Abstract
The ability of insects to detect and avoid ingesting naturally occurring repellents and insecticides is essential for their survival. Nevertheless, the gustatory receptors enabling them to sense toxic botanical compounds are largely unknown. The only insect gustatory receptor shown to be required for avoiding noxious compounds is the Drosophila caffeine receptor, Gr66a. However, this receptor is not sufficient for the caffeine response, suggesting that Gr66a may be a subunit of a larger receptor. Here, we report that mutations in the gene encoding the gustatory receptor, Gr93a, result in a phenotype identical to that caused by mutations in Gr66a. This includes an inability to avoid caffeine or the related methylxanthine present in tea, theophylline. Caffeine-induced action potentials were also eliminated in Gr93a-mutant animals, while the flies displayed normal responses to other aversive compounds or to sugars. The Gr93a protein was coexpressed with Gr66a in avoidance-gustatory receptor neurons (GRNs), and functioned in the same GRNs as Gr66a. However, misexpression of both receptors in GRNs that normally do not express either Gr93a or Gr66a does not confer caffeine sensitivity to these GRNs. Because Gr93a- and Gr66a-mutant animals exhibit the identical phenotypes and function in the same cells, we propose that they may be caffeine coreceptors. In contrast to mammalian and Drosophila olfactory receptors and mammalian taste receptors, which are monomeric or dimeric receptors, we propose that Drosophila taste receptors that function in avoidance of bitter compounds are more complex and require additional subunits that remain to be identified.
Keywords: Gr93a, gustatory receptor neuron, taste, repellent, chemosensation
The sense of taste enables insects to sample their environment and identify nutrient-rich botanical sources. Consequently, as part of a protective mechanism, many plants produce toxic compounds to avoid consumption. In turn, insects have developed receptors to detect these noxious chemicals and prevent the deleterious effects resulting from ingestion.
In the fruit fly, Drosophila melanogaster, the detection of attractive and aversive tastants appears to be encoded primarily by a family of 68 7-transmembrane gustatory receptors (Grs) (1–4). The Drosophila Grs have virtually no sequence similarity with mammalian taste receptors. Rather, these receptors are conserved among distantly related insects, as related Grs are encoded in mosquitoes, such as Anopheles gambiae (5), which diverged from flies ≈250 million years ago (6). Thus, characterization of Drosophila Grs offers a genetically tractable model for dissecting the sense of taste common to a variety of insect pests.
Three receptors essential for sugar detection have been identified. The first is Gr5a, which is necessary for the response to trehalose (7–9). Gr5a and other Drosophila taste receptors are expressed in gustatory receptor neurons (GRNs) rather than neuroepithelial cells, as in mammals. The GRNs are housed in chemosensory bristles distributed in several body locations, including the main taste organ, the proboscis, which is the functional homolog of the mammalian tongue (10). Gr5a is expressed in most sugar-responsive GRNs (7–9, 11, 12). Gr64a is essential for the detection of multiple other sugars, including sucrose, glucose, and maltose (13, 14). A third receptor, Gr64f, is a coreceptor, which is broadly required for the detection of most sugars (15). Gr64f functions in combination with Gr5a for trehalose detection, and in concert with Gr64a for sensing sucrose, maltose, and glucose. However, it appears that the Gr64f/Gr5a and Gr64f/Gr64a receptor pairs are not sufficient for eliciting responses to sugars (15). Thus, detection of a single sugar receptor may require more than 2 receptors.
Many Grs are expressed in all or subsets of GRNs, which function in the detection of noxious compounds (11, 12); however, the only Gr shown to be required for the response to aversive compounds is Gr66a (16). This receptor, which is expressed in most avoidance GRNs, appears to be a subunit of a caffeine receptor, as mutation of Gr66a eliminates caffeine-avoidance behavior and caffeine-induced action potentials in the GRNs. However, misexpression of Gr66a is not sufficient to produce caffeine sensation, suggesting that a minimum of 2 Grs is required for caffeine detection.
Here, we show that mutations in Gr93a result in a phenotype identical to Gr66a-mutant animals. Both mutants are unable to respond to caffeine and the related methylxanthine, theophylline. In addition, Gr93a and Gr66a are expressed in the same GRNs. However, misexpression of these 2 receptors is not adequate to recapitulate caffeine sensation. These data indicate that the ability to sense caffeine requires at least 2 Grs in addition to other subunits.
