Abstract
Junctions between ssDNA and dsDNA sequences are important in many cellular processes, including DNA replication, transcription, recombination, and repair. Significant transient conformational fluctuations (“DNA breathing”) can occur at these ssDNA–dsDNA junctions. The involvement of such breathing in the mechanisms of macromolecular complexes that operate at these loci is not well understood, in part because these fluctuations have been difficult to measure in a position-specific manner. To address this issue we constructed forked or primer-template DNA constructs with 1 or 2 adjacent 2-aminopurine (2-AP) nucleotide residues (adenine analogues) placed at specific positions on both sides of the ssDNA–dsDNA junction. Unlike canonical DNA bases, 2-AP absorbs, fluoresces, and displays CD spectra at wavelengths >300 nm, where other nucleic acid and protein components are transparent. We used CD and fluorescence spectra and acrylamide quenching of these probes to monitor the extent and nature of DNA breathing of A-T base pairs at specific positions around the ssDNA–dsDNA junction. As expected, spectroscopically measurable unwinding penetrates ≈2 bp into the duplex region of these junctions under physiological conditions for the constructs examined. Surprisingly, we found that 2-AP bases at ssDNA sites directly adjacent to ssDNA–dsDNA junctions are significantly more unstacked than those at more distant ssDNA positions. These local and transient DNA conformations on both sides of ssDNA–dsDNA junctions may serve as specific interaction targets for enzymes that manipulate DNA in the processes of gene expression.
Keywords: 2-aminopurine, circular dichroism, DNA unwinding, fluorescence, forked DNA
Double-stranded DNA molecules are stabilized by a network of interstrand hydrogen bonds between complementary A-T and G-C base pairs and by intrastrand base stacking. These DNA duplexes exist primarily in the Watson–Crick B-form conformation in aqueous solution at physiological temperature and salt concentration. Heating dsDNA induces a cooperative transition to single-stranded components; the midpoint of this transition is defined as the melting temperature (Tm), which depends on base composition, base sequence, and solvent conditions. dsDNA molecules also experience small thermal “breathing” fluctuations that transiently break hydrogen bonds and unstack bases at temperatures well below Tm. These fluctuations may play a role in the initial exposure of specific ssDNA binding targets that are otherwise buried in the duplex DNA (promoters, recombination or repair sites, etc.) to various regulatory protein complexes. However, this “interior DNA breathing” involves a very small fraction (≈10−6) of the total DNA at any one time and consequently cannot be observed by normal spectroscopic techniques. Methods such as hydrogen exchange (1–3) or chemical probes that interact irreversibly with and trap ssDNA sequences (for example, formaldehyde or dimethyl sulfate (4) can be used to monitor this interior breathing by accumulating the signal from these infrequent and otherwise reversible fluctuations (for a review see ref. 5).
Base pairs (especially A-T pairs) that are located at the ends of dsDNA molecules or at ssDNA–dsDNA junctions fluctuate into open states much more frequently than do those in the dsDNA interior. Depending on the local DNA sequence, such partially open base pairs may comprise significant fractions of the total conformation at these “end” positions, even at temperatures well below Tm. Many enzymes and protein assemblies that manipulate DNA act specifically at ssDNA–dsDNA boundaries (6–8). These include primer-template (P/T) junctions where polymerases extend newly-synthesized DNA and replication forks where helicases open the dsDNA to expose ssDNA templates for further DNA synthesis. To understand the functional roles of base pair “fraying” in such processes, it is important to be able to monitor the conformations of such ssDNA–dsDNA junctions.
We have shown that the low-energy (>300 nm) CD and fluorescence spectra of 2-aminopurine (2-AP) bases, which can replace adenine bases in nucleic acids without significant structural or biological perturbation, can be used (either as isolated substituents or as dimer pairs) to probe the conformations of nucleotide residues at specific positions within single-stranded and double-stranded sequences of DNA and RNA (9, 10). In this article, we use 2-AP spectral probes to study the breathing of adenine bases at and near ssDNA–dsDNA junctions within model DNA constructs. These probes reveal unusual DNA conformations at these loci that are likely to serve as interaction targets for the macromolecular machines of gene expression.
Results
The extent of thermal fluctuations at any particular dsDNA position depends on the overall sequence and stability of the DNA, the temperature, the ionic environment, and the proximity of DNA ends. Here, we have studied AT-rich sequences, in which breathing would be expected to be most apparent, at P/T junctions and DNA forks. The DNA constructs used are shown in Fig. 1. In replacing adenine bases with the 2-AP probes we took care to maintain the DNA sequence context of the probes to eliminate the dependence of the observed breathing and conformational changes on changes in base and base pair sequence. Two types of model DNA constructs were used in these studies. “Forked” constructs that carry noncomplementary ssDNA tails on both DNA strands at the ssDNA–dsDNA junction were used to model the leading edges of replication forks at which helicase activity opens duplex DNA. P/T constructs were used to model ssDNA–dsDNA elongation sites used in leading- and lagging-strand DNA replication.
