Abstract
For most dsRNA viruses, the genome-enclosing capsid comprises 120 copies of a single capsid protein (CP) organized into 60 icosahedrally equivalent dimers, generally identified as 2 nonsymmetricallyinteracting CP molecules with extensive lateral contacts. The crystal structure of a partitivirus, Penicillium stoloniferum virus F (PsV-F), reveals a different organization, in which the CP dimer is related by almost-perfect local 2-fold symmetry, forms prominent surface arches, and includes extensive structure swapping between the 2 subunits. An electron cryomicroscopy map of PsV-F shows that the disordered N terminus of each CP molecule interacts with the dsRNA genome and probably participates in its packaging or transcription. Intact PsV-F particles mediate semiconservative transcription, and transcripts are likely to exit through negatively charged channels at the icosahedral 5-fold axes. Other findings suggest that the PsV-F capsid is assembled from dimers of CP dimers, with an arrangement similar to flavivirus E glycoproteins.
Keywords: capsid assembly, mycovirus, Partitiviridae, partitivirus
Most known viruses with dsRNA genomes appear to share structural similarity by having a characteristic, 120-subunit T=1 capsid surrounding the genome (1). Such a capsid, sometimes nicknamed ″T=2″, provides protection for the genome and enzymes involved in RNA synthesis and may also contribute to genome packaging. Additional layers may build on this shell, depending on the complexity of each virus.
The encapsidated dsRNA viruses are currently classified into 7 families (Reoviridae, Chrysoviridae, Cystoviridae, Birnaviridae, Picobirnaviridae, Partitiviridae, and Totiviridae) with hosts ranging from bacteria to humans. Among these are pathogens of health and agricultural importance such as rotavirus, bluetongue, and rice dwarf viruses. The genomes of dsRNA viruses comprise a varying number of RNA segments, ranging from 1 in totiviruses up to 12 in some reoviruses. The intact virus particles are transcriptionally active, and newly synthesized plus-strand RNA transcripts have been shown to extrude from specific channels in the T=2 shell (2). The viral RNA-dependent RNA polymerase (RdRp) molecules, which are likely to be tethered to the capsid through noncovalent interactions (3), also catalyze the synthesis of minus-strand RNA from a plus-strand precursor to generate the dsRNA genome in assembling particles. How the viral genome segments, RdRp molecules, and capsid protein (CP) molecules interact to ensure asymmetric transcription and extrusion of the nascent transcripts is an intriguing question, and detailed structural studies have begun to shed some light on these processes (2–6).
Transmission electron cryomicroscopy (cryoEM) and 3D image reconstructions have been reported to date for particles from 6 of the 7 families of encapsidated dsRNA viruses (all but Picobirnaviridae) (7–17). In addition, several crystal structures have been reported, including ones for 3 reoviruses [mammalian orthoreovirus, bluetongue orbivirus, and rice dwarf phytoreovirus (18–20)], 1 birnavirus [infectious bursal disease virus (21)], and 1 totivirus [Saccharomyces cerevisiae virus L-A (22)]. Except for birnaviruses, these structures show that the basic building blocks of the T=2 capsid are CP dimers formed from nonsymmetrically-related CP molecules, A and B, that interact laterally near the icosahedral 5-fold (5f) axes. Although the A and B subunits have similar structural folds, they are distinguished by differences in secondary structures and domain hinge angles, attributable to different bonding interactions with their respective neighbors. The CP molecules of dsRNA viruses are usually large (>60 kDa), and form a thin and spherically-shaped capsid shell, with no large channels. In viruses with high genomic RNA densities, the genome is packed into liquid crystalline arrays that give rise to concentric shells of density, 25–30 Å apart, in the particle interiors (6, 11, 12, 18).
Penicillium stoloniferum viruses F and S (PsV-F and PsV-S) are members of the family Partitiviridae. This family includes members with the smallest CP molecules of any dsRNA virus known to date (e.g., only 420 aa for PsV-F and 434 aa for PsV-S), making them excellent models for studying dsRNA virus assembly. Both PsV-F and PsV-S have 2 essential genome segments, dsRNA1 and dsRNA2, the first encoding the RdRp and the second the CP (23). Each of these segments is packaged in a separate particle. PsV-F, but not PsV-S, contains a satellite segment, dsRNA3. PsV-F dsRNA3 has an unrelated sequence to the other 2 segments and putatively contains only 1 short ORF of 54 aa with unknown function (24). PsV-F and PsV-S can coinfect P. stoloniferum (25) but are distinguishable in many ways, including in their low sequence identities (29% for RdRp and 22% for CP) and in not packaging each others' genomes. The structure of PsV-S virions has been determined to 7.3-Å resolution (14), and the capsid shows 60 unusual, arch-like protrusions, each formed by a quasisymmetric CP dimer.
