Abstract
Adaptation to hypoxia is mediated through a coordinated transcriptional response driven largely by hypoxia-inducible factor 1 (HIF-1). We used ChIP-chip and gene expression profiling to identify direct targets of HIF-1 transactivation on a genome-wide scale. Several hundred direct HIF-1 targets were identified and, as expected, were highly enriched for proteins that facilitate metabolic adaptation to hypoxia. Surprisingly, there was also striking enrichment for the family of 2-oxoglutarate dioxygenases, including the jumonji-domain histone demethylases. We demonstrate that these histone demethylases are direct HIF targets, and their up-regulation helps maintain epigenetic homeostasis under hypoxic conditions. These results suggest that the coordinated increase in expression of several oxygen-dependent enzymes by HIF may help compensate for decreased levels of oxygen under conditions of cellular hypoxia.
Keywords: ChIP-chip, hypoxia, jumonji protein, dioxygenase, epigenetics
Adequate oxygenation is essential for normal physiology and functioning of all cells. In response to decreased oxygen tension, a coordinated transcriptional response is activated to maintain cellular homeostasis. This transcriptional program is mediated, at least in part, by activation of the heterodimeric transcription factors hypoxia-inducible factor 1 and 2 (HIF-1 and HIF-2). The HIFs are essential for several physiological processes, including normal development, erythropoietin (EPO) production, and wound healing. Unfortunately, HIF signaling also contributes to the pathophysiology of tumors by facilitating metabolic adaptation and by promoting angiogenesis, invasion, and metastasis (1).
The activity of HIF-1 and HIF-2 are controlled at multiple levels. The primary point of regulation is at the level of abundance of the α-subunits. Under normal oxygen (normoxic) conditions, HIF-1α and HIF-2α are hydroxylated by the HIF prolyl hydroxylases EGLN1–3, which target the proteins for binding to the von Hippel–Lindau (vHL) ubiquitin E3 ligase complex and rapid proteosomal degradation (2–6). As oxygen levels drop, prolyl-hydroxylation decreases, resulting in accumulation of HIF α-subunits and heterodimerization with ARNT (HIF-1β). As a consequence, HIF heterodimer levels are nominal under physiologic oxygen levels and increase exponentially with decreasing oxygen tension (7). HIF heterodimers then recruit transcriptional coactivator complexes (8, 9), and transactivate target genes containing the cognate hypoxia-response element (HRE) (1).
The direct transcriptional targets of HIF-1 play important roles in facilitating both short-term and long-term adaptation to hypoxia (1). Metabolic homeostasis is achieved by shifting from oxidative phosphorylation to anaerobic glycolysis through increased expression of the glycolytic enzymes and glucose transporters, inhibition of the TCA cycle, and induction of pH-regulating systems. A second HIF-1-mediated program increases oxygen delivery by inducing vasodilatation, increased vascular permeability, enhanced erythropoiesis and angiogenesis. Specific sites of HIF-1 binding have been validated within the promoter or enhancers of ≈50 genes (10, 11). Alignment of the sequences encompassing these well-characterized functional HREs (transcriptionally activated by hypoxia) has revealed a consensus HIF-1-binding motif (the core HRE) of 5′-RCGTG-3′ (R = A or G).
Because the core HRE is too promiscuous to accurately predict binding a priori, we used ChIP-chip to define HIF-1 chromatin binding on a genome-wide level. We integrated these results with gene expression profiling to interrogate mechanisms regulating hypoxia-induced gene expression and to more comprehensively identify direct targets of HIF-1 transactivation. We found that the family of 2-oxoglutarate-dependent dioxygenases are coordinately targeted by HIF, and up-regulated expression helps maintain global levels of histone methylation under hypoxic conditions.
Results
Identification of HIF-1-Binding Sites by ChIP-chip.