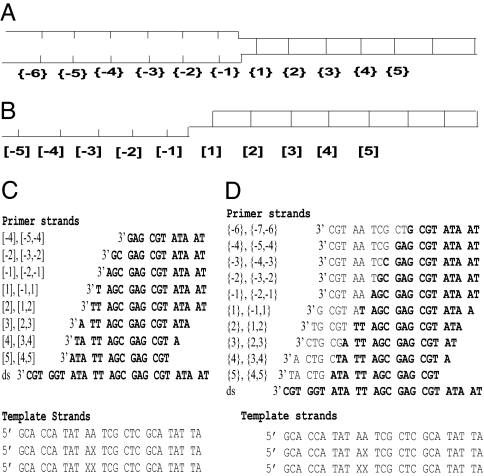
Fig. 1.
Nomenclature and sequences of the DNA constructs used. (A and B) The overall design of the forked (A) and the P/T (B) constructs. Negative numbers designate residue positions in ssDNA regions, and positive numbers correspond to dsDNA base pair positions. {} designate positions within the forked constructs; [] designate positions within the P/T constructs. (C and D) The sequence of the template (Lower) and corresponding primer (Upper) strands for P/T (C) and forked (D) constructs. X represents the position of 2-AP residue. Complementary base pairs in the various top strands are in bold. Each construct is designated by construct type (P/T or forked) and the position of the probe residues with respect to the single-stranded/double-stranded junction; the nomenclature used to designate the various duplex constructs is shown at the left of each nontemplate sequence.
Negative and positive integers in Fig. 1 designate nucleotide positions within the ssDNA and dsDNA components, respectively. The first duplex position at the ssDNA–dsDNA junction is designated 1, and the first unpaired position is −1. Positions within forked constructs are shown in curly brackets ({}), and positions within P/T constructs are in square brackets ([]). X represents the positions of 2-AP probes in the template strand (Fig. 1 C and D). Duplex constructs are designated by a single number (e.g., [4] or {4}) corresponding to the position within the construct that contains a single 2-AP residue or by double numbers (e.g., [−5,−4] or {−5,−4}), designating the positions of 2 adjacent 2-AP residue probes.
Local Melting at ssDNA–dsDNA Junctions.
UV and CD spectra of nucleic acids at wavelengths ≤300 nm arise from contributions from all of the base chromophores and reflect the overall conformation of the DNA constructs. In contrast, spectra of 2-AP at wavelengths >300 nm, where the canonical DNA bases are essentially optically transparent, give information only about DNA conformations at the probe positions (9–11).
UV spectral intensity changes measured at 260 nm (ΔA260) as a function of temperature for forked constructs {−2,−1} and {1,2} displayed similar cooperative melting properties (Fig. 2A) that showed, as expected, that the duplex portions of these constructs have the same overall stability. The absorbance of the {1,2} construct at 320 nm decreased on melting, as reported for 2-AP probes (10, 11). The cooperative transition at this wavelength occurred over the same temperature range as at 260 nm (although with a slightly lower Tm), showing that the local melting of the base pairs at positions 1 and 2 and global melting of the dsDNA portion of the forked construct occurs over the same temperature range and thus that the entire 14-bp dsDNA component melts cooperatively. In contrast, the 2-AP absorbance peak of the {−2,−1} construct at 320 nm decreased monotonically over the entire temperature range, with no indication of a cooperative transition. This behavior resembles the noncooperative melting curves for ssDNA containing a 2-AP dimer probe, measured either at 260 or 320 nm (see Fig. S1 and ref. 10). Hence the noncooperative thermal denaturation of bases in the single-stranded region of the forked construct appeared to be unperturbed by the melting of the adjacent duplex. The global and local Tm measured for the various constructs are summarized in Table S1.
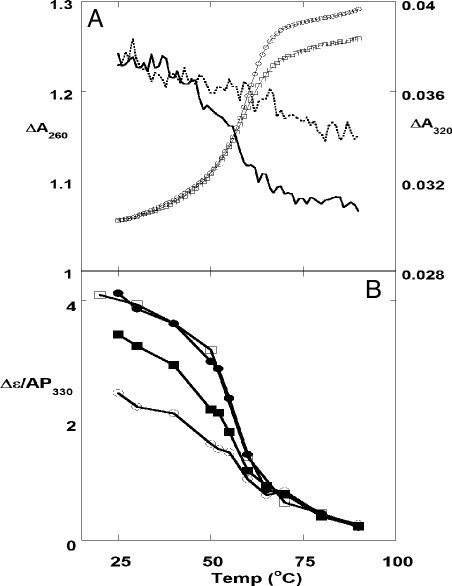
Fig. 2.
Melting curves of forked DNA constructs. (A) UV melting curves at 260 and 320 nm for forked constructs containing site-specific pairs of 2-AP probes. □, 260 nm, {1,2}; ○, 260 nm, {−2,−1}; solid line, 320 nm, {1,2}; dashed line, 320 nm, {−2,−1}. (B) CD melting curves at 330 nm for forked constructs containing 2-AP dimer probes. ○, {−1,1}; ■, {1,2}; ●, {3,4}; □, {4,5}.
