Abstract
Protein folding often competes with intermolecular aggregation, which in most cases irreversibly impairs protein function, as exemplified by the formation of inclusion bodies. Although it has been empirically determined that some proteins tend to aggregate, the relationship between the protein aggregation propensities and the primary sequences remains poorly understood. Here, we individually synthesized the entire ensemble of Escherichia coli proteins by using an in vitro reconstituted translation system and analyzed the aggregation propensities. Because the reconstituted translation system is chaperone-free, we could evaluate the inherent aggregation propensities of thousands of proteins in a translation-coupled manner. A histogram of the solubilities, based on data from 3,173 translated proteins, revealed a clear bimodal distribution, indicating that the aggregation propensities are not evenly distributed across a continuum. Instead, the proteins can be categorized into 2 groups, soluble and aggregation-prone proteins. The aggregation propensity is most prominently correlated with the structural classification of proteins, implying that the prediction of aggregation propensity requires structural information about the protein.
Keywords: cell-free translation, protein aggregation, protein folding
The unique native structure of a protein is encoded in its amino acid sequence (1). However, protein folding is often hampered by protein aggregation, which is generally prevented by a variety of chaperone proteins in the cell (2). Despite the presence of chaperones, a certain level of aggregation still occurs in cells. For example, aggregates commonly form upon the heterologous expression of recombinant proteins, as exemplified by the formation of inclusion bodies (3). In special cases, protein aggregation could lead to the formation of ordered aggregates, known as amyloid fibrils, which are closely associated with many severe neurodegenerative diseases in mammals (4, 5).
Understanding the mechanism underlying aggregate formation is required for the development of a wide variety of protein sciences. However, the relationship between the protein aggregation propensities and the primary sequences remains poorly understood. Because it is empirically known that some proteins tend to aggregate, several groups systematically studied the effects of mutations in on proteins of interest that caused the formation of insoluble aggregates (6–9). Subsequently, the information on the mutations has been used to build prediction tools for protein aggregation, and most of them were developed for amyloid formation (10–13). However, the application of the prediction tools has currently been restricted to a narrow range of proteins because of the lack of sufficient data on the aggregation. To overcome this limitation, a database on the propensity of a given protein to aggregate would be an invaluable resource to understand the nature of protein aggregation.
In our previous studies, we developed a method to evaluate the solubility of individual proteins using a cell-free translation system (14–16). The cell-free translation system, named PURE, is a reconstituted system that only contains the essential Escherichia coli factors responsible for protein synthesis (17, 18). In this study, we performed a comprehensive analysis, in which the complete E. coli ORF library (ASKA library) (19) was translated in the PURE system under the same conditions. Because the PURE system is chaperone-free (14, 17), we could evaluate the inherent aggregation propensities of thousands of proteins in a translation-coupled manner.
Results
Comprehensive Aggregation Analysis of the Entire Ensemble of E. coli Proteins Using an in Vitro-Reconstituted Translation System.
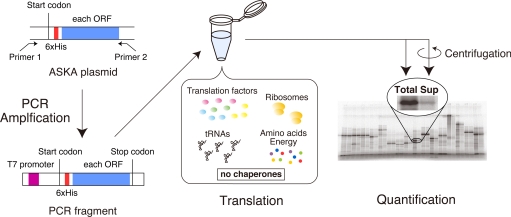
The ASKA library consists of all predicted ORFs of the E. coli genome, including membrane proteins (19). A total of 4,132 ORFs were individually amplified by PCR using a common primer set (Fig. 1) and then were used for protein synthesis in the PURE system at 37 °C for 60 min.
Fig. 1.
Schematic illustration of the experiment. Each ORF in the ASKA library, which has all of the E. coli ORFs, was amplified by PCR using 2 common primers to translate the gene in the cell-free translation system. The reconstituted cell-free translation system (the PURE system) contains no chaperones. After the 60-min translation, an aliquot of the translation mixture was centrifuged to obtain the soluble fraction. The uncentrifuged (Total) and supernatant (Sup) fractions were subjected to SDS/PAGE, and the translated products were quantified by autoradiography.