Results
Generation of Gr93a Mutants.
To characterize new requirements for Grs for Drosophila taste, we focused on Gr93a, because it was included in 1 of the 2 branches of the Gr phylogenetic tree (1) most distantly related to those that contain the Grs known to be required for the attractive (Gr5a, Gr64a, and Gr64f) and aversive (Gr66a) responses to tastants. To disrupt Gr93a, we obtained 2 transposable element lines, one of which had a Piggybac insertion (f01688) in the Gr93a intron (Gr93a1), while the other contained a P-element insertion (EY11817) in the 3′ noncoding exon (Fig. 1A). We mobilized the P-element, resulting in a 2.4-kilobase deletion, which removed the entire 419-aa coding region [see Fig. 1 A and B; Gr93a2 and supporting information (SI) Fig. S1]. Thus, Gr93a2 was a null allele. Gr93a1 appeared to be a strong allele as the Gr93a RT-PCR product was greatly reduced relative to an internal control (tubulin) (Fig. 1C, Upper). However, Gr93a1 may not be a null allele, as we detected a low-level of a Gr93a RT-PCR product after additional PCR amplification cycles (Fig. 1C, Lower) (see Methods). To create a third allele, we used ends-out homologous recombination (17). We deleted a 539 base-pair region encoding the N-terminal 180 residues and first 3 transmembrane segments, and inserted the GAL4-coding region at the site corresponding to the initiation codon in Gr93a (Fig. 1 A and D).
Fig. 1.
Generation of Gr93a alleles. (A) Schematic of the Gr93a locus and alleles. The 2 Gr93a exons are indicated by rectangles. The insertion sites of the Piggybac transposon (Gr93a1) and the P-element, EY11817 are indicated. The deletion in Gr93a2 created by imprecise excision of EY11817 is indicated. Gr93a3 was generated by ends-out homologous recombination. The gray and striped boxes indicate the GAL4 and miniwhite genes, respectively. The bracket below the representation of the complete Gr93a gene indicates the deletion in Gr93a3, which removes the 5′ end of the protein-coding region. The arrowheads indicate the primers used for the PCR analyses in (B) and (D). The arrows indicate the orientation of the GAL4 and miniwhite genes. (B) Confirmation of the deletion in Gr93a2 by PCR (using primers a and b; see A) and by DNA sequencing. (C) Analyses of Gr93a and tubulin RT-PCR products from Gr93a1/Gr93a2 and w1118 flies. The Gr93a RT-PCR products in the top and bottom panels were obtained after 35 and 37 PCR cycles, respectively. (D) Confirmation of the Gr93a3 mutation by PCR analyses using the indicated primer pairs.
Gr93a Is Required for Caffeine-Avoidance Behavior.
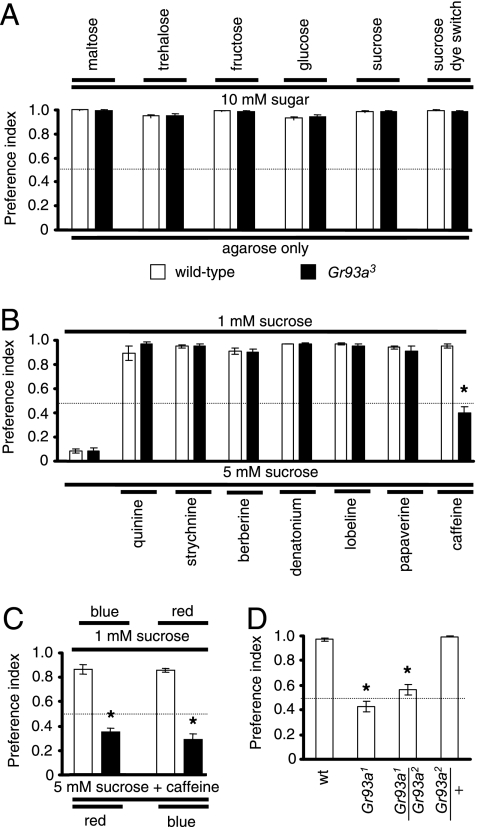
To address whether Gr93a was required for the behavioral responses to either aversive or attractive tastants, we used a variation of the binary food-choice assay (18). We allowed starved flies to feed in a microtiter dish with wells alternating between agar mixed with sugar or agar alone. The 2 alternatives were mixed with either red or blue food dyes and the colors of the abdomens were assessed (red, blue, or purple). Preference indexes (PI) of 1 or 0 indicated that all of the flies consumed either the sugar or the agar alone, respectively, while a PI of 0.5 resulted if there was no bias. We found that the Gr93a-mutant flies displayed similar preferences for sugars as the wild-type controls (Fig. 2A and Table S1).