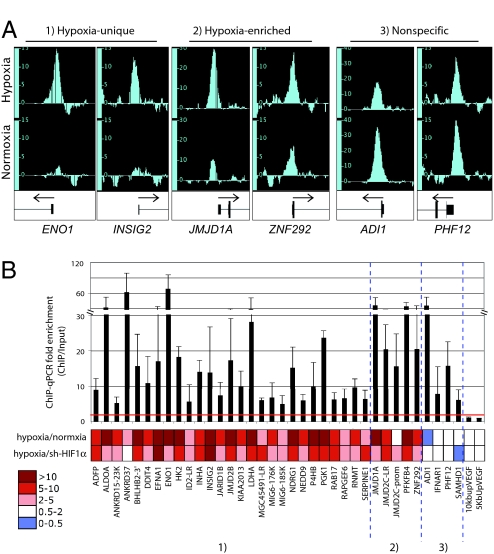
To identify HIF-1-binding sites across the genome, HepG2 cells were grown under both normoxic (ambient) and hypoxic (0.5% O2) conditions, and ChIP was performed by using a HIF-1α polyclonal antibody without appreciable cross-reactivity to HIF-2α (Fig. S1A). ChIP and input DNA were hybridized to Affymetrix GeneChip Human Tiling 2.0R Array Sets, consisting of probes covering the entire nonrepetitive human genome at 35-bp resolution. The model-based analysis for tiling arrays (MAT) algorithm (12) was used to identify probe signal peaks comparing triplicate biological replicates of HIF-1 ChIP DNA to their matched inputs by using an initial P value cutoff of 1E−5. Consistent with the fact that HIF-1α protein levels increase dramatically in hypoxic compared with normoxic samples (Fig. S1B), 91% of the putative HIF-1-binding sites were characterized by a positive peak call only under hypoxic conditions (e.g., ENO1 and INSIG2; Fig. 1A). However, because HIF-1α is not entirely absent in normoxia, peaks in which probe intensities were significantly increased under hypoxic conditions by comparison with normoxia were also retained (e.g., JMJD1A and ZNF292; Fig. 1A). A minority of peaks (7%) had peak intensities that did not increase in hypoxic compared with normoxic samples (e.g., ADI1 and PHF12; Fig. 1A), and these were suspected to be nonspecific.
Fig. 1.
HIF-1 ChIP-chip analysis and validation. (A) Representative integrated genome browser (IGB; Affymetrix) tracks showing peaks of probe intensities (vertical bars) arrayed by chromosomal position. For each locus, tracks are scaled identically. ChIP peaks were identified by using the MAT algorithm and were classified as hypoxia-unique, hypoxia-enriched, or nonspecific. (B) ChIP-chip results were validated by ChIP-qPCR using primer pairs surrounding the putative binding sites in the indicated loci. For each locus, the fold enrichment comparing HIF-1 ChIP DNA to input is represented in the bar graph (mean ± SD). To determine specificity for HIF-1, the bottom heat map depicts the fold enrichment comparing hypoxic to normoxic cells and control hypoxic cells to cells in which HIF-1α was specifically depleted with a lentiviral shRNA (sh-HIF1α). Primer pairs locate 5 and 10 kb upstream of the VEGF gene were used as controls.
To validate the ChIP-chip analysis pipeline, we used quantitative PCR (qPCR) to quantify HIF-1 binding to chromatin. Loci with a positive call on ChIP-chip analysis also had enrichment by ChIP-qPCR, with most loci having at least 8-fold enrichment of ChIP DNA compared with input (Fig. 1B, bar graph). To establish the specificity of the ChIP for HIF-1, we compared ChIP-qPCR results in hypoxic and normoxic samples and also compared control hypoxic samples to hypoxic samples in which HIF-1α had been specifically depleted by shRNA knockdown (sh-HIF1α) (Fig. S1B). These studies confirmed that at loci in which ChIP-chip identified hypoxia-unique or hypoxia-enriched peaks (Fig. 1A), there was specific enrichment of HIF-1 binding by ChIP-qPCR analysis based on increased enrichment under hypoxic conditions and decreased binding when HIF-1α was specifically depleted (Fig. 1B, heat map). Loci in which there was no increase in peak intensity in hypoxia by ChIP-chip (Fig. 1A) likewise showed no enrichment under hypoxic conditions by ChIP-qPCR (Fig. 1B). Importantly, specific knockdown of HIF-1α did not alter binding at these loci (Fig. 1B), verifying that these peaks were nonspecific and likely due to antibody cross-reactivity. This minority (7%) of nonspecific ChIP fragments were therefore excluded from the analysis pipeline.
We used ChIP-qPCR to evaluate a total of 130 loci consisting of HIF-1-binding regions identified by ChIP-chip, negative regions as well as some previously reported HIF-1 targets (Table S1). Using the ChIP-qPCR results as the benchmark, we used receiver operating characteristic (ROC) curve analysis (Fig. S1C) to establish optimized thresholds for the ChIP-chip analysis (MAT score >5.77 and P value < 2.5 × 10−7) that together resulted in high specificity (95.5%) and sensitivity (82.5%) (Fig. S1D). Within a subset of 40 well-validated HIF-1 targets evaluated by ChIP-qPCR (Table S2), these thresholds produced an empiric sensitivity of 75% and specificity of 100%. With these optimized thresholds, a total of 377 HIF-1-binding sites were identified across the genome.
The canonical motif that is bound by HIF-1, the core HRE, is well established (5′-RCGTG-3′). Despite there being >2.5 million occurrences of this promiscuous motif in the human genome, we observed only a very small minority bound by HIF-1. To determine whether this heightened specificity was due to a previously unrecognized extended binding preference, we used several de novo motif search tools, including Weeder (13) and MEME (14, 15), to search for overrepresented motifs within our list of HIF-1-bound fragments. De novo motif search of our expanded list of putative HIF-1 bound sites revealed overrepresentation of motifs that were highly similar to a position-weighted matrix (PWM) generated based on the alignment of 68 well-characterized HIF-1-binding sites previously reported in the literature (10) (Fig. S2 A–C). Although we did note a stronger bias for an adenine in the 5′ position (5′-ACGTG-3′, Fig. S2B), we did not identify extended sequence preferences beyond the core HRE that would explain the lower-than-predicted number of observed HIF-1-bound sites.