We also monitored the CD changes for replication fork constructs at 330 nm (Δε330) as a function of temperature (Fig. 2B). The 2-AP dimer probe was first placed at the ssDNA–dsDNA junction ({−1,1}) and then moved progressively deeper into the dsDNA component (to positions {1,2}, {3,4}, and {4,5}). The signal for the {−1,1} probe showed essentially no cooperative melting. The {1,2} probe did display cooperative melting, although with a lowered melting amplitude and a slightly lowered Tm, presumably reflecting significant thermal fraying at these positions (see below). The melting curves for the {3,4} and the {4,5} probes displayed cooperative melting transitions centered on the global Tm, which showed that optically-measurable DNA breathing does not penetrate >2–3 bp into the duplex interior under the experimental conditions used. The ΔA320 and the Δε330 melting curves were superimposable (Fig. S2), confirming that both fluorescence and CD monitor the same melting transitions.
Local Conformations at ssDNA–dsDNA Junctions.
The spectra of 2-AP residues inserted into DNA reflect base conformations at the sites of the probes. The fluorescence of a 2-AP probe decreases significantly relative to that of free 2-AP as a result of stacking interactions with the neighboring bases, and the fluorescence of 2-AP residues located in dsDNA is more quenched than in ssDNA (12–14); we note that the intensity of the fluorescence signal changes without alteration of the shape or position of the emission spectrum (Fig. S3), permitting the intensity changes that accompany the opening and closing of base pairs to be monitored at a single wavelength. The fluorescence signals for ssDNA molecules containing 2-AP dimer probes are approximately half as intense (per 2-AP base) as that for ssDNA containing a single 2-AP. This “self-quenching” probably reflects better orbital mixing between a pair of adjacent 2-AP bases than between a single 2-AP base and canonical neighboring bases, although this property has not been studied in detail. We note that similar self-quenching of adjacent probe residues has also been seen with pyrrolocytosine bases in DNA (15).
Low-energy CD signals of 2-AP can also be used to characterize local conformations and conformational changes within nucleic acid constructs (9, 10, 16). Fig. 3 shows representative CD spectra obtained for 2-AP monomer and dimer probes incorporated into forked constructs at the indicated positions. The low-energy CD spectrum of an oligonucleotide containing an isolated 2-AP probe showed a single peak located near 320 nm, which is the absorbance maximum for 2-AP (10). In Fig. 3A this peak is present as a shoulder superimposed on the signal intensity from the <300-nm region of the CD spectra. It is important to note that this “spillover” may vary from one spectrum to the next and can be significant at wavelengths up to ≈315 nm. Spillover depends on the sequence of the construct and G residues, in particular, increase this effect (Fig. S4). This point is important in interpreting some of the CD results that follow.
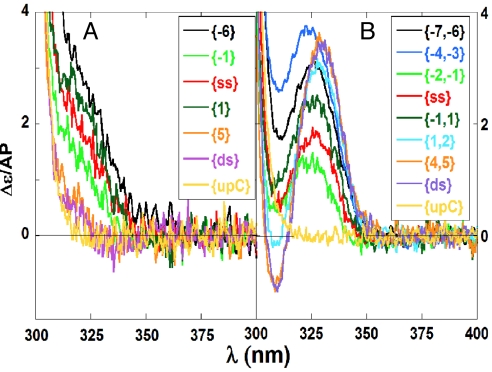
Fig. 3.
Representative CD spectra of forked DNA constructs containing 2-AP bases at defined positions. Constructs contained either single 2-AP probes (A) or 2-AP dimer probes (B) at the indicated positions. Spectra labeled upC correspond to dsDNA control constructs containing no 2-AP probes.
The low-energy CD of dimer 2-AP probes (Fig. 3B) displayed a peak and a trough centered around the 2-AP absorption maximum, which is characteristic of exciton coupling between adjacent 2-AP residues stacked in a double helical B-form DNA conformation (9, 10). Because dimer probes are moved into duplex DNA the exciton coupling becomes stronger, presumably reflecting reduced thermal fluctuations and the increased stability of the stacked 2-AP dimers. Careful inspection of these CD spectra showed that a small red shift of the peak accompanied this increased stacking and stabilization, which when the bases were fully stacked the CD peak shifted no further. Based on these observations we present our CD results as ellipticity per mol 2-AP at 330 nm for 2-AP dimers and 320 nm for 2-AP monomers.
Forked Constructs.
Fluorescence and CD signals of the 2-AP monomer probe varied as a function of position relative to the dsDNA–ssDNA junction of forked DNA. In Fig. 4 A and C we present the fluorescence and CD signal intensities for a single 2-AP probe as a function of probe position; comparable plots for DNA constructs containing pairs of 2-AP bases are shown in Fig. 4 B and D.
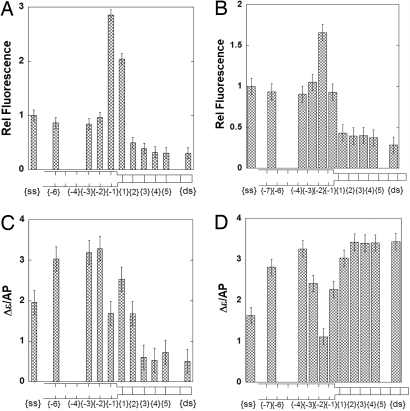
Fig. 4.