Fig. 2.
Binary food-choice assays. (A–C) Wild-type (white bars) and Gr93a3 (black bars). (A) Sugar preferences. Flies were allowed to choose between 1% agarose plus the indicated sugar (10 mM) or 1% agarose only. (B) Avoidance of noxious compounds. The 2 alternatives were 1-mM sucrose alone or 5-mM sucrose either alone or in combination with the following aversive compounds: 1-mM quinine, 0.5-mM strychnine, 0.1-mM berberine, 0.3-mM denatonium, 0.3-mM lobeline, 2-mM papaverine, or 10-mM caffeine. (C) The dye color did not affect caffeine avoidance. The red and blue dyes were added to 1-mM sucrose or 5-mM sucrose plus caffeine, as indicated. (D) Caffeine-avoidance behavior in Gr93a1 and Gr93a1/Gr93a2 flies. The error bars represent SEMs. The asterisks indicate significant differences from wild type (P < 0.01) using unpaired Student's t tests. See Tables S1–S4 for statistics.
To assess whether Gr93a was required for avoiding bitter compounds, we assessed the ability of mutant flies to choose between either 1-mM sucrose or 5-mM sucrose combined with aversive compounds. The bitter tastants were mixed with a higher concentration of sucrose (5 mM) because some compounds that elicit avoidance responses, such as caffeine, suppress the attractive response to sugars (18). We found that the Gr93a3 flies avoided quinine, strychnine, berberine, denatonium, lobeline, and papaverine to the same extent as wild-type (Fig. 2B and Table S2).
In contrast to the results with other aversive compounds, caffeine avoidance was impaired in the Gr93a3-mutant animals (see Fig. 2B and Table S2). These results were surprising, as the only Gr expressed in avoidance GRNs (Gr66a) and previously characterized functionally was also required exclusively for the caffeine response (16). However, not every Gr expressed in avoidance GRNs is essential for the caffeine response because we have recently generated mutations in Gr8a and Gr47a, which are expressed in Gr66a-expressing GRNs but are not required for aversive behavior to caffeine (S.J.M. and C.M., unpublished observations). The wild-type avoidance of caffeine and the defect in the Gr93a mutants was not a consequence of different responses to the red or blue food coloring because the same phenotypes were observed upon switching the tastants/dye combinations (Fig. 2C and Table S3). To provide additional evidence that Gr93a was required for caffeine sensation, we examined additional alleles and found that Gr93a1 and Gr93a1/Gr93a2 transheterozygous flies displayed defects in caffeine avoidance similar to Gr93a3 (Fig. 2D and Table S4). The caffeine response in Gr93a2/+ was indistinguishable from wild-type, demonstrating that loss-of-function mutations in Gr93a were recessive (see Fig. 2D and Table S4).
Gr93a Functions in Aversive GRNs.
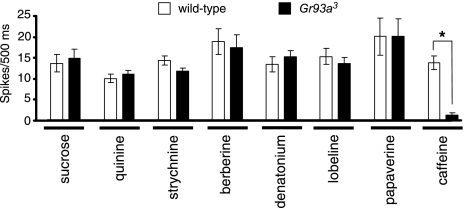
Because both Gr66a and Gr93a are required for the aversive responses to caffeine, the question arises as to whether they function together in the same GRNs. As a first step in addressing this question, we performed tip recordings, which measure action potentials in the GRNs in response to tastants. Consistent with the behavioral assays, the frequencies of action potentials stimulated by application of sucrose and most bitter tastants were similar in Gr93a3 and wild-type (Fig. 3 and Table S5). However, caffeine-induced action potentials were virtually eliminated (see Fig. 3 and Table S5). These results indicate strongly that Gr93a is required for the caffeine response in GRNs. The combination of the Gr93a3 and Gr66aex83 mutations does not appear to cause a more severe gustatory defect than the single mutations, as Gr66aex83,Gr93a3 flies show wild-type electrophysiological responses to sucrose and quinine, in addition to the expected deficit in the caffeine-induced action potentials (Fig. S2).