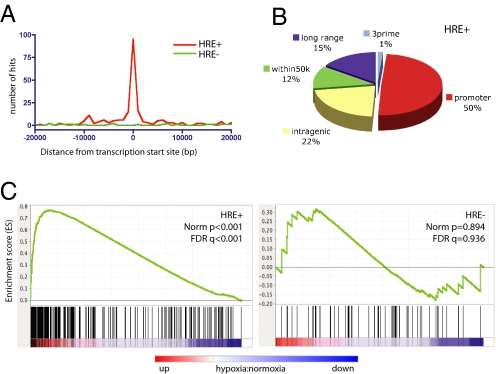
We used the Match algorithm (16) to identify core HREs in the 377 HIF-1 ChIP-chip fragments. A total of 283 (75%) contained at least 1 core HRE (HRE+ hits). When mapped to the human genome, HRE+ hits were found to be tightly centered around the transcription start sites (TSS) of genes (Fig. 2A), with a majority of binding sites located either within promoters (50%) or intragenic regions (22%) (Fig. 2B) and only 15% located in long-range (>50 kb) relationship to genes (Fig. 2B).
Fig. 2.
Distribution of HIF-1-bound sites and association with transactivation. (A) Distribution of HIF-1 ChIP hits relative to the TSS of associated genes. (B) Distribution of core HRE-containing HIF-1 hits (HRE+) relate to the annotated structure of associated genes. (C) GSEA analysis of mRNA expression profiles for hypoxic (0.5%O2, 12 h) vs. normoxic cells. Relative expression was rank-ordered by signal-to-noise (S2N) ratios of triplicate hypoxic samples vs. triplicate normoxic samples. Genes associated with HRE+ HIF-1-binding sites were strongly correlated with the hypoxic phenotype. In contrast, no such enrichment was evident for the ChIP-chip fragments without identifiable core HREs (HRE−). The color bar indicates up-regulated (red, positive S2N) and down-regulated (blue, negative S2N) genes.
HIF-1 Binding Is Correlated with Up-Regulated Expression.
To investigate the relation of HIF-1 binding to hypoxia-induced gene expression, we performed mRNA profiling of cells under normoxic and hypoxic conditions. Gene set enrichment analysis (GSEA) (17) was used to determine whether HIF-1 binding was functionally associated with hypoxia-induced changes in mRNA expression. Genes within 50 kb of a HIF-1-binding site were significantly enriched for up-regulated mRNA expression under hypoxic conditions (Fig. 2C, HRE+). The association between HIF-1 binding and increased gene expression was most apparent for binding sites located within the promoter of genes but was also highly significant when binding was within intragenic or enhancer regions (Fig. S2D). By comparison, the set of ChIP fragments without identifiable core HREs had no demonstrable enrichment in gene expression under hypoxic conditions (Fig. 2C, HRE−). Likewise, fragments excluded above as “nonspecific” (e.g., Fig. 1) did not have enriched expression under hypoxia (FDR q value 0.23). The average MAT score (ChIP enrichment) of HRE− hits was also significantly lower than HRE+ hits (Fig. S2E), and de novo motif search did not identify any overrepresented motifs that might suggest an alternative HIF-1 binding motif. Together, these results suggested that HIF-1 ChIP fragments without identifiable core HREs were not direct HIF-1-binding sites and may represent indirect binding, such as through bridging interactions with coactivator complexes such as p300/CBP (8, 9). Furthermore, none of the core HRE-negative hits have been previously reported as hypoxia-responsive genes. Therefore, to focus our analysis on direct HIF-1-binding sites and transactivation targets, all subsequent analysis focused on the 283 high-confidence core HRE-containing HIF-1-binding sites (Table S3).
Dioxygenases Are HIF-1 Targets.