Spectroscopic properties of forked DNA constructs as a function of position relative to the ssDNA–dsDNA junction. (A) Fluorescence intensity changes at 370 nm for constructs containing single 2-AP probes. (B) Fluorescence intensity changes at 370 nm for constructs containing 2-AP dimer probes. (C) Ellipticity changes at 320 nm for constructs containing single 2-AP probes. (D) Ellipticity changes at 330 nm for constructs containing 2-AP dimer probes. Intensities for probes in ssDNA or dsDNA are at the left and right of each panel, respectively. Ellipticity changes are per mol 2-AP. Fluorescence intensity changes have been normalized to the intensity of the probe signal in the control ssDNA. The bars corresponding to the single 2-AP probes are centered on the probe position. The bars corresponding to the 2-AP dimer probes are broader and bracket the 2 positions involved.
DNA constructs with a single 2-AP residue placed at position {3} or beyond showed CD and fluorescence signal intensities characteristic of a stable interior probe in dsDNA (Fig. 4 A and C). In contrast, thermal fluctuations were observed at the first and second duplex base pairs adjacent to the ssDNA–dsDNA junction. The relative fluorescence intensity of a construct containing a single 2-AP residue at position {1} was ≈2-fold greater than that of a probe residue in ssDNA, and the fluorescence intensity of a probe at position {−1} was almost 3-fold greater. We have used these intensity changes to calculate the fraction of the base pairs that are “open” at each position adjacent to the duplex termini of the ssDNA–dsDNA junction constructs that we have studied. These calculations are summarized in Discussion and presented in Tables S2 and S3.
The CD signal for a single 2-AP at position {−1} also differed markedly from that for a single 2-AP probe in ssDNA. The fluorescence intensity reached a plateau at approximately the same level seen for a 2-AP probe in ssDNA. In contrast, we note that the CD intensities for these probes did not correspond to the signal seen for probes in ssDNA, presumably because of sequence-dependent spillover of the higher-energy CD signals (Fig. S4). The low-energy CD and fluorescence signals of a 2-AP dimer probe also varied within the forked DNA constructs as a function of distance from the ssDNA–dsDNA junction (Fig. 4 B and D). Spectral intensities corresponded to those of the fully double-stranded construct from positions {2,3} onwards. At positions {−2,−1} the relative fluorescence signal was ≈1.5 times higher than the signals seen in ssDNA regions. This increase was smaller than the enhancement seen with the monomer probe, perhaps as a result of self-quenching of the 2-AP dimer mentioned above. Measurements were also conducted with constructs in which the 2-AP dimer was placed at different positions in the ssDNA region of the template. CD signals at positions near the fork junction did not become comparable to those of a dimer probe in a fully ssDNA region until approximately positions {−3,−2} or beyond have been reached.
P/T Junction Constructs.
We also studied fluorescence and CD intensity changes as a function of probe position for single and dimer 2-AP probes located at and near P/T junctions that carry only 1 noncomplementary strand, rather than the 2 noncomplementary strands characteristic of a forked junction. The results were much like those seen with the forked junction constructs and are presented in Fig. S5. As with the forked constructs the fluorescence and CD intensities from single 2-AP probes in P/T junction constructs changed progressively from position [1] to [3] of the DNA duplex region and thereafter the values were also identical with the signals observed for dsDNA (Fig. S5 a and c). The fluorescence intensity increased at the [−1] position by a factor of 1.2 relative to ssDNA, which is a smaller fluorescent enhancement than seen at this position in forked DNA. Isolated 2-AP residues in the ssDNA portion of the P/T construct showed spectral properties characteristic of fully ssDNA sequences at and beyond position [−3]. Similar results were obtained for 2-AP dimer probes (Fig. S5 b and d).
Taken together, these findings show that for the P/T constructs, as for the forked constructs, optically significant DNA unwinding of A-T base pairs penetrates only ≈2 bp into the double-helical side of the construct. Furthermore the proximity of the ssDNA–dsDNA junction prevents adenine (2-AP) bases in the template strand from achieving fully single-stranded properties, certainly at the −1 position and perhaps at the −2 position as well. Such conformations at ssDNA–dsDNA junctions may play a role in controlling the DNA processing enzymes that operate at these loci.
Acrylamide Quenching of 2-AP Probes Located Near the ssDNA–dsDNA Junction.
