted in a second RNC1 peak that coeluted with MBP-WTF1 (box in Fig. 6B). When material in this latter peak was cleaved with TEV protease to separate MBP from WTF1 and then applied again to the column, RNC1 and WTF1 coeluted at a position corresponding to that expected for a heterodimer (Fig. 6B Bottom). The genetic and biochemical data together provide strong evidence that WTF1 and RNC1 form a heteromultimer, most likely a heterodimer, that associates with and promotes the splicing of most group II introns in chloroplasts.
Fig. 6.
WTF1 forms a stable complex with RNC1. (A) Amylose pull-down assay. Recombinant RNC1, MBP, and/or MBP-WTF1 were combined as indicated (Input), and incubated with amylose affinity beads. Material retained on the beads after washing (Pulldown) was assayed by SDS/PAGE. (B) Gel filtration interaction assays. Elution of proteins from a Superdex 200 column was monitored by SDS/PAGE and Coomassie staining. Shown from top to bottom are the elution of: TEV protease-cleaved MBP-WTF1; TEV-protease cleaved MBP-RNC1; a preincubated mixture of MBP-WTF1 and RNC1; the pooled fractions indicated in the box, cleaved with TEV-protease and then applied again to the column. Fraction numbers and the positions of size standards are indicated at top.
Discussion
Our results show that the DUF860 protein WTF1 functions in concert with RNC1 to promote the splicing of group II introns in chloroplasts, that WTF1 interacts in vivo with those introns whose splicing it facilitates, and that both WTF1 and its DUF860 in isolation have RNA-binding activity. These results assign an RNA-binding function to DUF860, add to our understanding of the chloroplast splicing machinery, and provide insight into the molecular innovations that emerged during the coevolution of nuclear and organellar genomes.
DUF860 Is a Plant-Specific RNA-Binding Domain Found in a Family of Predicted Organelle-Localized Proteins.
DUF860 is found in a protein family comprising 14 orthologous groups (Fig. S2). All members of this family contain a single DUF860 and lack other discernable functional motifs. DUF860 has been placed under the umbrella of the PF01088/ubiquitin C-terminal hydrolase domain at Pfam (http://pfam.sanger.ac.uk/family?acc = PF01088). The characterized members of PF01088 are deubiquitinating enzymes found in a clade that includes proteins from all eukaryotic branches. Proteins with DUF860 form a separate clade comprised only of plant proteins. The similarity between DUF860s and UBH is very weak, and amino acids that are essential for UBH activity are not conserved in DUF860s (Fig. S6). Thus, DUF860s are unlikely to have peptidase activity.
DUF860 clearly merits a name distinct from PF01088/ubiquitin C-terminal hydrolase, because it is distinct from UBH proteins in function (RNA binding versus peptidase) and phylogenetic distribution (plant-specific versus all eucaryotes). We propose that DUF860 be renamed the PORR (plant organelle RNA recognition) domain. Structural models of DUF860 generated by the highly-rated I-TASSER algorithm (20, 21) suggest that DUF860 adopts a different structure than UBH proteins (Fig. S7). Unlike the globular α+ß structure of UBH enzymes (18, 19), models of 2 DUF860s representing the most divergent branches of the DUF860 family are characterized by a broad surface composed of arrayed alpha helices. This surface is reminiscent of the proposed RNA-binding surface of PPR tracts (6).
Most members of the PORR/DUF860 family are predicted to localize to either mitochondria or chloroplasts (Fig. S2). Considering the RNA-binding and RNA-splicing activities of WTF1, it seems likely that most PORR/DUF860 proteins function in organellar RNA metabolism. The one prior report concerning a member of this family, rpd1 in Arabidopsis, is consistent with this view (22). The authors concluded that defects in root and embryo development in rpd1 mutants result from the failure to maintain rapid cell divisions at critical developmental stages. In light of our results and the fact that RPD1 is predicted to localize to mitochondria, it seems likely that RPD1 functions in mitochondrial gene expression, and that the developmental defects in rpd1 mutants result from an energy deficit.
Complexity of the Chloroplast Splicing Machinery.