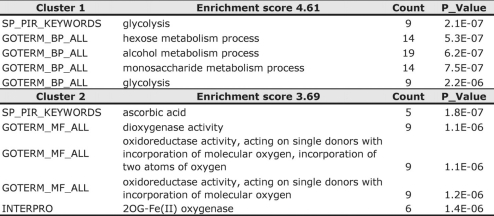
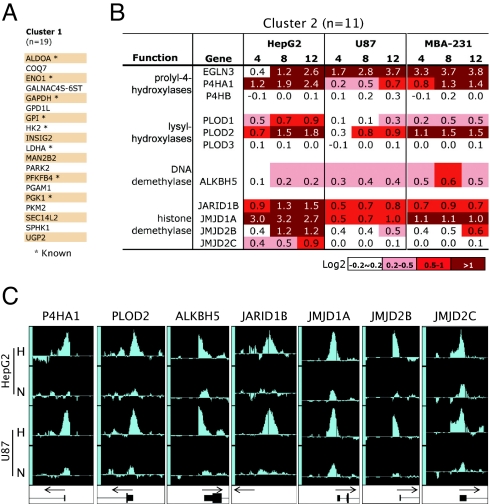
Only a minority of the genes (≈1/6) associated with HIF-1 ChIP hits have been previously described as hypoxia-responsive genes (Table S3). To determine whether this larger set of HIF-1 target genes could provide insight into functional groups that are coordinately regulated by HIF-1, we performed functional annotation clustering using the DAVID Bioinformatics Resource (http://david.abcc.ncifcrf.gov/). As expected, DAVID identified the glycolytic pathway enzymes (e.g., ENO1, GAPDH, HK2), which are well-characterized HIF-1 targets (1), as the most highly enriched subsets in our HIF-1 ChIP hits (Table 1). Within our list of direct HIF-1 targets (Table S3), there were 19 genes (Fig. 3A) associated with various glycolysis functional annotation groups (Table 1). Surprisingly, several GO annotations for 2-oxoglutarate and ferrous iron-dependent dioxygenases (2-OG-dioxygenases), were also highly enriched in the DAVID analysis (Table 1). We found 11 2-OG-dioxygenase genes with direct HIF-1 binding (Fig. 3B and Table S3), and these were associated with several functional annotation groups that in some cases were as significantly enriched as that for the glycolytic enzymes (Table 1).
Table 1.
Functional annotation clustering of HIF-1 ChIP hits
Fig. 3.
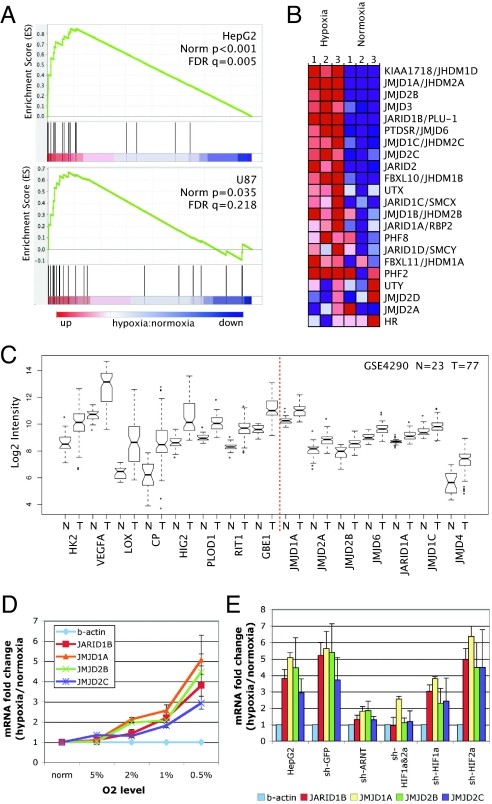
Overrepresentation of 2-OG-dioxygenases among HIF-1 ChIP hits. (A) HIF-1 target genes in Cluster 1, with previously described HIF-1-bound targets indicated with an asterisk. (B) HIF-1-targeted 2-OG-dioxygenases in Cluster 2. Average change (Log2) in mRNA expression levels at 4, 8, and 12 h of hypoxia in HepG2, U87 and MDA-MB231 cells are indicated by using the assigned color scale. (C) Representative IGB tracks showing HIF-1 binding at promoter regions of the 2-OG-dioxygenase family genes in HepG2 and U87 cell lines. For each locus, tracks are scaled identically between hypoxia (H) and normoxia (N) samples.
The 2-OG-dioxygenases catalyze oxidation–reduction reactions and can be subgrouped based on specific enzymatic activities. HIF-1 was found to bind to genes within most enzymatic subgroups within the family (Fig. 3B), including the prolyl-hydroxylases, procollagen lysyl-hydroxylases, DNA demethylase, and Jumonji-domain (JmjC)-containing demethylases. To verify the binding of HIF-1 to these 2-OG-dioxygenases, we performed HIF-1 ChIP-chip in a second cell type, U87 glioma cells. High-affinity binding of HIF-1 was verified at all loci under hypoxic conditions (Fig. 3C). Furthermore, the abundance of mRNA encoding most of these 2-OG-dioxygenases was induced under hypoxic conditions in 3 different cell types (Fig. 3B), with some cell-type variation in the degree of induction. Together, these results demonstrate that multiple members of the 2-OG-dioxygenase family are direct HIF-1 targets and are coordinately up-regulated under hypoxic conditions.
JmjC-Containing Histone Demethylases (JHDMs) Are Direct HIF-1 Targets.