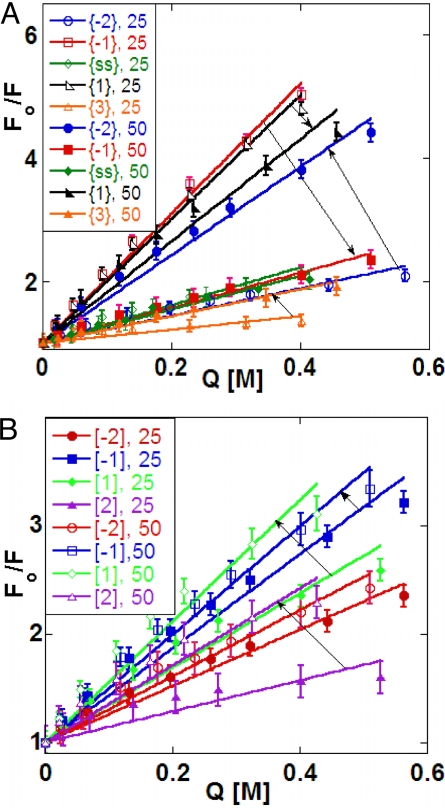
Acrylamide quenching of the fluorescence of 2-AP residues in DNA depends on the access of the fluorophore to the solvent (17) and provides additional insight into the structural and dynamic aspects of DNA breathing near these single stranded–double stranded junctions (Fig. 5). Quenching data were fit to the Stern–Vollmer (S-V) equation:
where Fo is the fluorescence intensity in the absence of quencher, F is the observed fluorescence at specific acrylamide concentrations [Q], and KSV is the quenching coefficient. S-V plots of Fo/F against [Q] were linear for all of the DNA constructs and probe positions examined (Fig. 5), showing quenching to be dynamic at all construct positions (17). Increasing values of KSV as a function of temperature generally correspond to increased solvent (and quencher) access to the probe bases.
Fig. 5.
Acrylamide quenching of 2-AP probes in DNA constructs. (A) Forked DNA. (B) P/T DNA. Open symbols correspond to quenching at 25 °C, and closed symbols correspond to quenching at 50 °C. The arrows point from the 25 ° to the 50 °C S-V plots for the same construct in each case.
Probes within the dsDNA region of the constructs (position {3}) showed a small increase in KSV in going from 25 °C to 50 °C, reflecting the expected decrease in the stability of base pairs within the dsDNA component over in this temperature range; we note that 50 °C is close to the Tm for these constructs (Table S1). KSV for a probe at position 2 in a P/T construct increased somewhat more at the higher temperature than did this parameter at the equivalent position within a forked construct. We found that acrylamide quenching of 2-AP probes in ssDNA was approximately the same at 25 °C and 50 °C, suggesting that quencher access to a 2-AP base in the control ssDNA sequences was little changed over this temperature range. In contrast, probes located in ssDNA sequences closer to a P/T junction (position [−2]) showed somewhat more exposure with increased temperature, whereas those near a fork ({−2}) showed an even greater increase in quencher access at higher temperature. This observation suggests that fluctuations in the ssDNA component near the ssDNA–dsDNA junction reduces not only local stacking, but may also increase the temperature-dependent flexibility of the chain near the junction, permitting greater access.
Quenching results were quite different for ssDNA positions {−1} and {1} directly adjacent to a fork junction. Here, the S-V slope (KSV) became smaller as the temperature increased. These results suggest that in these constructs 2-AP probes located directly adjacent to the fork are unusually exposed to the solvent at 25 °C, so much that the increased flexibility of the construct at higher temperature decreases access to the quencher (perhaps by permitting stacking to increase), especially at position {−1}. This effect is less evident, but still present, at comparable positions in the more open P/T junction constructs (Fig. 5B).
These results suggest that at 25 °C the base at the {−1} position of the forked construct is in a conformation that is more exposed to solvent than in either ssDNA or dsDNA at the same temperature. This position seems to partially lose its unique character as Tm is approached, reverting to a conformation more comparable to that of a “standard” 2-AP probe in a ssDNA construct. However, the Tm values for these constructs (Table S1) showed that the complementary bases at position {1} were still largely base-paired at 50 °C; as a result the decrease in quenching that was observed relative to 25 °C was smaller than that for a 2-AP probe at the {−1} position in a forked construct.
Discussion
DNA breathing at ssDNA–dsDNA junctions should be most apparent in AT-rich sequences. We have therefore studied the thermal fluctuations of the adenine base analogue, 2-AP, located at and near the single-stranded—double-stranded boundaries of DNA models of the P/T junction and replication fork structures. The low-energy CD and fluorescence spectra of site-specifically positioned 2-AP probes were used to monitor these fluctuations. Results show that the first A-T base pair in the duplex region of a fork or ssDNA–dsDNA junction is significantly destabilized at temperatures well below Tm, and that the penultimate base pair is also perturbed, although to a lesser extent. Base pairs located 3 or more positions away from the junction are spectroscopically comparable with those deeply buried in dsDNA. We also found that 2-AP bases located in the ssDNA immediately vicinal to the junction displayed an unusually “open” conformation relative to bases located further away on the ssDNA side. We suggest that these unusual conformations on both sides of the ssDNA–dsDNA boundary may be physiologically important, given that P/T junctions and ssDNA–dsDNA forks comprise the “operational venues” of many of the macromolecular machines of gene expression.