With the characterization of WTF1, each of the group II introns in angiosperm chloroplasts is known to require at least 1 nucleus-encoded protein for its splicing (Fig. 7). The most complex intron RNPs characterized thus far involve the ndhB, petB, petD, and trnG introns, with 5 splicing factors known for each. Each of these proteins is necessary for splicing in vivo, and all of them coimmunoprecipitate and cosediment with their cognate intron RNAs. The combined molecular mass of these proteins (≈300 kDa) exceeds that of the introns with which they associate (≈250 kDa). Therefore, these intron RNPs are as much protein as RNA, resembling ribosomes and spliceosomes in this respect. This situation superficially resembles that in Chlamydomonas reinhardtii chloroplasts, whose 2 group II introns require multiple proteins for their splicing (23–26). However, the C. reinhardtii introns are transcribed in fragments that are spliced in trans, possibly necessitating specialized factors for intron assembly. By contrast, the complexity of intron RNPs in plant chloroplasts cannot be attributed to intron fragmentation, because only the rps12-1 intron is trans-spliced. The complexity of chloroplast group II intron RNPs also has no precedent in bacteria or fungi (27). Proteins involved in chloroplast splicing are highly conserved among monocots and dicots (9, 10, 28), but are distinct in C. reinhardtii, correlating with the independent origin of chloroplast introns in land plants and chlorophytes. Therefore, it appears that there are many evolutionary routes to the acquisition of protein interactions that are permissive for the decay of self-splicing ribozymes into protein-dependent enzymes. Dissecting the specific contributions of each protein to intron recognition, RNA folding, splicing catalysis, and intron turnover presents a challenge for the future.
Fig. 7.
Nucleus-encoded proteins that promote chloroplast group II intron splicing in angiosperms. Introns are designated as subgroup IIA or IIB, according to ref. 35. Introns found in Arabidopsis but not in maize are marked with asterisks. Splicing factors are shown to the outside, annotated with their conserved domains. Where analyzed (9, 10, 28), functions are conserved between monocots and dicots. Results are summarized from this work and from refs. 7–12, 14, 16, 17, and 36. Not shown are WHY1, which associates with and stimulates splicing of the atpF intron (15), and HCF152, which is required for the accumulation of spliced petB RNA but not its excised intron (37). In addition to its primary role in ycf3-2 splicing, OTP51 stimulates the splicing of the atpF, trnV, and trnK introns (8).
Organelles As a Vessel for the Evolution of Protein Families Harboring Noncanonical RNA-Binding Domains.
Genetic screens have consistently identified novel proteins that are characteristic neither of the nuclear/cytosolic compartment nor of bacteria as having critical roles in organellar gene expression. For example, the CRM domain, initially revealed through the genetic analysis of chloroplast splicing, serves as an RNA-binding module specifically in plants (29). The ≈14 member CRM family in angiosperms appears to be dedicated to promoting the splicing of group I and group II introns in mitochondria and chloroplasts (9–12, 29). The PPR family illustrates similar themes, as most members of this large, eukaryote-specific family influence RNA metabolism in organelles (6). As for the CRM and PPR families, the first member of the PORR/DUF860 family to be molecularly characterized (WTF1) influences organellar RNA metabolism, and most other PORR/DUF860 proteins are predicted to localize to mitochondria or chloroplasts. Thus, it seems likely that other members of the PORR/DUF860 family have RNA-related roles in both organelles.
The CRM, PPR, and PORR/DUF860 proteins characterized so far influence aspects of organelle gene expression that are not characteristic of the nuclear/cytosolic compartment (e.g., group II intron splicing and RNA editing). Thus, the acquisition of these processes by organelles appears to have been accompanied by the evolution of new families of RNA-binding proteins that are dedicated to mediating these processes. Once invented, each new RNA-binding motif was exploited repeatedly to perform related functions in both organelles. The similar repertoire of RNA metabolism processes in plant mitochondria and chloroplasts, and similarities between the transit peptides for mitochondrial and chloroplast protein import likely facilitated the sharing of novel RNA-binding protein families between the 2 organelles.
Materials and Methods
Details of experimental procedures are available in SI Materials and Methods.
Identification of WTF1 in CAF1 and CAF2 Immunoprecipitates.
The immunoaffinity purification of CAF1 and CAF2 RNPs, and the identification of coimmunoprecipitating proteins by mass spectrometry were described previously (7).
Plant Material.
Insertion alleles of wtf1 were identified in a PCR-based reverse-genetic screen of a collection of Mu-transposon induced maize mutants (http://chloroplast.uoregon.edu) by using methods analogous to those described previously (30). The duplicated sequences at each insertion site, its coordinates with respect to the start codon, and the type of Mu insertion are: wtf1-1 (−34) gcctaccac MuDR gcctaccac (−25); wtf1-3 (+505) catcgagaa (Mu1) catcgagaa (+514); wtf1-4 (+560) gtctgcctc Mu1 gtctgcctc (+569); hcf7 mutants are pale green with reduced chloroplast polysome assembly (31), whereas iojap mutants are albino and lack plastid ribosomes (32). Seedlings were grown under a 16-h light/8-h dark cycle at 26 °C, and harvested between 7 and 9 days after planting.
Antibodies.
A WTF1 fragment (amino acids 101–270) with a 6× histidine tag was expressed in E. coli, purified and used for immunization of rabbits. Antisera to RPL2 and MDH were generously provided by A. Subramanian (University of Arizona, Tucson, AZ) and K. Newton (University of Missouri, Columbia, MO), respectively. Other antibodies have been described previously (7, 9–12, 33).