Here, we report a 3.3-Å crystal structure of the PsV-F virion for family Partitiviridae. The capsid shows a surprisingly different protein organization compared with other dsRNA viruses. In particular, the CP dimers exhibit almost-perfect local 2-fold (2f) symmetry and form prominent surface arches. Moreover, these 2 CP molecules interact extensively by structure swapping, a feature not yet seen in other dsRNA viruses. The first 41 aa of CP are disordered in the crystal structure, but an 8.0-Å cryoEM map suggests that this region forms a separate domain that interacts with the genome. PsV-F virions can synthesize transcripts from each of the 3 dsRNA segments via a semiconservative mechanism, and these transcripts are likely to be extruded through negatively charged channels at the icosahedral 5f axes. The results additionally provide strong evidence that assembly of the PsV-F capsid involves dimers of CP dimers, with an arrangement similar to flavivirus E glycoproteins (26).
Results and Discussion
Overall Architecture of the PsV-F Capsid.
Purified PsV-F virions were examined by cryoEM, and a 3D reconstruction at 8.0-Å resolution was computed from 2,605 particle images. Like in PsV-S, the most prominent features of the PsV-F structure are 60 arch-like protrusions that decorate the spherical shell (Fig. 1A). Closer inspection, however, reveals numerous differences between these 2 partitiviruses. For example, the arches are thinner and oriented distinctly in PsV-F, and the distribution of the genomic dsRNA differs in the 2 viruses.
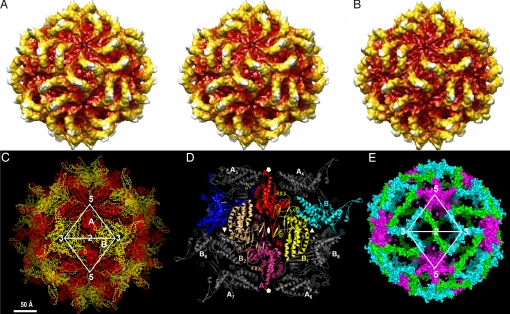
Fig. 1.
Structure of the PsV-F capsid. (A) Radially color-coded surface view of 8.0-Å cryoEM structure, in stereo. (B) Radially color-coded surface view of X-ray crystal structure calculated at 8.0-Å resolution. (C) The Cα trace of the PsV-F capsid structure viewed along an icosahedral 2f axis. The CPA and CPB molecules are colored red and yellow, respectively. Icosahedral 2f, 3f, and 5f symmetry axes are marked. (D) A CP dodecamer with the 4 possible CPA–CPB dimers (A1–B1, A1–B2, A1–B3, and A1–B5). The biologically relevant one, A1–B1, is shown in red and yellow, whereas its 2f-related counterpart, A2–B2, in shown in magenta and light orange. (E) Molecular contacts made by CPA–CPB dimers at the icosahedral 2f (green), 3f (cyan), and 5f (purple) axes. The colored volume shows only structure elements buried at the intermolecular interfaces; thus, the size of the surface shown is directly proportional to the size of buried surface.
To provide further insights into partitivirus structure and assembly, both PsV-F and PsV-S virions were subjected to crystallization. Only PsV-F produced crystals suitable for X-ray diffraction studies. A 3.3-Å structure of PsV-F was then determined by using its cryoEM reconstruction as a phasing model (Table S1). As seen clearly in the crystal structure, the PsV-F capsid consists of 120 CP molecules arranged with icosahedral symmetry (Fig. 1 B and C). Two nonsymmetrically related molecules, A and B, form each asymmetric unit of the icosahedron. Sixty CPA molecules surround the icosahedral 5f axes and form 12 flower-shaped pentamers that contact only at the icosahedral 2f axes. Sixty CPB molecules pack into 20 trimeric clusters around the icosahedral 3-fold (3f) axes. These CPB trimers are fully isolated from one another. The maximum outer and minimum inner diameters of the capsid are ≈370 and ≈250 Å, respectively. The capsid has a rough outer surface featuring 60 arch-like protrusions, each ≈45 Å tall (Fig. 1 B and C). Small pores penetrate this shell at the icosahedral 5f and 3f axes and may play important roles in the export of RNA transcripts as discussed below.
Structure of PsV-F CP.