Fig. 3.
Gr93a is required for caffeine-induced action potentials. Tip recordings were carried out on S6 bristles on the labella. Shown are average frequencies of action potentials (spikes/500 ms) after application of 50-mM sucrose, 1-mM quinine, 0.1-mM berberine, 1-mM denatonium, 1-mM lobeline, 5-mM papaverine, 1-mM strychnine, or 10-mM caffeine. The error bars represent SEMs. The asterisk indicates a significant difference from wild type (P < 0.00001) using the unpaired Student's t test. See Table S5 for statistics.
Corequirements for Gr93a and Gr66a.
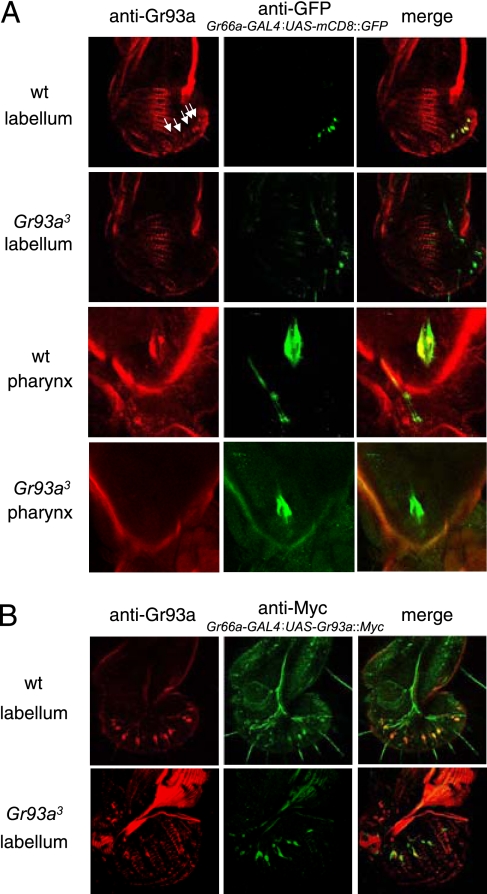
Loss of Gr66a results in the same deficit in the caffeine response as in Gr93a-mutant animals, raising the possibility that the 2 Grs are functionally coexpressed. To detect Gr93a expression, we attempted to use the GAL4/UAS system to generate a Gr93a reporter. However, the GAL4 reporter inserted in Gr93a3 was not expressed (data not shown). We also obtained 2 transgenes, 1 of which fused the GAL4 gene to 9 kb of the Gr93a 5′ flanking sequence, and the other of which included 2 kb of 5′ flanking sequence and 7 kb of 3′ flanking sequence. Neither of these latter GAL4 drivers was expressed (data not shown). Therefore, we attempted to raise anti-Gr93a polyclonal antibodies, although previous attempts to detect Grs with antibodies have not been successful. We found that the anti-Gr93a antibodies stained dendrites, axons, and cell bodies of a subset of GRNs in wild-type labella (Fig. 4A and Fig. S3A). Although the antibodies were not effective on Western blots, the immunostaining in the labellum appeared to be specific for Gr93a because the GRNs were not stained in Gr93a3-mutant labella (see Fig. 4A and Fig. S3C). We did not detect anti-Gr93a expression in 2 other tissues examined: legs and antennae (data not shown). Because Gr66a antibodies were unavailable, we costained labella from Gr66a-GAL4;UAS-mCD8::GFP flies with anti-GFP and anti-Gr93a. The anti-Gr93a and anti-GFP signals overlapped extensively, if not completely (see Fig. 4A and Fig. S4). In addition, 2 internal taste neurons from the pharynx expressed the Gr93a protein and the Gr66a reporter (see Fig. 4A). The Gr expression in these latter neurons, which are situated between the mouth and digestive system, suggest that flies may evaluate food quality after it is ingested and before it is transmitted to the gut. We introduced the Gr66a-GAL4;UAS-mCD8::GFP transgenes into the Gr93a3 background and found that the anti-GFP signals were indistinguishable between wild-type and Gr93a3 flies (see Fig. 4A). These data, combined with the observation that the anti-GFP signal was present in Gr93a3 labella expressing the mCD8::GFP reporter (see Fig. 4A), indicate that the Gr93a3 mutation did not result in loss of the GRNs that express Gr93a and Gr66a.