Breathing at the ends of duplex DNA has, of course, been studied by other techniques including, in particular, imino proton exchange NMR spectroscopy (18). Base pairs near duplex termini are known to be more mobile than interior base pairs, reflecting the reduced structural constraints present at the end of a DNA duplex. Increased mobility at these regions has also been demonstrated with X-ray crystallography (increased B factors), chemical probes, and computer simulation. Spectroscopic methods are most suitable for monitoring equilibrium breathing at ssDNA–dsDNA junctions at levels that might be used in the processes of DNA manipulation by proteins. CD measurements reflect the chirality of the ground state of the chromophore and in favorable cases the CD signal can be unambiguously assigned to a conformation.
Previously (9, 10) and in this study, we have shown that pairs of 2-AP probes embedded in the interior of a B-form dsDNA sequence display strong exciton coupling between the 2-AP bases and that the resulting CD signal (see Fig. 3B) can be taken as evidence that these base pairs are stacked in a B-form conformation. CD and absorbance reflect time scales close to 10−15 seconds. As a consequence conformational distributions do not significantly change during a CD measurement, whereas a chromophore can undergo numerous vibrations and translational displacements during a fluorescence emission process. This simple consequence of the Franck–Condon principle means that CD (and UV absorbance), unlike fluorescence, measure the equilibrium distribution of local conformations, whereas this distribution can change significantly during a fluorescence measurement, depending on the lifetime of the excited state. Fluorescence measurements with 2-AP probes have also been used to study local conformational changes of P/T DNA constructs during DNA replication (6, 12) and transcription (19). Reported signal changes at the corresponding ssDNA–dsDNA junctions are consistent with our results. It is difficult to assign fluorescence quenching unambiguously to a particular conformational change (such as a change in base stacking) without independent structural measurements. CD signals provide an “instantaneous snapshot” and thus direct information about the distribution of conformations at the 2-AP probe.
Our CD and fluorescent measurements with both single and dimer 2-AP probes have shown that, as expected, large-scale conformational fluctuations extend only ≈2 bp into the dsDNA sequence of both forked and P/T junctions at 25 °C (Fig. 4 and Fig. S5). The 14-bp dsDNA component of our constructs melts cooperatively (Fig. 2), and Tm values measured with 2-AP probes located on the dsDNA side of the fork were about the same as those for the overall melting of the duplex sequence (Table S1), although the amplitude of the melting transition decreased as the probes were moved toward the junction and the open fraction of the base pairs increased. Hence Fig. 2B provides a measure of the extent of fraying at each position of the dsDNA portion of the construct. Beyond position 2 the A-T base pairs of the junction melted with essentially the same stability as those located deep in the dsDNA interior. Furthermore we found that (2-AP)-T base pairs in dsDNA were not subject to significant fluorescent quenching by acrylamide (Fig. 5), consistent with their fully stacked and hydrogen-bonded conformations within the DNA duplex. Although we know on both theoretical (DNA melting is a first-order phase transition) and experimental (hydrogen exchange) grounds that small-scale thermal fluctuations must extend much further into dsDNA, the fraction of base pairs open beyond the 2 position is clearly much less than the 3–5% that represents the limits that can be detected of our spectral measurements.
The relative levels of breathing of specific base pairs on the double-stranded side of the ssDNA–dsDNA junction that we observed are expected on general thermodynamic grounds. Thus, the thermodynamic “cost” of opening an average single base pair at the end of a DNA double helix is 1–2 kcal/mol bp (7), which corresponds to a thermal energy of ≈2 kT at room temperature. Assuming a normal Boltzmann distribution of DNA conformations, one would expect to find the first double-stranded base pair at the ssDNA–dsDNA junction to be significantly unwound, the second base pair unwound much less, and base pairs located further into the dsDNA interior to be open much <1% of the time and thus not detectable by our measurements. We have used the changes in fluorescence and CD signal intensities monitored here to estimate the fraction to which the base pairs at positions 1 and 2 of the dsDNA sequences of our constructs are open at 25 °C for the sequences we have examined here using 2-AP probes. Assuming a 2-state transition between discrete open and closed forms of each base pair, our calculations show that in the constructs investigated here the base pairs in position 1 were open ≈50% of the time, whereas the base pairs in position 2 were ≈10% open. Additional estimates (with similar results) were made at various temperatures by using the CD melting curve data of Fig. 2B (see Tables S2 and S3). We note also that the open base pair fractions obtained independently from the CD and fluorescent signal intensity change data are very similar, consistent with the 2-state assumption for local individual base pair breathing fluctuations made in these calculations.