The atomic model of PsV-F CP includes residues 42–420. The first 41 aa, the only disordered region, appear to extend into the interior of the particle for both CPA and CPB. The overall structure of CP has an unusual reclining-V shape (Fig. 2A, C, and D). The lower arm (amino acids 42–192 and 315–420) is the basic building block of the capsid and is referred to as the shell domain (Fig. 2D). The upper arm (amino acids 193–314) forms the arch-like protrusion and is referred to as the arch domain. The shell domain can be further divided into elbow, wrist, and finger subdomains. The elbow is located at the proximal end of the lower arm and is largely α-helical. It is organized by a long, central helix, α2 (amino acids 101–126), surrounded by 6 additional helices: α1, α3, α4, α5, α7, and α14. The wrist and fingers are mostly β-stranded and have important roles in mediating intermolecular interactions within the shell. The arch domain has a mixed α/β structure. It emanates from the shell domain through 2 antiparallel β-strands (β5 and β8) and ends in a β-hairpin (β6 and β7) at the tip of the arch.
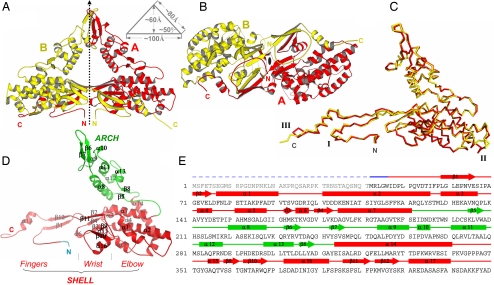
Fig. 2.
Structure of PsV-F CP. (A) Side view of the icosahedral asymmetric unit of the virus particle. The local 2f axis that relates the CPA (red) and CPB (yellow) molecules is shown. This axis is normal to the capsid shell. Dimensions of the dimer are shown in a schematic diagram. (B) CPA–CPB dimer viewed along the local 2f axis from the interior of the virus particle. (C) Superposition of CPA and CPB using all Cα atoms. The 3 most structurally divergent regions are labeled I, II, and III. (D) Structure of a CPA molecule. Secondary structure elements are labeled. (E) Secondary structure assignments in CPA. Rods, arrows, and lines represent α-helices, β-strands, and coils/turns, respectively. The disordered N-terminal region is shown by a dashed line. The color scheme follows that in D.
Among the 4 unique types of CPA–CPB pairs in the PsV-F capsid (Fig. 2B), the one connected by an arch has the largest buried surface area (≈9,900 Å2), suggesting that this type of CP dimer is the likely assembly precursor. The arch affords the dimer a striking profile: a 100 × 80 × 80-Å isosceles triangle with protein densities along the sides and a central solvent space (Fig. 2A). Most of the A–B interactions within this dimer are made by the shell domain (Fig. 2A). In particular, the fingers subdomain, which contains both N- and C-terminal regions of the polypeptide, reaches over and grabs the wrist and elbow of the other molecule. Notable structural features at the dimer shell interface are four 4-stranded β-sheets. Two of these sheets consist of β-strands from the same molecule (β2, β11, β3, and β4), whereas the other 2 sheets are formed by β-strands between the 2 molecules (β9 and β10 from one and β1 and β12 from the other). Additional A–B interactions within this dimer are made by the arch domain (Fig. 2A). In the surface arch, a β-hairpin (β6 and β7) and a connecting helix (α9) from each of the 2 CP molecules wrap around each other. The arch-like protrusions are fairly flexible as suggested by their atomic temperature factors (≈90 Å2 for the tip of the arch vs. ≈52 Å2 for the capsid overall) (see Fig. 4A). Hinge motions in the 2 loops that connect arch and shell presumably give rise to this high mobility.
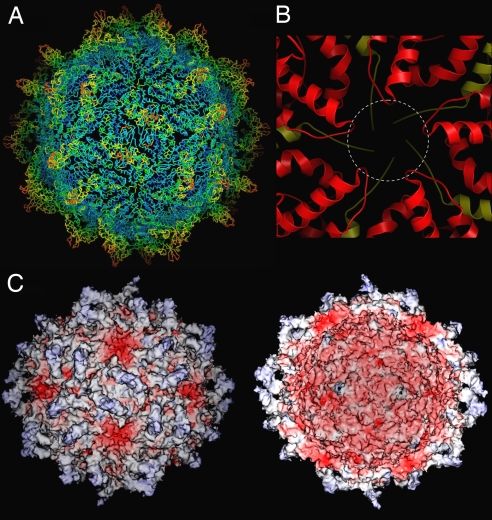
Fig. 4.