Chloroplast Fractionation and Protein Analyses.
Preparation of leaf extracts and immunoblotting were performed as described previously (34). Chloroplast subfractions were those described in Williams and Barkan (30). Stromal extracts were fractionated by sedimentation through sucrose gradients as described in ref. 13.
RNA Analyses.
RNA coimmunoprecipitations and analysis by microarray and slot-blot hybridizations, as well as poisoned-primer extension and ribonuclease protection assays were performed as described previously (7). Arrays contained 248 PCR fragments that tile the chloroplast genome, each in 6 replicate spots.
RNA-Protein and Protein–Protein Interaction Assays.
Recombinant RNC1 was expressed and purified as described previously (7). Mature WTF1 (starting at amino acid 56) and its DUF860 (starting at residues FDTV and ending at residues PRRA) were expressed as MBP fusion proteins from pMAL-TEV. They were purified by amylose-affinity and size-exclusion chromatography, essentially as described for RNC1 (7). Gel mobility shift assays were performed as previously described (7). Binding reactions contained 20 mM Tris·HCl pH 7.5/180 mM NaCl/2 mM DTT/17 μg/μL BSA/0.5 mM EDTA/8.3% glycerol/20 μg/mL heparin. Details of protein–protein interaction assays are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank John Robbins for informing us about the appearance of the Arabidopsis WTF1 ortholog in TAIR release 8.0, and Rosalind Williams-Carrier and Susan Belcher for expert technical assistance. This work was supported by National Science Foundation Grants DBI-0421799 and MCB-0744960 (to A.B.), by U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2008-35301-19038 (to A.B.), and by National Science Foundation Grant DBI-0211935 (to K.J.v.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FJ264201).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812503106/DCSupplemental.
References
- 1.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 2.Marchfelder A, Binder S. In: Molecular Biology and Biotechnology of Plant Organelles. Daniell H, Chase C, editors. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. pp. 261–294. [Google Scholar]
- 3.Zerges W. In: Molecular Biology and Biotechnology of Plant Organelles. Daniell H, Chase C, editors. Dordrecth, The Netherlands: Springer; 2004. pp. 347–383. [Google Scholar]
- 4.Bollenbach TJ, Schuster G, Stern DB. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog Nucleic Acid Res Mol Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz-Linneweber C, Barkan A. In: Cell and Molecular Biology of Plastids. Bock R, editor. Vol 19. Berlin and Heidelberg: Springer; 2007. pp. 213–248. [Google Scholar]
- 6.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 7.Watkins K, et al. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falcon de Longevialle A, et al. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008;56:157–168. doi: 10.1111/j.1365-313X.2008.03581.x. [DOI] [PubMed] [Google Scholar]
- 9.Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asakura Y, Bayraktar O, Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA. 2008;14:2319–2332. doi: 10.1261/rna.1223708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostheimer G, et al. Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7:1227–1238. doi: 10.1017/s1355838201010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins B, Barkan A. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 2001;20:872–879. doi: 10.1093/emboj/20.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz-Linneweber C, et al. A Pentatricopeptide Repeat Protein Facilitates the trans-Splicing of the Maize Chloroplast rps12 Pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prikryl J, et al. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins B, Kulhanek D, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel J, Boerner T, Hess W. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucl Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston SC, et al. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69:108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 22.Konishi M, Sugiyama M. A novel plant-specific family gene, ROOT PRIMORDIUM DEFECTIVE 1, is required for the maintenance of active cell proliferation. Plant Physiol. 2006;140:591–602. doi: 10.1104/pp.105.074724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivier C, Goldschmidt-Clermont M, Rochaix J. Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 2001;20:1765–1773. doi: 10.1093/emboj/20.7.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perron K, Goldschmidt-Clermont M, Rochaix J-D. A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 1999;18:6481–6490. doi: 10.1093/emboj/18.22.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merendino L, et al. A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 2006;34:262–274. doi: 10.1093/nar/gkj429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldschmidt-Clermont M, Girard-Bascou J, Choquet Y, Rochaix J-D. Trans-splicing mutants of Chlamydomonas reinhardtii. Mol Gen Genet. 1990;223:417–425. doi: 10.1007/BF00264448. [DOI] [PubMed] [Google Scholar]
- 27.Fedorova O, Zingler N. Group II introns: Structure, folding and splicing mechanism. Biol Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- 28.Asakura Y, Barkan A. Arabidopsis Orthologs of Maize Chloroplast Splicing Factors Promote Splicing of Orthologous and Species-specific Group II Introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barkan A, et al. The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams P, Barkan A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003;36:675–686. doi: 10.1046/j.1365-313x.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 31.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walbot V, Coe EH. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc Natl Acad Sci USA. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 1998;297:38–57. [Google Scholar]
- 35.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns - A review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meierhoff K, et al. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.