We noted that 4 JmjC-containing proteins (JARID1B, JMJD1A, JMJD2B, and JMJD2C) were direct HIF-1 target genes with robust HIF-1 binding within their promoters and up-regulated expression under hypoxic conditions. An expanded analysis of the JmjC family revealed strong enrichment for up-regulated mRNA expression under hypoxic conditions in both HepG2 and U87 cells (Fig. 4 A and B). This included both the direct HIF-1 targets (JARID1B, JMJD1A, JMJD2B, and JMJD2C) and other family members without evidence of direct HIF-1 binding. In HepG2 cells, 17 of the 22 JmjC family members had significantly increased mRNA abundance under hypoxic conditions (Fig. 4B). These results are supported by recent studies demonstrating that HIF-1 up-regulates JMJD1A (18) and JMJD2B (19) in vitro.
Fig. 4.
Hypoxia- and HIF-dependent up-regulation of JHDMs. (A) GSEA analysis of mRNA expression profiles for hypoxic (12 h) vs. normoxic cells with gene set containing all known JmjC histone demethylases in HepG2 and U87 cells. (B) Gene expression changes under hypoxia in HepG2 cells. (C) Expression dataset for Grade IV glioblastoma was interrogated for changes in mRNA levels of the JmjC family. The most highly up-regulated JmjC family member is shown for normal tissue (N) vs. tumor (T) samples. Known HIF-1 targets are on the left. Data represented as the median (bar), 25–75th percentiles (box), and range (whiskers). (D) Quantitative RT-PCR verification of up-regulated expression of HIF-1-bound JmjC genes under different levels of hypoxia. Results are expressed as mean ± SD after normalization to β-actin. (E) shRNA depletion of HIF-1α, HIF-2α, HIF1α and 2α, or ARNT was used to determine HIF-dependent up-regulation of JmjC proteins. Data represent fold change comparing hypoxic with normoxic samples (mean ± SD).
To determine the potential relevance of these findings in vivo, we also examined the expression of JmjC proteins in human tumors, because most solid tumors are hypoxic compared with normal tissues (1). In glioblastoma multiforme, a disease in which hypoxia is a prominent feature (20), the expression of multiple JmjC proteins was significantly increased in hypoxic tumor samples compared with corresponding normal brain (Fig. 4C and Fig S3). The significance of increase was comparable with other well-characterized HIF-1 targets, such as hexokinase 2 and VEGF (Fig. 4C). Thus, the expression of multiple JmjC proteins is increased not only in vitro in hypoxic cell lines but also in vivo within hypoxic human tumors.
Quantitative RT-PCR was used to verify increased mRNA abundance of HIF-1-targeted JmjC proteins under hypoxic conditions (Fig. 4D). We used shRNA knockdown of HIF-1α and HIF-2α to determine the specificity of hypoxia-induced expression. Individually, shRNA depletion of HIF-1α or HIF-2α did not significant affect hypoxia-induced expression, whereas knockdown of both HIF-1α and HIF-2α abolished hypoxia inducibility of JARID1B, JMJD1A, JMJD2B, and JMJD2C (Fig. 4E). The hypoxia inducibility of these JHDMs was also abolished by knockdown of ARNT (Fig. 4E), which is the common β-subunit for both HIF-1 and HIF-2. These results demonstrate that these JmjC proteins are redundantly transactivated by both HIF-1 and HIF-2 under hypoxic conditions.
Up-Regulation of JHDMs Maintains Histone Methylation Homeostasis.
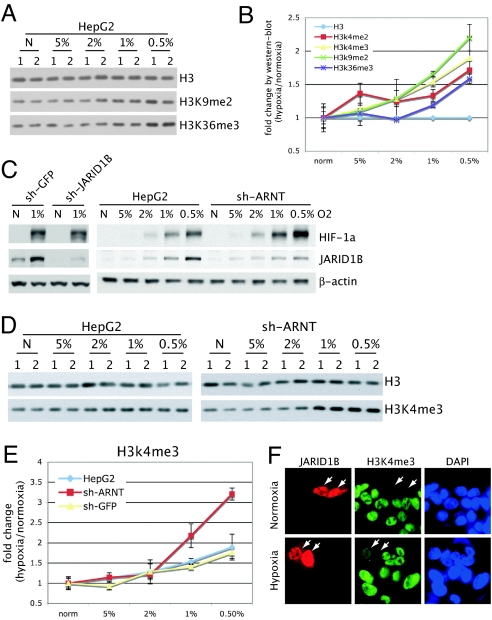
The JmjC-containing proteins function as histone demethylases (JHDMs) (21–26). Because the hydroxylation and demethylation reactions catalyzed by 2-OG-dioxygenases require molecular oxygen, we hypothesized that the up-regulated expression of these enzymes may be a compensatory mechanism in response to decreasing oxygen tension. That is, with decreasing levels of molecular oxygen, increasing the concentration of the enzyme may help partially compensate by maintaining forward drive of the chemical reaction. To experimentally test this hypothesis, we first assessed the effect of hypoxia on global histone methylation. Relative to normoxic conditions, overall methylation of histone H3 lysines 4, 9, and 36 were found to increase with decreasing oxygen tension, especially at 0.5–1% oxygen (Fig. 5 A and B). These results are consistent with prior studies (27, 28) and suggest that histone demethylation decreases as oxygen levels drop, analogous to the decrease in prolyl-hydroxylation of HIF-1α by EGLN1–3 under subphysiological oxygen tensions.