Temperature factors and electrostatic potentials of PsV-F capsid. (A) Temperature factors were mapped to the Cα trace. Residues with temperature factors ranging from high (124 Å2) to low (22 Å2) are, respectively, colored by using a red-to-blue spectrum. The virus particle is oriented as in Fig. 1A. (B) Close-up view of areas around an icosahedral 5f axis viewed from the particle exterior, with CPA and CPB colored in red and yellow, respectively. The dotted circle shows an 18-Å channel that may form through structural rearrangement during transcription. (C) Electrostatic potentials on the outer (Left) and inner (Right) surfaces of the virus particle. The molecular surfaces are colored continuously from −74 to 24 kT/e with negative potential in red and positive potential in blue.
The 2 molecules in a CPA–CPB dimer assume nearly identical conformations and are related by almost-perfect local 2f symmetry (1.2 Å rmsd for 379 Cα atoms with a 180° rotation and a 0.2-Å translation). Careful inspection reveals that major differences between the 2 molecules are restricted to 3 small regions (Fig. 2C). Region III, which is formed by the C-terminal region of the CP polypeptide, shows the most pronounced difference. The C terminus of CPB is located near the icosahedral 5f axis (Fig. 1 C and D). In contrast, the C terminus of CPA is located between the 5f and 3f axes and adopts a bent conformation by contacting a neighboring CPA molecule (Fig. 1 C and D). Continuation of the CPA C terminus without bending would result in steric clash with a neighboring CPB molecule at the 3f axis. The rmsd is reduced to 0.7 Å if only 6 residues are removed from these 3 regions.
With only 420 aa (46.5 kDa), PsV-F CP is the smallest among dsRNA viruses for which crystal structures are available. The shell domain is somewhat reminiscent of the apical domain, or domain II, of reovirus CPs in terms of the arrangement of α-helices (18–20), but the overall structure is much smaller (Fig. S1). The arch structure has not been seen in other dsRNA virus families. Indeed the analogous capsid in those other dsRNA viruses are generally thin, smooth (18–20, 22), and in the case of Reoviridae family members, requires additional proteins for stabilization (17–20). Saccharomyces cerevisiae virus L-A of family Totiviridae does not have a stabilizing protein, but faces fewer environmental challenges than do reoviruses because it lacks an extracellular phase to its life cycle. Members of family Partitiviridae likewise do not transmit extracellularly, and their CP molecules are unusually small in comparison with those of other dsRNA viruses. Thus, the presence of surface arches in partitiviruses may serve to enhance capsid stability, which is likely to be a problem for thin capsid shells built from small CP subunits. The arch, shell, and disordered N-terminal domains of PsV-F CP are respectively analogous to the protruding (P), shell (S), and RNA-binding (R) domains of other viral CP molecules as first described in tomato bushy stunt virus (27).
Role of CP Dimers in Organizing the PsV-F Capsid.
In reoviruses and others, the A–B dimer that encompasses the most intermolecular contacts has its 2 subunits lying side-by-side with a 50–60° angle between them (Fig. S1). The A and B subunits from these other viruses also superimpose poorly because of large domain movements. For instance, superposition of the A and B subunits from S. cerevisiae virus L-A or rice dwarf virus yields a respective rmsd of 1.9 or 4.9 Å, and this value is even larger, >10 Å, for the A and B subunits of mammalian orthoreovirus or bluetongue virus.
In PsV-F, such a side-by-side A–B dimer does not exist, because the 2 A and B molecules occupying similar locations do not interact. Instead, the most stable dimer appears to be the one related by the protruding surface arch with almost-perfect 2f symmetry. In each capsid, 60 CPA–CPB dimers interact around 2f, 3f, and 5f axes to form an icosahedral particle. The strongest interactions between these dimers appear to occur at each icosahedral 2f axis, where a surface area of ≈3,500 Å2 is buried. As noted above, these interactions are made entirely between the 2 dimer-associated CPA molecules (Fig. 1 D and E). In comparison, interactions between dimer pairs related by the icosahedral 3f or 5f axes bury respective surface areas of only ≈2,000 and ≈1,900 Å2 (Fig. 1 D and E).