Fig. 4.
Expression of the Gr93a protein in the labellum and pharynx. (A) Coexpresson of Gr93a in Gr66a-expressing GRNs. GFP was expressed using the UAS-mCD8::GFP and the Gr66a-GAL4 transgenes (anti-Gr93a antibodies, red; anti-GFP antibodies, green). The right panels show the merged anti-Gr93a and anti-Gr66a signals. The genotypes and tissues are indicated to the left side. Examination of multiple stacks of confocal optical sections indicates that the Gr93a protein and the Gr66a reporter are coexpressed (Fig. S4). Expression of the Gr66a reporter in Gr93a3 labella (Second row) indicates that the Gr66a- and Gr93a-expressing GRNs are present in Gr93a3. Gr93a is expressed in the pharynx in Gr66a-expressing GRNs (Third row, left), but not in Gr93a3 pharynx (Fourth row, left). (B) The Gr93a::Myc protein was expressed in flies harboring the Gr66a-GAL4 and UAS-Gr93a::Myc transgenes. Labella were stained (anti-Gr93a, red; anti-Myc, green) and the signals were detected by confocal microscopy. The merged images are shown to the right.
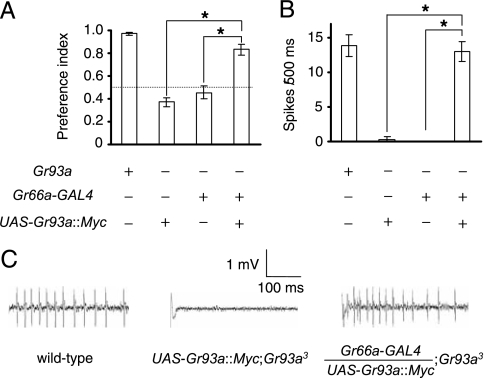
To address whether Gr66a and Gr93a both function in the same GRNs, we first tested whether the behavioral and electrophysiological deficits in Gr93a-mutant animals were rescued by expression of a wild-type Gr93a transgene (UAS-Gr93a::Myc) under the control of the Gr66a promoter (Gr66a-GAL4). We found that introduction of these transgenes into Gr93a mutant flies (see Fig. 4B) significantly suppressed the impairment in caffeine avoidance (Fig. 5A; Table S6) and caffeine-induced action potentials (Fig. 5 B and C; Table S7). Because Gr66a and Gr93a are both required for caffeine avoidance, it is possible that they are sufficient for detecting caffeine in GRNs. However, coexpression of the 2 Grs in Gr5a-expressing GRNs (Fig. S5) did not elicit behavioral or electrophysiological responses to caffeine (Fig. S6). The Gr93a and Gr66a::Myc proteins localized normally in Gr66a and Gr93a mutants, respectively, indicating that 1 receptor did not affect trafficking of the other receptor (see Fig. S3).
Fig. 5.
Rescue of the caffeine sensation defect in Gr93a3 using the Gr66a-GAL4 and UAS-Gr93a::Myc transgenes. (A) Binary food-choice assay using either Gr93a+ flies or Gr93a3 mutant flies harboring the Gr66a-GAL4 and/or UAS-Gr93a::Myc transgenes. (B) Average frequencies of action potentials (spikes/500 ms) induced by presentation of 10-mM caffeine using the indicated fly lines. (C) Sample tip recordings. The asterisks indicate significant differences (P < 0.01) from wild-type. See Tables S6 and S7 for statistics.