Fig. 5.
Histone hypermethylations from loss of JHDM induction in hypoxia. (A) A representative immunoblot analysis of total histone H3 and the specified modifications in biological replicates (1 and 2) under normoxic (N) and hypoxic conditions (O2 levels indicated). (B) Quantitation of a single representative study is shown, with results for each modification normalized to total histone H3 within each sample (mean ± SD). (C) Accumulation of JARID1B protein under hypoxia was verified by Western blot analysis with a JARID1B-specific antibody, with specificity verified by shRNA depletion of JARID1B (Left). Knockdown of ARNT (sh-ARNT) (Right) reduces accumulation of JARID1B in hypoxia. (D) Global histone H3 and H3K4me3 levels were determined in unmodified HepG2 cells, control shRNA cells (sh-GFP), and ARNT depleted cells (sh-ARNT). Western blot analysis for histone H3 and H3K4me3 of duplicate biological replicates (1 and 2) is shown for a representative experiment. (E) Quantitated results represented as mean ± SD. (F) A HA-JARID1B expression construct was transfected into HepG2 sh-ARNT cells. Normoxic and hypoxic cells were stained with anti-HA (red) and anti-H3K4me3 (green) antibodies. In both conditions, overexpression of JARID1B reduced the H3K4me3 level by comparing transfected cells (with arrows) to nontransfected cells.
To evaluate the role of HIF-induced expression of JHDMs in maintaining histone methylation under hypoxic conditions, we focused on JARID1B, which has a robust HIF-1-binding site within its promoter (Fig. 3C), has up-regulated mRNA expression in multiple cell types (Fig. 3B), and is a well-defined demethylase for trimethylated histone H3 lysine 4 (H3K4me3) (29). We used immunoblots to first verify increased protein abundance in hypoxia. Normal tissues have oxygen levels of ≈3–5%. Stabilization of HIF-1α and increased abundance of JARID1B was apparent starting at 2% oxygen, and significantly increased at 1% and 0.5% oxygen (Fig. 5C). Because HIF-1 and HIF-2 appear to transactivate JARID1B (Fig. 4E), we assessed the effects of shRNA depletion of ARNT on JARID1B protein levels. Consistent with reduced accumulation of JARID1B mRNA (Fig. 4E), shRNA knockdown of ARNT reduced JARID1B protein accumulation under hypoxic conditions (Fig. 5C).
If up-regulated expression of JARID1B is a compensatory mechanism for maintaining H3K4 methylation levels under conditions of decreasing oxygen, we predicted that decreasing the induction of JARID1B by HIF would result in hypermethylation of H3K4 under hypoxic conditions. We compared control HepG2 cells with cells in which ARNT was depleted by shRNA to block both HIF-1 and HIF-2 activity. Knockdown of ARNT had no appreciable effect on basal H3K4 methylation under normoxic conditions (Fig. 5D). With increasing hypoxia, there was a global increase in H3K4me3 levels (Fig. 5 D and E). Inhibiting HIF transactivation through knockdown of ARNT resulted in an increase in global H3K4me3 levels at specific levels of hypoxia (Fig. 5 D and E). To determine whether ectopic expression of JARID1B could reverse hypoxia-induced H3K4 hypermethylation, an expression plasmid was transfected into HepG2 cells in which ARNT was depleted by shRNA. Under conditions of hypoxia, cells ectopically expressing JARID1B had decreased levels of H3K4 methylation (Fig. 5F). Together, these results support the hypothesis that up-regulated expression of JARID1B helps compensate for decreased levels of molecular oxygen in maintaining H3K4 methylation.
Discussion
Adequate oxygenation is essential for normal cell physiology, and a coordinated transcriptional response to hypoxia is critical for maintaining cellular homeostasis under conditions of decreased oxygen tension. To achieve a fuller understanding of hypoxia-induced gene expression, we used ChIP-chip and mRNA expression profiling to define HIF-1 chromatin binding and gene transactivation on a genome-wide scale. Given the promiscuity of the established core HIF-1-binding motif, it is of interest that our analysis revealed only a few hundred high-confidence HIF-1-binding sites across the human genome. This list of HIF-1-binding sites is certainly not comprehensive, because it was generated from a single time point (the peak of HIF-1 protein abundance), and it is likely that additional HIF-1 sites are occupied after prolonged hypoxia. There are also clearly cell type-specific differences in HIF-1 binding. Furthermore, to increase the specificity of our analysis, we imposed a stringent analysis pipeline that decreased the overall sensitivity of the methodology. Nonetheless, the number of HIF-1-binding sites observed is of similar magnitude to the ≈540 sites acutely bound by p53 (30), and the ≈1,200 loci bound by Foxp3 in mouse T cells (31). Genome-wide analysis of estrogen receptor (ER) revealed ≈3,600 binding sites (32). By comparison, E2F1 and MYC bind to >15,000 sites each (33), and the insulator protein CTCF binds to >13,000 sites (34). Together, these studies suggest that DNA-binding proteins may partition into those with higher promiscuity (E2F1, MYC, CTCF) and lower promiscuity (HIF-1, p53, ER, Foxp3). It is also relevant to note that in the case of HIF-1, p53, and ER, the studies were driven by specific stimuli (hypoxia for 4 h, 5-FU for 6 h, and estrogen for 45 min, respectively). Thus, the distinction between these 2 groups may also reflect a difference between acute versus tonic chromatin binding.