We thus consider it likely that assembly of PsV-F particles proceeds from dimers of CP dimers. Indeed, the extent of protein–protein interaction within a viral capsid has been used to predict the most likely pathway of particle assembly (28). In PsV-F, 30 such dimers of dimers, each of which has a nearly perfect diamond shape with smooth edges (Fig. 1D), are then likely to interact further via 3f and 5f symmetry contacts, with a buried area of ≈3,900 Å2, to complete assembly of the 120-subunit icosahedron. Interestingly, similar dimer–dimer interactions are observed for the E glycoprotein in flavivirus virions (26), even though flavivirus E and PsV-F CP have very different structural folds. Our assembly pathway involving dimers of CP dimers for PsV-F is distinct from that involving pentamers of dimers as proposed for other dsRNA viruses (29), and indeed dimers of dimers and pentamers of dimers represent mutually exclusive assembly pathways.
Internal Densities and Genome Organization.
The N-terminal region of PsV-F CP makes a sharp turn at Ile-47 after extending up from the particle interior. These visible tips of the CPA and CPB N termini are spatially close, with their Cα atoms being only ≈15 Å apart. Because the first well-ordered residue in the CP crystal structure is Met-42, we recognized that it, instead of Met-1 in the ORF of dsRNA2, might be the actual start of CP. To address this possibility, we performed mass spectrometry on particle-associated CP, with results indicating that CP is indeed full length (46.5 kDa, consistent with amino acids 1–420) relative to its ORF (24).
The disordered sequence contains several basic residues. For example, there are 8 Arg or Lys residues in amino acids 11–43 (24% compared with 11% in CP overall). This greater preponderance of basic residues suggests that the N-terminal region might bind to RNA and participate in RNA packaging during particle assembly. It might also play a role during viral transcription. Indeed, some densities for the CP N terminus appear to be present in the 8.0-Å cryoEM map of PsV-F virions (Fig. 3) and in the X-ray crystallography map calculated at lower resolution. Both of these maps suggest interactions between the N-terminal region and the underlying dsRNA. An interesting possibility is that the 2 disordered, spatially close N termini from the same CPA–CPB dimer may form a dimeric structure with RNA-binding activity.
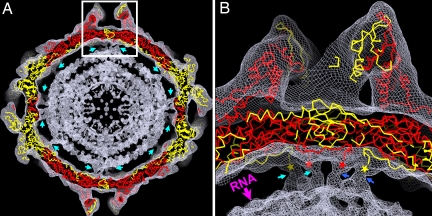
Fig. 3.
Densities of the disordered N-terminal regions and genomic dsRNA. (A) Superposition of the virus atomic model onto the cryoEM reconstruction at 8.0-Å resolution. Only the Cα trace is shown for the atomic model (CPA, red; CPB, yellow). Densities corresponding to the disordered N termini of CPA and CPB are indicated by cyan arrows. (B) Close-up view of the boxed region from A. The disordered N-terminal region, indicated by cyan and blue arrows for densities from 2 different A–B dimers, clearly extends into the underlying RNA (indicated by a pink arrow) through a tube-like cryoEM density. The ordered N-terminal ends of CPA and CPB are indicated by red and yellow stars, respectively.
In each particle, 60 such dimeric structures would be evenly distributed on the inner surface of the capsid shell. The spacing between the shell and the first spherically-ordered RNA density layer is larger than between other concentric RNA density layers (Fig. 3), perhaps because of the presence of these N-terminal structure elements. The presence of these N-terminal peptides increases the mass density in the virion interior by ≈100 mg/mL over the packaged genome. Indeed, at least 3 layers of RNA density are discernible in the cryoEM reconstruction of PsV-F, with an interlayer spacing of ≈25 Å, indicating tight, liquid crystalline packing of the dsRNA.
RNA synthesis activity has been demonstrated for PsV-S (30, 31) and for intact virions of PsV-F in this study (see Fig. 5). The PsV-F capsid structure shows no pores or channels large enough to accommodate an RNA transcript (Fig. 4 A and C). However, structural elements surrounding the 5f axes appear to be flexible. For example, the 5f axes are surrounded by the C-terminal ends of 5 CPB molecules on the inner side and a helix–loop–helix motif from each of 5 CPA molecules on the outer side (Fig. 4B), both of which have rather high temperature factors (Fig. 4A), and may therefore be able to adopt an alternative conformation, opening a channel as much as 18 Å wide. There is also an ≈5-Å pore at each 3f axis, but temperature factors for this region are low (Fig. 4A), suggesting less flexibility. These 3f pores might be used instead for nucleotide substrate or pyrophosphate byproduct diffusion.