The [35S]methionine-labeled proteins were quantified after electrophoresis of the translation products. We successfully quantified ≈70% of the E. coli ORFs (3,173 proteins of 4,132). The remainder was not quantified, because of insufficient translation and trouble during the electrophoresis (translated proteins were stuck in the gel, several protein bands were detected, and so on). The unquantifiable group contained ≈60% of the inner membrane proteins (435 of 754), whereas >80% of the cytoplasmic proteins (2,277 of 2,688) were quantified [supporting information (SI) Fig. S1]. The yield of the quantified proteins was 33 μg/mL, on average, but ranged broadly from the detection limit to ≈100 μg/mL as the maximum (Fig. S2A), although we used common primers, which resulted in a common N-terminal flanking sequence in all of the ORFs, and performed the translation under the same conditions.
The propensity for protein aggregation was examined by a centrifugation assay (14, 15). An aliquot of the translation mixture was centrifuged. The proportion of the supernatant fraction, which was obtained after the centrifugation of the translation mixture, to the uncentrifuged total protein was defined as the solubility, the index of the aggregation propensity (a representative experiment is shown in Fig. 1). The SD of the solubilities was 8.8% on average, and the highest SD was 25%, based on data from 33 randomly chosen proteins (Fig. S3).
Bimodal Distribution of Protein Solubility.
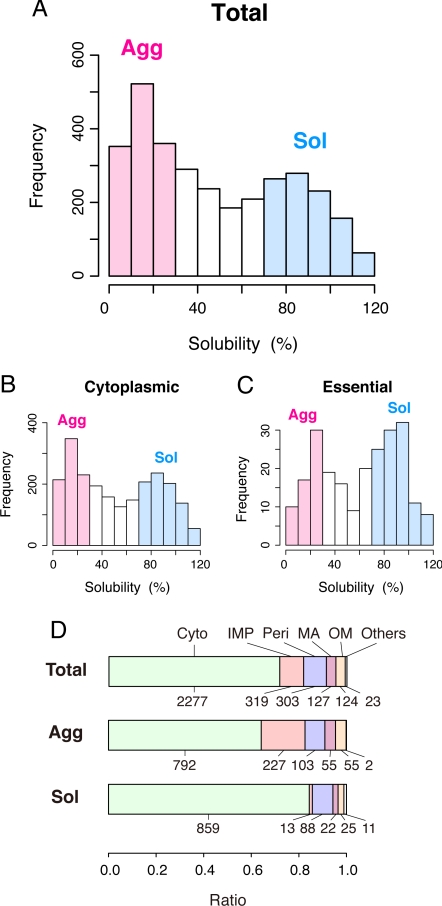
A histogram of the individual solubilities, based on data from 3,173 translated proteins, showed a clear bimodal, rather than normal Gaussian, distribution (Fig. 2A), indicating that the aggregation propensities are not evenly distributed across a continuum. Subtraction of the predicted integral membrane proteins (IMPs) from the data did not change the bimodal distribution (Fig. 2B), suggesting that the cytoplasmic proteins can be categorized into an aggregation-prone group and a highly soluble one. To elucidate which characteristics of the protein influence this bimodality, we compared a variety of protein properties in the aggregation-prone (Agg, defined as <30%) and highly soluble (Sol, defined as >70%) groups. Because all of the translated proteins contain the common short flanking peptides at the N and C termini, including the N-terminal 6× histidine tag, the solubilities of 120 randomly chosen cytoplasmic proteins were analyzed with their endogenous ORF sequences, without the additional flanking peptides (Fig. S4). Only 2 proteins shifted from the Agg to the Sol group, indicating that the influence of the common N-terminal extension with the histidine tag is only marginal.
Fig. 2.
Solubility distribution for quantified proteins. (A) Histogram of solubility for the 3,173 quantified proteins. The proteins with solubilities <30% and >70% were defined as the aggregation-prone (Agg, colored pink) and soluble (Sol, colored blue) groups, respectively. (B) Histogram of solubility for 2,277 predicted cytoplasmic proteins. (C) Histogram of solubility for essential proteins. (D) The ratio of subcellular location (predicted) in all quantified (Total), Agg, and Sol groups. Cyto, cytoplasmic proteins; IMP, integral membrane proteins; Peri, periplasmic proteins; MA, membrane-anchored proteins; OM, outer membrane lipoproteins and β-barrel proteins.