Quite unexpectedly, our CD and fluorescent measurements showed that 2-AP probes located at single-stranded positions directly adjacent to the ssDNA–dsDNA junction appeared more open or unstacked than these probes located in a fully ssDNA position (Figs. 3–5 and Fig. S5). These bases were also more accessible to acrylamide quenching than 2-AP bases in ssDNA. This was especially true at temperatures far removed from Tm, whereas at higher temperatures this unusual solvent accessibility was less prominent. The unstacking observed on the ssDNA side of the junction could reflect a number of influences. Although the thermodynamic forces that drive base stacking are not completely understood, it is clear that interactions with the aqueous solvent are important. Base stacking minimizes the size of the cavity produced in the solvent by a base, and thus reduces the cost of solvation compared with unstacked bases (20). Within duplex DNA both surfaces of the base are buried, whereas at the ends of the double-helix one face must be exposed to the aqueous environment. This “end effect” could contribute to the stability of unusual structures in the ssDNA immediately adjacent to the junction. In addition ssDNA–dsDNA junctions are also likely to involve an unusual local ionic environment. The charge of the phosphate groups of the DNA backbones is largely neutralized by counterion condensation that produces a high local concentration of cations, independent of the bulk salt concentration of the solution (20). The local concentration of this ionic “cloud” is less for ssDNA than for dsDNA (21). Thus, single-stranded residues close to the junction may be exposed to higher counterion concentrations than residues in “normal” ssDNA positions, and this difference could also stabilize unusual conformations.
Finally, the ssDNA–dsDNA junction sterically constrains the neighboring ssDNA compared with the situation at “interior” ssDNA sequences. Two ssDNA chains emerging from the fork may further restrict one another's conformational possibilities, thus making stacking at such positions more difficult and thus less likely. Unstacking of the single-stranded edge of the junction is significantly greater for forked DNA than for P/T DNA constructs, which have only 1 ssDNA chain at the junction (Fig. 4 and Fig. S5). This difference is consistent with the steric interference of 1 ssDNA chain at a fork with the stacking interactions of the bases of the other.
Single nucleotide overhangs at either the 3′ or 5′ termini of dsDNA, dsRNA, or hybrid DNA–RNA molecules generally increase the overall stability of a nucleic acid duplex (22, 23). The molecular mechanisms by which these so-called dangling ends affect stability are thought to involve stacking between the unpaired base and the terminal base pair of the duplex. Position −1 of our P/T and forked constructs correspond to the locus of a 5′-terminal dangling base in the above studies. Fluorescence and CD measurements using our 2-AP probes (Fig. 4 and Fig. S5) indicate that, unlike the situation with a single nucleotide overhang, the bases and base pairs at positions 1 and −1 in our constructs are highly unstacked. In addition, we show that the presence of the noncomplementary strand of the forked construct further unstacks these residues. These results suggest that base stacking interactions at the ssDNA–dsDNA junction may be sensitive to the context of the “dangling base.” A long “dangling sequence” may offer an alternative stacking arrangement that is not observed with single base overhangs that stack with the residue at position 1. The presence of a noncomplementary strand may further favor interactions between the base at position 1 and the adjacent ssDNA.
In conclusion, we have shown that breathing at the single-stranded–double-stranded interface of P/T and forked DNA can be quantified by the methods reported here. The possible roles of such breathing in the activities of DNA processing enzymes require additional investigation. We speculate that the more open state of the ssDNA bases immediately adjacent to the junction may increase the specific binding of the DNA enzymes that operate at these loci, perhaps helping to target them to these functional positions.
Materials and Methods
DNA Constructs.
ssDNA oligonucleotides containing site-specific 2-AP substitutions were purchased from Operon; unlabeled complementary ssDNA oligonucleotides were obtained from IDT. DNA concentrations were determined by UV absorbance at 260 nm (25 °C), based on extinction coefficients furnished by the manufacturer. Base-paired structures were formed by heating equimolar (3 μM) concentrations of the appropriate 2-AP-labeled ssDNA strand with the complementary unlabeled ssDNA at 90 °C for 5 min, and then cooling to room temperature over a period of 2 h. Thermal melting monitored by ΔA260nm was used to confirm the double-stranded character of the resulting dsDNA. Unless stated otherwise, experiments were performed at 25 °C in a buffer containing 20 mM Hepes (pH 7.5), 100 mM NaOAc, 10 mM Mg(OAc)2, and 0.1 mM EDTA.
Spectroscopic Procedures.
CD spectra were measured at wavelengths from 230 to 400 nm by using a JASCO model J-720 CD spectrometer equipped with a temperature-controlled sample holder. Ten to 15 spectra were scanned, averaged, and plotted as graphs of Δε (the difference in the molar extinction coefficient of left and right circularly polarized light) per mol residue versus wavelength. Fluorescence spectra were measured by using either a Jobin-Yvon Flourolog or a Flouromax spectrophotometer. Samples were excited at 315 nm, and emission spectra were recorded from 300 to 450 nm. Reported fluorescence intensities presented are values measured at 370 nm, normalized to the fluorescence of the corresponding ssDNA sequence. Thermal melting experiments were monitored by UV absorbance at 260 nm (or 320 nm for 2-AP residues) and at 330 nm for CD. Melting was performed at a temperature “ramp rate” of 0.1 °C/min. The error bars shown for the fluorescence measurements represent standard deviations for 2–4 independent experiments. The reported errors for the CD measurements correspond to the measured noise levels for each experiment.
Supplementary Material
Acknowledgments.