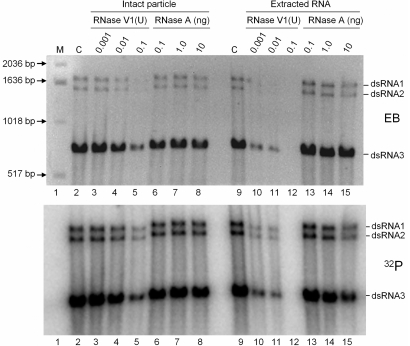
Fig. 5.
RNA synthesis activity of PsV-F virions. RNA synthesis was performed by using purified PsV-F virions in a standard reaction buffer including [α-32P]GTP. Reaction products were treated with various amounts of RNase V1 (lanes 3–5 and 10–12, in units) or RNase A (lanes 6–8 and 13–15, in ng) for 20 min at room temperature, before (lanes 2–8) or after (lanes 9–15) the total RNA was extracted by using phenol:chloroform. The control samples (lanes 2 and 9) had no RNase treatment. All RNA samples were reextracted and subjected to agarose gel electrophoresis, and the resulting gel was stained with ethidium bromide (EB; Upper) and photographed before drying for autoradiography (32P; Lower).
Calculation of electrostatic potential shows that the PsV-F capsid is negatively charged on its inner surface (Fig. 4C), a feature common to many dsRNA viruses (18). Because the genome is confined to the particle interior during transcription, these negative charges on the inner capsid surface may facilitate the movement of template and/or product RNA molecules by repulsion. In contrast, the electrostatic potential of the outer capsid surface is more variable, ranging from slightly positive to highly negative (Fig. 4C). Notably, the highly negative areas on the outer surface immediately surround the icosahedral 5f axes and indeed spill onto the inner surface through the 5f channels. These observations suggest that specific mechanisms, rather than random diffusion, may be in place to guide RNA transcripts in exiting the particle interior.
RNA Synthesis by PsV-F Virions.
PsV-F virions, the same as used for crystallization, were used in an in vitro assay for RNA synthesis. Genome segments dsRNA1 (1,677 bp) and dsRNA2 (1,500 bp), and satellite segment dsRNA3 (677 bp), were found to be transcribed by the particle-associated RdRp molecules. Transcript yields from each of the 3 segments appeared to be directly related to its relative amount in the virion sample, with dsRNA3 and its transcript being most abundant (Fig. 5). Because the reported terminal sequence of dsRNA3 is different from that of dsRNA1 and dsRNA2 (24), it seems that the RdRp may not require a specific terminal sequence for transcription initiation.
RNA digestion experiments showed that the nascent RNA transcribed from each template is in the form of dsRNA, in that the radiolabeled products were sensitive to RNase V1, but not RNase A (Fig. 5). This result is consistent with the conclusions that transcription by PsV-F particles proceeds through a semiconservative mechanism as noted (30) and that only 1 transcription cycle occurred within the 2-h incubation used in our experiments. Additional factors may thus be needed for efficient reinitiation of transcription by PsV-F particles. Transcription was mediated by intact virions, because nascent RNA products were resistant to RNase V1 if not first extracted with phenol:chloroform (Fig. 5, lanes 3 and 4 vs. 10 and 11). However, the PsV-F capsid appears not to be wholly impervious, because prolonged treatment with RNase V1 led to degradation of the genome segments (Fig. 5, lanes 5 and 12).
What are the structural implications for semiconservative transcription by PsV-F virions? During each transcription cycle, the duplex-associated plus-strand RNA must be unwound from the minus strand and directed to exit the particle. But is the plus-strand RNA directed first into the particle interior during transcription and only later finds its way to an exit channel, or is it immediately directed to an exit channel while being nascently unwound from the minus strand? The answer is not known, but negatively charged surface features and flexible structural elements near the 5f axes of the PsV-F capsid suggest these channels as the probable sites of transcript exit.
Previous stoichiometric estimates are for 1, or perhaps 2, copies of RdRp per particle (32). The RdRp molecules are presumed to be noncovalently anchored to the inner surface of the PsV-F capsid, although that has yet to be demonstrated. No strong evidence for internal densities attributable to the RdRp molecules was obtained in this study, which is not surprising given their low copy number. Future studies of transcribing PsV-F particles to localize the RdRp and transcripts should provide new insights into the RNA packaging and transcription mechanisms of this and related viruses.
Materials and Methods
Sample Preparation.
PsV-F virions were grown and purified as described (14). Mass spectrometry (MALDI-TOF; Tufts University Core Facility, Boston) indicated a molecular mass of 46,586.0 kDa, close to the calculated value of 46,538.4 kDa for full-length CP. N-terminal sequencing (also performed at Tufts) showed that the N terminus is blocked and therefore likely intact.