One might expect that the bimodal distribution in the histogram is simply due to the difference in the synthesized yield of proteins, because it has been generally believed that higher protein concentrations generate more protein aggregates. However, this is not the case, because there is no apparent correlation between the solubilities and the yields (Fig. S2B).
We then extracted the essential proteins for cell viability (Fig. 2C). The bimodality in the distribution was the same as those in the total and cytoplasmic protein groups (Fig. 2 A and B), but we found that the essential proteins tended to be enriched in the high solubility group (Fig. 2C). This result suggests that the essential proteins might have evolved to be soluble for their irreplaceable properties. In addition to the essentiality, we categorized the data according to the protein functions and ranked them with the solubilities (Fig. S5A). We found that the solubilities depend strongly on the functions. For example, the Structural component group, which is mainly composed of ribosomal proteins, and the Factor group, which includes transcription or translation factors, chaperones, and proteases, showed a strong bias to the high-solubility group. In contrast, the proteins in the Transporter group tended to be aggregation-prone. Regarding the oligomeric states of the proteins, preliminary analysis shows that heterooligomers seem to be aggregation-prone (Fig. S5B), although we cannot say the tendency is statistically significant because of the incomplete database on the oligomeric states.
Regarding the subcellular locations, the ratio of IMPs in the Agg group (227 of 1,234) was much larger than that in the Sol group (13 of 1,018) (Fig. 2D). Although the IMPs translated under membrane-free conditions were expected to form insoluble aggregates, it is noteworthy that some portion of the IMPs was soluble. There was no remarkable difference in the other locations. Because the subtraction of IMPs from the histogram did not change the bimodality (Fig. 2B), further analyses were performed only with the cytoplasmic proteins.
Relationship Between Solubility and Physicochemical Properties.
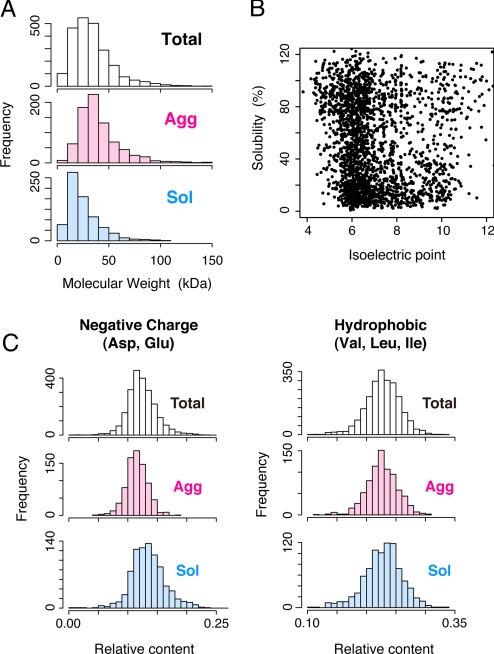
Next, we compared the physicochemical properties of the proteins, such as the molecular mass, the deduced isoelectric points (pI), and the amino acid residue content, to address the relationship between solubility and amino acid sequence (Fig. 3). The distribution of molecular mass in the Sol group was shifted to smaller sizes compared with the total histogram, whereas the Agg group was slightly larger than the total distribution (P < 0.01, Fig. 3A and Table S1). Regarding the isoelectric points, we observed an enrichment of low-pI (5–7) proteins in the high-solubility distribution, whereas the aggregation-prone proteins showed a somewhat broader pI distribution (ranging from 5 to 10) (Fig. 3B). We then tested whether the amino acid residue content affected the solubility and found that higher contents of negatively charged residues (Asp and Glu) tended to be soluble (Fig. 3C and Table S1). Higher contents of aromatic residues (Phe, Tyr, and Trp) were slightly biased to be aggregation-prone (Fig. S6 and Table S1). The differences in the histograms suggested that Asp/Glu-rich and/or aromatic-poor proteins tend to be soluble. In contrast, no significant difference was observed in the contents of hydrophobic residues (Val, Leu, and Ile) and positively charged residues (Lys, Arg, and His) (Fig. 3C, Fig. S6, and Table S1). Because it has been believed that the hydrophobic interaction is a critical driving force in aggregate formation, the lack of an apparent correlation in the hydrophobic residue content was unexpected. Other attempts to detect a bias between the solubility and the hydrophobicity, including a well-known hydropathy plot analysis (20, 21), which shows clusters of hydrophobic residues in the primary amino acid sequences, or several hydrophobic-polar alternates analyses also failed. We note that Gln/Asn-rich sequences including polyglutamine repeats, which tend to form amyloid fibrils, are very rare in the E. coli ORFs (22).