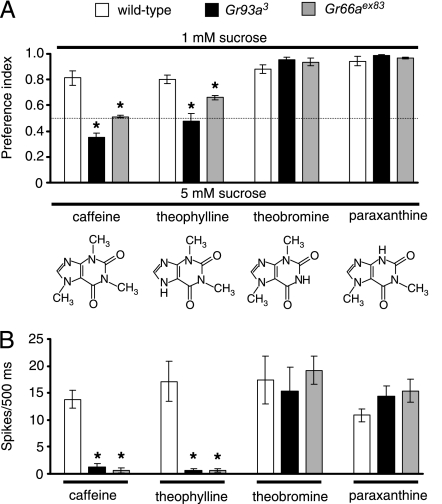
To test further whether there are identical requirements for Gr66a and Gr93a for Drosophila taste, we assayed the responses of the mutant flies to a variety of methylxanthines related to caffeine. Wild-type flies avoid all methylxanthines tested. In addition to caffeine (1,3,7-trimenthylxanthine), this includes theophylline (1,3-dimethylxanthine) and theobromine (3,7-dimethyxanthine), which are produced in tea and cocoa, respectively, and paraxanthine (1,7-dimethyxanthine) (Fig. 6A). Presentation of these methylxanthines also produced action potentials in the GRNs (Fig. 6B). As we have reported previously, Gr66aex83 mutant flies displayed deficits in the behavioral and electrophysiological responses to caffeine and theophylline, but not theobromine (see Fig. 6) (16). However, in the present study, we found that paraxanthine induced action potentials and aversive behavior in Gr66aex83 (see Fig. 6). Of significance here, we found that these 4 methyxanthines elicited the same behavioral and electrophysiological profiles in the Gr93a and Gr66a mutants (see Fig. 6; Tables S8 and S9).
Fig. 6.
Gr93a and Gr66a are required for detection of the same methylxanthines. (A) Binary food-choice assays were conducted with 1-mM sucrose versus 5-mM sucrose plus 6-mM of each methylxanthine. (B) The average frequencies of action potentials (spikes/500 ms) were collected upon application of 10-mM of each methylxanthine. Error bars represent SEMs. The asterisks indicate significant differences from wild type using the unpaired Student's t test. See Tables S8 and S9 for statistics.
Discussion
The ability to avoid ingestion of noxious botanical compounds is crucial for insect survival. However, before the current study, only 1 receptor, Gr66a, which is essential for the caffeine response, was associated with a specific bitter tastant (16). Because Gr66a appears to be insufficient for generating a caffeine response, a critical question concerns the molecular complexity of the receptors that operate in detecting deleterious nonvolatile substances.
We propose that Gr93a might be a coreceptor required in concert with Gr66a for sensing caffeine. In support of this conclusion, mutation of Gr93a and Gr66a results in identical phenotypes. Both Grs are required for avoiding caffeine and for caffeine-induced action potentials in GRNs, but not for the responses to any other unrelated compound tested. Moreover, the 2 Grs are expressed in the same GRNs and function in the same cells, as introduction of a wild-type Gr93a transgene under control of the Gr66a transcriptional control rescued the Gr93a-mutant phenotype. However, unlike the odorant receptor (Or), Or83b, which is required for the spatial localization of other Ors (19), neither Gr66a nor Gr93a appeared to impact of the cellular distribution of the other receptor.
We suggest that the composition of the Drosophila taste receptors is more complex than for other types of chemosensory receptors in flies or in mammals. In mammals, taste receptors are either homo- or heterodimers, while olfactory receptors are homomeric proteins (20, 21). Drosophila Ors appear to heterodimers comprised of Or83b in combination with one additional Or (19, 22). The Drosophila CO2 receptor is a heterodimer consisting of Gr21a and Gr63a (23, 24). Misexpression of these 2 receptors is sufficient to confer CO2 sensitivity to neurons that do not normally respond to CO2 (23, 24). In contrast, cointroduction of the 2 taste receptors Gr66a and Gr93a in Gr5a-expressing GRNs does not confer caffeine sensitivity to these cells. Although we cannot exclude that there are critical downstream-signaling molecules missing in Gr5a-expressing GRNs, we suggest that functional bitter taste receptors are not heterodimers, but are higher order assemblies consisting of additional subunits. Because caffeine and many other botanical compounds function as naturally occurring insect repellents and pesticides, the identification of the composition of the Grs that are sufficient for avoiding toxic chemicals offers potential for devising mechanisms for reducing interactions between insect pests and plants.
Methods
Genetics, Fly Stocks, and Constructs.