CryoEM and Image Reconstruction.
PsV-F sample preparation and data collection were performed essentially as described for PsV-S (14). A total of forty 4K × 4K CCD images, exhibiting minimal specimen drift and image astigmatism and recorded at underfocus settings of between 1.58 and 2.88 μm, were selected for boxing images of individual particles. The random model method (33) was used to generate an initial map from 150 particle images, and this map then served as the starting model to initiate full orientation and origin determinations for a set of 2,605 particle images by using AUTO3DEM (34). The resolution of the final reconstruction was estimated at 8.0 Å by Fourier-shell correlation analysis [0.5 threshold criterion (35)].
Crystallization, Data Collection, and Data Processing.
Crystals were obtained by hanging drop vapor diffusion. Crystallization solution consisted of a 1:1 mixture of virus suspension (4 mg/mL) and well solution [0.1 M Tris (pH 8.5), 1.98 M ammonium sulfate, and 0.1 M potassium fluoride]. Crystals generally appeared after 4 days and grew to full size (0.05–0.15 mm) after 2 weeks at 10 °C.
Cryogenic cooling under either ambient or high pressure resulted in severe crystal damage and limited diffraction to ≈8-Å resolution. Crystals were therefore either premounted in quartz capillary tubes (Hampton Research) or mounted on site in MicroMount loops (MiTeGen). X-ray diffraction data were collected at 1 °C from crystals mounted in MicroMount loops (MiTeGen) at the MacCHESS F1 beamline (Cornell University, Ithaca, NY). A 0.2° oscillation angle and a 6-s exposure time were used for each frame. Serious decay in diffraction patterns often occurred after 4 exposures. The data were indexed and integrated by using Denzo (36). Because of the alternative indexing problem, frames from each space group were divided into 2 separate sets with consistent hands and then merged together by using ScalepackLvirus (36). A total of ≈100 crystals, yielding 125 frames, were used to generate the final dataset (Table S1). Self-rotation function and packing consideration showed 4 particles in each F23 unit cell. There was 1 pentamer, or 1/12 of the particle, in every crystallographic asymmetric unit.
Crystal Structure Determination.
The 8.0-Å cryoEM reconstruction of PsV-F virions was used as phasing model. AVE, part of the RAVE software package (37), was used to create a crystal unit cell using the proper F23 symmetry. The best initial model (R factor = 49.3%, correlation coefficient = 44.5% for data from 50 to 10 Å) was obtained when the cryoEM map was scaled down to 94.7% of the original size. Using 5f noncrystallographic symmetry, phases were gradually extended (1 reciprocal lattice interval per step) from 10 to 3.3 Å using the RAVE and CCP4 programs (38). Fifteen averaging cycles were performed at each phase extension step.
The final map at 3.3-Å resolution was of excellent quality with continuous main-chain and clearly defined side-chain densities. Small bumps corresponding to main-chain carbonyl groups could be frequently seen. Atomic models were built in O (39). All refinements were carried out by using CNS (40) with 5f noncrystallographic symmetry restraints. Ramachandran plot (41) showed 84% of non-Gly residues in the most-favored regions with none in disallowed regions.
RNA Synthesis.
PsV-F virions (100 μg) were used in an in vitro assay (100 μL) for RNA synthesis in a standard buffer [20 mM Tris (pH 8.0), 5 mM MgCl2, 5 mM KCl, 5 mM NaCl, and 4 mM DTT] containing 0.4 μM [α-32P]GTP (specific activity 3,000 Ci/mmol), 10 μM GTP, and 500 μM each of CTP, ATP, and UTP. After incubation at 37 °C for 2 h, reaction products were purified by using G-50 spin columns (Roche) and aliquoted into 20 fractions. Each aliquot was treated for 20 min at room temperature in a total volume of 10 μL with either RNase V1 [in 10 mM Tris (pH 7), 0.1 M KCl, and 10 mM MgCl2] or RNase A [in 20 mM Tris (pH 7.0), 0.5 M NaCl, and 10 mM MgCl2] before or after phenol:chroloform extraction. RNase-treated samples were reextracted and subjected to 1.5% agarose gel electrophoresis in Tris-borate-EDTA buffer, and the gel was then stained with ethidium bromide and photographed before drying for autoradiography.
Figure Preparation.
Ribbon diagrams and Cα traces were prepared by using Molscript (42) and PyMOL (www.pymol.org). Molecular surfaces were calculated and colored by using SPOCK (http://quorum/tamu/edu).