We thank John Schellman, Walt Baase, and members of our laboratory at the University of Oregon for suggestions and advice and Drs. Craig Martin (University of Massachusetts, Amherst) and Ken Breslauer (State University of New Jersey, Rutgers) for helpful comments on the manuscript. This work was supported in part by National Institutes of Health Grant GM-15792 (to P.H.v.H.) and the Centre National de la Recherche Scientifique (N.P.J.). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Note Added in Proof.
We have also recently used low energy CD and fluorescence measurements of a cytosine derivative, pyrrolocystosine, to characterize the formation of a 3′-TGC loop in oligonucleotides that model a 3-nt frameshift deletion (24).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900803106/DCSupplemental.
References
- 1.McConnell B, von Hippel PH. Hydrogen exchange as a probe of the dynamic structure of DNA. I. General acid-base catalysis. J Mol Biol. 1970;50:297–316. doi: 10.1016/0022-2836(70)90194-4. [DOI] [PubMed] [Google Scholar]
- 2.Gueron M, Leroy J. Nucleic Acids and Molecular Biology. New York: Springer; 1992. Base-pair opening in double-stranded nucleic acids; pp. 1–22. [Google Scholar]
- 3.Every AE, Russu IM. Probing the role of hydrogen bonds in the stability of base pairs in double-helical DNA. Biopolymers. 2007;87:165–173. doi: 10.1002/bip.20811. [DOI] [PubMed] [Google Scholar]
- 4.McGhee JD, von Hippel PH. Formaldehyde as a probe of DNA structure. III Mechanism of the initial reaction of formaldehyde with DNA. Biochemistry. 1977;16:3276–3293. doi: 10.1021/bi00634a002. [DOI] [PubMed] [Google Scholar]
- 5.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q Rev Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 6.Hochstrasser RA, Carver TE, Sowers LC, Millar DP. Melting of a DNA helix terminus within the active site of a DNA polymerase. Biochemistry. 1994;33:11971–11979. doi: 10.1021/bi00205a036. [DOI] [PubMed] [Google Scholar]
- 7.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 8.Wong CJ, Lucius AL, Lohman TM. Energetics of DNA end binding by E. coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J Mol Biol. 2005;352:765–782. doi: 10.1016/j.jmb.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 9.Datta K, Johnson NP, von Hippel PH. Mapping the conformation of the nucleic acid framework of the T7 RNA polymerase elongation complex in solution using low-energy CD and fluorescence spectroscopy. J Mol Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 10.Johnson NP, Baase WA, Von Hippel PH. Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA. Proc Natl Acad Sci USA. 2004;101:3426–3431. doi: 10.1073/pnas.0400591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eritja R, et al. Synthesis and properties of defined DNA oligomers containing base mispairs involving 2-aminopurine. Nucleic Acids Res. 1986;14:5869–5884. doi: 10.1093/nar/14.14.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 13.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: Role of base stacking. Proc Natl Acad Sci USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law SM, Eritja R, Goodman MF, Breslauer KJ. Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry. 1996;35:12329–12337. doi: 10.1021/bi9614545. [DOI] [PubMed] [Google Scholar]
- 15.Johnson NP, Baase WA, von Hippel PH. Investigating local conformations of double-stranded DNA by low-energy circular dichroism of pyrrolo-cytosine. Proc Natl Acad Sci USA. 2005;102:7169–7173. doi: 10.1073/pnas.0502359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee BJ, Barch M, Castner EWJ, Völker J, Breslauer KJ. Structure and dynamics in DNA looped domains: CAG triplet repeat sequence dynamics probed by 2-aminopurine fluorescence. Biochemistry. 2007;46:10756–10766. doi: 10.1021/bi7005674. [DOI] [PubMed] [Google Scholar]
- 17.Lakowicz JR. Principles of Flourescence Spectroscopy. 2nd Ed. New York: Kluwer/Plenum; 1999. [Google Scholar]
- 18.Andreatta D, et al. Ultrafast dynamics in DNA: “Fraying” at the end of the helix. J Am Chem Soc. 2006;128:6885–6892. doi: 10.1021/ja0582105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Martin CT. Promoter clearance by T7 RNA polymerase. Initial bubble collapse and transcript dissociation monitored by base analog fluorescence. J Biol Chem. 2002;277:2725–2731. doi: 10.1074/jbc.M108856200. [DOI] [PubMed] [Google Scholar]
- 20.Cantor CR, Schimmel PR. Biophysical Chemistry. San Francisco: Freeman; 1980. [Google Scholar]
- 21.Ballin JD, Shkel IA, Record MT., Jr Interactions of the KWK6 cationic peptide with short nucleic acid oligomers: Demonstration of large Coulombic end effects on binding at 0.1–0.2 M salt. Nucleic Acids Res. 2004;32:3271–3281. doi: 10.1093/nar/gkh646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bommarito S, Peyret N, SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JD, Zhao L, Xia T. The dynamic structural basis of differential enhancement of conformational stability by 5′- and 3′-dangling ends in RNA. Biochemistry. 2008;47:5962–5975. doi: 10.1021/bi800210t. [DOI] [PubMed] [Google Scholar]
- 24.Baase WA, et al. DNA models of trinucleotide frameshift deletions: The formation of loops and bulges at the primer–template junction. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.