The f01688, EY11817, and UAS-mCD8::GFP fly lines were from the Exelixis Collection at the Harvard Medical School or from the Bloomington Stock Center. The Gr66a-GAL4 line was provided by H. Amrein (11). The f01688 Piggybac line was semilethal; however, after 5 out-crossings to w1118, the line (Gr93a1) was fertile and viable. EY11817 inserted in the 3′ untranslated region 670 base pairs 3′ to the Gr93a stop codon. Caffeine avoidance in EY11817 was similar to wild-type: PI was 0.86 ± 0.04 and 0.91 ± 0.02, respectively. We mobilized the EY11817 P element by crossing them to a genetic source of transposase: w; Sp/Cyo; ry Sb1 P{ry[+t7.2] = Δ2–3}99B/TM6B. The imprecise excision deleted the entire Gr93a coding region (Gr93a2). To identify the Gr93a2 deletion, we screened ≈200 lines by PCR using the following primers: a, 5′-AAATTTAATGGCGATACTTGTT-3′; and b, 5′-ACATATTGTAGCTACCTCACCC-3′. Gr93a2 had a 2,357 base-pair deletion extending from 365 nucleotides upstream of the Gr93a start codon to the P-element-inserted site. This deletion removed the entire Gr93a coding region. The wild-type control for all experiments was w1118.
Homologous Recombination and Generation of Transgenic Flies.
We generated Gr93a3 by ends-out homologous recombination (17). We subcloned 2 PCR-amplified genomic fragments (3.16 and 3.03 kb) into a modified pw35 vector containing the yeast GAL4 gene, obtained nonhomologous insertions by germline transformation, and generated homologous recombinants as described (17). The Gr93a3 mutation was verified by PCR and out-crossed 5 times to w1118.
To obtain the UAS-Gr93a::Myc transgene, we amplified the full-length Gr93a cDNA by RT-PCR using fly head mRNA (Stratagene), and inserted the DNA coding for a Myc tag in place of the Gr93a stop codon. The DNA sequence of the construct was verified by sequencing and subcloned into the pUAST vector.
RT-PCR Analyses of Gr93a1/Gr93a2.
Whole-fly mRNA from wild-type and Gr93a1/Gr93a2 flies were extracted (Stratagene) and AMV reverse transcriptase was used to generate cDNAs (Promega). For quantitative RT-PCR, we used the following primers: 5′-TGGGATAAGAGTGTTGAAA-3′ and 5′-CTGTAAGTAGCTTAATCA-3′ with tubulin primers as an internal control.
Chemicals.
Caffeine, quinine hydrochloride, denatonium benzoate, papaverine hydrochloride, strychnine nitrate salt, sucrose, glucose, maltose, fructose, tricholine citrate, and sulforhodamine B were from Sigma-Aldrich. Lobeline hydrochloride, and trehalose were from Fluka, and berberine sulfate trihydrate, and brilliant blue FCF were from Wako Chemical.
Immunohistochemistry and Generation of Anti-Gr93a Antibodies.
Antibody stainings were performed as described (25). Briefly, we placed freshly dissected tissue (e.g., labella) from 3- to 7-day-old flies into wells of 48-well cell-culture cluster plates (Costar Corp.) maintained on ice and containing 940 μl of Fix Buffer (0.1M Pipes pH6.9, 1 mM EGTA, 1% TritonX-100, 2 mM MgSO4, 150 mM NaCl) and 60 μl of 37% formaldehyde. The formaldehyde was added to the wells containing the Fix Buffer and mixed immediately before adding the tissue. We transferred as many dissected tissues into the Fix Buffer-containing formaldehyde as we could dissect in 30 min. The tissues were incubated for another 30 min, washed with Wash Buffer (1× PBS, 0.2% saponin) and blocked for 4 to 8 h at 4 °C with 1 ml of Blocking Buffer (1× PBS, 0.1% saponin, 5 mg/ml BSA). The tissues were transferred and incubated into the primary antibody mixture overnight at 4 °C, washed 3 times with Wash Buffer for 15 min each, incubated in the secondary antibody mixture for 4 h at 4 °C, and washed 3 times with Wash Buffer for 15 min each on ice. The tissues were transferred into 1.25× PDA Dilution Buffer (37.5% Glycerol, 187.5 mM NaCl, 62.5 mM Tris pH8.8), incubated >1 h at 4 °C, and mounted and analyzed using a Carl Zeiss confocal microscope.