Supplementary Material
Acknowledgments.
We thank Y. Zhou, B. V. V. Prasad, J. Tang, C. U. Kim, and S. M. Gruner for valuable discussions. This work was supported by the National Institutes of Health (Y.J.T. and T.S.B.), U.S. Department of Agriculture (S.A.G.), the Welch Foundation (Y.J.T.), and the Kresge Science Initiative Endowment Fund. The San Diego Supercomputer Center provided access to TeraGrid computing, and support from the University of California-San Diego and the Agouron Foundation (to T.S.B.) were used to establish and equip cryoEM facilities at the University of California-San Diego.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ES5). The cryoEM map has been deposited in the Electron Microscopy Database at the European Bioinformatics Institute, www.ebi.ac.uk.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812071106/DCSupplemental.
References
- 1.Harrison SC. Principles of virus structure. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 59–98. [Google Scholar]
- 2.Lawton JA, Estes MK, Prasad BVV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage: Structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 5.Prasad BVV, et al. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 6.Gouet P, et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 7.Bottcher B, et al. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J Virol. 1997;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caston JR, et al. Structure of L-A virus: A specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caston JR, et al. Three-dimensional structure of Penicillium chrysogenum virus: A double-stranded RNA virus with a genuine T=1 capsid. J Mol Biol. 2003;331:417–431. doi: 10.1016/s0022-2836(03)00695-8. [DOI] [PubMed] [Google Scholar]
- 10.Dryden KA, et al. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: Analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 12.Huiskonen JT, et al. Structure of the bacteriophage φ6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure (London) 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Jaalinoja HT, Huiskonen JT, Butcher SJ. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages φ6 and φ8. Structure (London) 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Ochoa WF, et al. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure (London) 2008;16:776–786. doi: 10.1016/j.str.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad BVV, Wang GJ, Clerx JP, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou ZH, et al. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat Struct Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- 18.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6-Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 19.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa A, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure (London) 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Coulibaly F, et al. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4-Å resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 23.Ghabrial SA, Ochoa WF, Baker TS, Nibert ML. Partitiviruses: General features. In: Mahy BWJ, van Regenmortel MHV, editors. Encyclopedia of Virology. 3rd Ed. Vol 4. Oxford: Elsevier; 2008. pp. 68–75. [Google Scholar]
- 24.Kim JW, Choi EY, Lee JI. Genome organization and expression of the Penicillium stoloniferum virus F. Genes. 2005;31:175–183. doi: 10.1007/s11262-005-1793-y. [DOI] [PubMed] [Google Scholar]
- 25.Bozarth RF, Wood HA, Mandelbrot A. The Penicillium stoloniferum virus complex: Two similar double-stranded RNA virus-like particles in a single cell. Virology. 1971;45:516–523. doi: 10.1016/0042-6822(71)90352-7. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn RJ, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopper P, Harrison SC, Sauer RT. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol. 1984;177:701–713. doi: 10.1016/0022-2836(84)90045-7. [DOI] [PubMed] [Google Scholar]
- 28.Reddy VS, Johnson JE. Structure-derived insights into virus assembly. Adv Virus Res. 2005;64:45–68. doi: 10.1016/S0065-3527(05)64003-1. [DOI] [PubMed] [Google Scholar]
- 29.Tang J, et al. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J Struct Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Buck KW. Semiconservative replication of double-stranded RNA by a virion-associated RNA polymerase. Biochem Biophys Res Commun. 1978;84:639–645. doi: 10.1016/0006-291x(78)90753-2. [DOI] [PubMed] [Google Scholar]
- 31.Chater KF, Morgan DH. Ribonucleic acid synthesis by isolated viruses of Penicillium stoloniferum. J Gen Virol. 1974;24:307–317. doi: 10.1099/0022-1317-24-2-307. [DOI] [PubMed] [Google Scholar]
- 32.Buck KW, Kempson-Jones GF. Capsid polypeptides of two viruses isolated from Penicillium stoloniferum. J Gen Virol. 1974;22:441–445. [Google Scholar]
- 33.Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan X, Sinkovits RS, Baker TS. AUTO3DEM: An automated and high-throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA. A set of averaging programs. In: Dodson EJ, Gover S, Wolf W, editors. Molecular Replacement. Warrington, UK: SERC Daresbury Laboratory; 1992. pp. 91–105. [Google Scholar]
- 38.Collaborative Computational Project No. 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, MacArthur MW, Moss DS, Thomton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 42.Kraulis P. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.