We have found that a majority of HIF-1-binding sites exist in close association with genes, predominantly within promoter and intragenic regions, similar to the binding pattern of p53, Foxp3, E2F1, and NF-κB (30, 31, 33, 35). Because many loci have alternative transcriptional start sites, many intragenic binding sites might in fact represent promoters for these alternative coding (or noncoding) transcripts. These results are in distinct contrast to ER binding (32, 36), where genome-wide analysis revealed that only a small minority of binding occurs within promoter regions and the vast majority of binding sites are long-range in relationship to known genes. Approximately half of all binding sites for MYC (33), CTCF (34), and Foxp3 (31) exist in long-range relationships to genes. Based on the conventional view of transcriptional transactivation as a promoter-driven process, many ChIP-chip studies have made use of microarrays focused on promoter regions. Even in the case of HIF-1, which shows a strong bias toward promoter regions, nearly half of the binding sites would have been missed if proximal promoter arrays had been used, underscoring the importance of comprehensive assessment across the entire genome.
In contrast to ER (36) and p53 (30), which are almost equally associated with gene up- and down-regulation, we find no statistical enrichment of HIF-1-binding sites in genes whose expression are down-regulated by hypoxia. Although the numbers of mRNAs that are up- and down-regulated under hypoxic conditions are approximately equal, HIF-1 binding is enriched only in genes whose expression increases. These results suggest that HIF-1 functions predominantly as a transcriptional activator and rarely as a transcriptional repressor (e.g., through recruitment of corepressor complexes), as is the case with other transcription factors such as p53 and ER (30, 36).
Even with a more comprehensive understanding of the direct targets of HIF-1 binding, only a minority of hypoxia-induced gene expression changes can be accounted for through the direct effects of HIF-1. It is likely that another portion of the transcriptional changes are due to HIF-2 mediated transactivation. However, recent studies suggest that HIF-1 and HIF-2 bind to the same HREs, and differential gene transactivation is conferred by interacting with distinct transcriptional cofactors (37). Thus, the HIF-1-binding sites defined in this study may largely encompass HIF-2-binding sites as well. It is also difficult to associate long-range binding sites with specific genes, and thus long-range transactivational mechanisms may account for another portion of hypoxia-inducible genes. However, because there are >5- to 10-fold more genes that are up-regulated by hypoxia compared with HIF-1-binding sites, it is likely that most transcriptional changes are secondary in nature. Several transcription factors are direct HIF-1 target genes, providing likely candidates for secondary transactivation.
Using the expanded list of direct HIF-1 targets, we noted that the family of 2-OG-dioxygenases are coordinately up-regulated by HIF. The diversity of known HIF-1 target genes can be broadly integrated into 2 major homeostatic programs that facilitate adaptation to hypoxia. The first encompasses a “metabolic program” in which metabolism is switched from predominantly oxidative phosphorylation to anaerobic glycolysis (i.e., through induction of glycolytic enzymes, pH regulating proteins and negative mitochondrial regulators). The second homeostatic program is an “oxygen-delivery program” that increases oxygen delivery acutely through vasodilatation (iNOS) and vascular permeability (VEGF), and long-term through induction of angiogenesis and erythropoiesis (VEGF, EPO). Our study suggests a third major adaptive program consisting of coordinated up-regulation of multiple members of the 2-OG-dioxygenase family. Although the specific enzymatic reactions catalyzed by individual family members differ, they all have in common a requirement for molecular oxygen. As oxygen levels drop, increased expression of the enzymes may help maintain forward drive of critical hydroxylation and demethylation reactions. For example, the striking enrichment for JHDMs suggests that coordinated up-regulation of these enzymes counterbalances decreased levels of oxygen to maintain global histone methylation patterns. Together, these results suggest that in the face of hypoxia, cellular homeostasis is maintained in part through a third major adaptive program—a “dioxygenase homeostasis program”—that helps maintain the forward drive of certain critical oxygen-dependent dioxygenases.
Methods
Detailed experimental methods are contained in SI Materials and Methods.
ChIP-chip.