We generated the polyclonal rabbit anti-Gr93a antibodies using the following peptide: [KLH]-CIESQDERYRNTKYRR-NH2 (Peptron). The antibodies were preabsorbed using fly embryos and used for staining at a 1:1,000 dilution. Other antibodies were used at the following dilutions: mouse anti-Myc (1:200, Santa Cruz), rabbit anti-GFP (1:500, Santa Cruz), mouse anti-GFP (1:1,000, Invitrogen–Molecular Probes), goat anti-mouse secondary antibodies (Alexa 488; 1:200, Invitrogen–Molecular Probes) and goat anti-rabbit secondary antibodies (Alexa 568; 1:200, Invitrogen–Molecular Probes).
Behavioral Assays.
The binary food-choice assays were performed as described (16). In short, we starved the flies (3–7 days old) for 18 h on 1% agarose. We then placed the flies into 72-well microtiter dishes, which contained wells filled with 1% agarose plus either red dye (sulforhodamine B, 0.2 mg/ml, Sigma-Aldrich) or blue dye (brilliant blue FCF, 0.125 mg/ml, Wako Chemical). Either the red or blue dye mixtures contained aversive compounds (1 mM quinine, 0.3 mM denatonium, 0.1 mM berberine, 0.3 mM lobeline, 2 mM papaverine, or 0.5 mM strychnine) or sugars at the indicated concentrations. The flies were placed in the microtiter dishes for 90 min at room temperature (in the dark and in a humidified chamber), and the numbers of the flies that were blue (NB), red (NR), or purple (NP) were ascertained by inspection of the abdomen color. The PIs were determined using the following equation: PI = (NB + 0.5 NP)/NTotal or (NR + 0.5 NP)/NTotal. The dyes did not influence the preferences. Every experiment was conducted ≥4 times.
Electrophysiology.
To determine the frequencies of action potentials in response to tastants, we performed tip recordings on taste sensilla as described (16). Briefly, we immobilized 3-day-old flies by puncturing the thorax with a glass capillary filled with Ringer's solution and sliding it into the head. This electrode also served as a reference electrode. We stimulated the labellar bristles with the recording electrode (10–20 μm tip diameter) using the indicated concentrations of aversive compounds and either 1-mM KCl or 50-mM sucrose plus 30-mM tricholine citrate as the electrolyte. We performed the recordings on S6 sensilla on the labial palp. We connected the recording electrode to a preamplifier (Taste PROBE, Syntech), amplified the signals 10× using a signal connection interface box (Syntech) in conjunction with a 100 to 3,000 Hz band pass filter, and recorded action potentials at a 12 kHz sampling rate. We performed all recordings 6 to 12 times.
Supplementary Material
Acknowledgments.
This work was partially supported by Postdoctoral Fellowship 2006-352-C00065 from the Korea Research Foundation (to Y.L.) and by National Institute on Deafness and Other Communication Disorders Grant DC007864 (to C.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811744106/DCSupplemental.
References
- 1.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA 100 Suppl. 2003;2:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- 3.Scott K, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 4.Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 5.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 6.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 7.Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- 8.Chyb S, Dahanukar A, Wickens A, Carlson JR. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc Natl Acad Sci USA. 2003;100:14526–14530. doi: 10.1073/pnas.2135339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno K, et al. Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Curr Biol. 2001;11:1451–1455. doi: 10.1016/s0960-9822(01)00450-x. [DOI] [PubMed] [Google Scholar]
- 10.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 11.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci USA. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon SJ, Kottgen M, Jiao Y, Xu H, Montell C. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meunier N, Marion-Poll F, Rospars JP, Tanimura T. Peripheral coding of bitter taste in Drosophila. J Neurobiol. 2003;56:139–152. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- 19.Larsson MC, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Imai T, Sakano H. Odorant receptor-mediated signaling in the mouse. Curr Opin Neurobiol. 2008;18:251–260. doi: 10.1016/j.conb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Scott K. Taste recognition: food for thought. Neuron. 2005;48:455–464. doi: 10.1016/j.neuron.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Neuhaus EM, et al. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- 23.Jones WD, Cayirlioglu P, Grunwald Kadow I, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 24.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci USA. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y, et al. Loss of spastic paraplegia gene atlastin induces age-dependent death of dopaminergic neurons in Drosophila. Neurobiol Aging. 2008;29:84–94. doi: 10.1016/j.neurobiolaging.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.