HepG2 and U87 cells (ATCC) were cultured under normoxic (ambient) or hypoxic (0.5%O2, 4 h) conditions. HIF-1α ChIP-chip was performed with a HIF-1α pAb (Table S4) as previously described (32). HepG2 HIF-1α ChIPed DNA replicates and inputs were amplified and hybridized onto the Affymetrix GeneChip Human Tiling 2.0R Array Set. U87 HIF-1 ChIP samples were hybridized onto Affymetrix GeneChip Human Promoter 1.0R Array. The MAT algorithm (12) was used to identify peaks of probe intensity (“hits”). ChIP hits were associated with RefSeq genes from the University of California Santa Cruz (UCSC) RefGene table for HG18 based on chromosomal position.
qPCR Validation of ChIP Hits.
Quantitative PCR primers were designed against regions of interest and also 2 negative control regions (5 and 10 kb upstream of the VEGF gene). All primer sequences are specified in Table S1. HIF-1-specific binding was defined as >2-fold enrichment when HIF-1 ChIP and matched input samples were compared, and a >2-fold greater binding in hypoxic samples relative to either normoxic cells or hypoxic HIF-1α knockdown (sh-HIF1α) cells.
Identification of Putative Core HREs and Motif Search.
A PWM was generated from 68 reported human functional HRE sequences (10). We used Match (16) to locate sites in our ChIP hits matching this PWM, using a core score cutoff of 0.9 and matrix score of 0.85.
mRNA Expression Profiling.
HepG2, U87, and MDA-MB231 cells were collected under normoxic conditions (0 h) and after 4, 8, and 12 h of hypoxia (0.5% O2). Triplicates were hybridize to Affymetrix HG-U133Plus2 arrays. For analysis of human tumor material, a dataset for Grade IV glioblastoma multiforme (38) was downloaded from the National Center for Biotechnology Information.
GSEA and Functional Annotation Clustering.
We created gene sets containing all genes that could be associated with a ChIP hit (HRE+ hits or HRE− hits, within 50 kb). These sets along with a gene set containing all known JHDMs were added to a file of gene sets (c5.mf.v2.5.symbols.gmt) downloaded from the GSEA web site at the Broad Institute (www.broad.mit.edu/). We used the command line version of GSEA2.0 with gene set permutation to derive significance, with signal-to-noise as the distance metric and maximum expression to collapse probe sets to genes. For functional annotation clustering, the gene sets containing all genes associated with a ChIP hit (with 50 kb) were uploaded onto the David Go Annotation site (http://david.abcc.ncifcrf.gov).
Western Blot Analyses and Immunofluorescence.
Histones were isolated by a standard acid extraction protocol and standardized amounts of protein were fractionated by SDS/PAGE followed by Western blot analysis. All antibodies used are specified in Table S3. Densitometric quantitation of each histone modification was normalized to total histone H3 to correct for loading. HepG2-shARNT cells were transfected with a JARID1B expression plasmid pCS2 + 3HA-JARID1B (a gift from Yang Shi, Harvard Medical School). Cells were incubated for 24 h under either normoxic or hypoxic (0.5%O2) conditions, fixed, and stained with anti-HA mAb and H3K4me3 pAb.
Supplementary Material
Acknowledgments.
We thank Yang Shi and Xiaodong Li (Harvard Medical School) for providing the JARID1B expression plasmid and protocols and Chris Fry (Cell Signaling Technology) for JARID1B antibody. This work was supported by the Sidney Kimmel Foundation, American Cancer Society, National Institutes of Health, and the DFCI-Novartis Drug Discovery Program (A.L.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810067106/DCSupplemental.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 3.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 6.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 7.Jiang BH, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol. 1996;271:C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 8.Arany Z, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ema M, et al. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 11.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavesi G, et al. MoD Tools: Regulatory motif discovery in nucleotide sequences from co-regulated or homologous genes. Nucleic Acids Res. 2006;34:W566–W570. doi: 10.1093/nar/gkl285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol ISMB. 1994;2:28–36. [PubMed] [Google Scholar]
- 15.Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kel AE, et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellmann S, et al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008;372:892–897. 2008. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- 19.Pollard PJ, et al. Regulation of Jumonji-domain containing histone demethylases by hypoxia inducible factor (HIF) 1-alpha. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 20.Brat DJ, Mapstone TB. Malignant glioma physiology: Cellular response to hypoxia and its role in tumor progression. Ann Intern Med. 2003;138:659–668. doi: 10.7326/0003-4819-138-8-200304150-00014. [DOI] [PubMed] [Google Scholar]
- 21.Cloos PA, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 22.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- 24.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 25.Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 26.Yamane K, et al. JHDM2A, a JmjC-Containing H3K9 demethylase, Facilitates Transcription Activation by Androgen Receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat Res. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane K, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 32.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martone R, et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 37.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia inducible factors HIF-1{alpha} and HIF-2{alpha} Mol Biol Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.