Fig. 3.
Correlation between solubility and physicochemical properties. (A) Histograms of molecular mass in the Total, Agg, and Sol groups. (B) Scatter plot of solubility versus isoelectric point. (C) Histograms of the relative contents of negatively charged residues (Asp and Glu) (Left) and hydrophobic residues (Val, Leu and Ile) (Right) in the Total, Agg, and Sol groups.
We subsequently conducted several analyses related to the secondary structures. We predicted the secondary structure contents by using popular prediction methods, such as Chou–Fasman (23) and PSIPRED (24, 25). However, we could not detect a notable correlation between the predicted secondary structure content and the solubility (Fig. S7 for the PSIPRED analysis.
Correlation Between Solubility and Tertiary Structure.
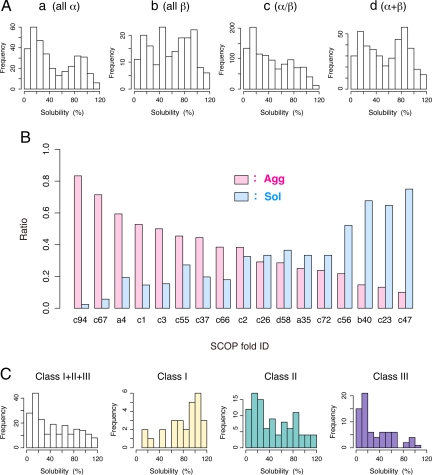
To address the correlation between the solubilities and the tertiary structures, we compared the solubilities with the Structural Classification of Proteins (SCOP) database, which is a comprehensive ordering of all proteins with known structures, according to their evolutionary and structural relationships (26). The classification is based on hierarchical levels: class, fold, superfamily, and family. Superfamilies and families are defined as having a common fold if their proteins have the same major secondary structures in the same arrangement and with the same topological connections. Most of the folds are assigned to one of the following structural classes: all-α (SCOP class a), all-β (class b), α/β (class c), and α+β (class d). Besides the all-β (class b) proteins, the bimodality of the histograms was maintained, although the distribution of class c was slightly biased to aggregation-prone (Fig. 4A), roughly confirming that the secondary structures did not correlate with the aggregation propensities. We then categorized the SCOP folds into solubility groups and found that some of the SCOP folds were extremely biased toward their solubilities (Fig. 4B and Table S2). For example, in the periplasmic binding protein-like II fold (SCOP fold: c94) group, which is largely dominated by DNA-binding transcriptional regulator proteins, 83% of the members were low-solubility proteins (35 of 42 assigned proteins), whereas only 1 protein was in a soluble group (Table S2). Other low-solubility folds included PLP-dependent transferases fold (c67), DNA/RNA-binding 3-helical bundle fold (a4), TIM β/α-barrel fold (c1), and P-loop containing nucleoside triphosphate hydrolases (c37) (P < 0.01, Table S2). For the highly soluble folds, we assigned Flavodoxin-like fold (c23), OB-fold (b40), and Thioredoxin fold (c47) (P < 0.01, Table S2).
Fig. 4.
Correlation between solubility and tertiary structure. (A) Histograms of solubility in the SCOP classes. SCOP class abbreviations: all α proteins (a); all β proteins (b); α and β proteins (α/β) (c); α and β proteins (α+β) (d). (B) The ratio of the Agg and Sol proteins in each SCOP fold. Details of each fold and the assigned number of proteins with statistical significance (P values) in each fold are described in Table S2. (C) Histograms of solubility for the GroEL substrate proteins. The classification of the substrates is according to Kerner et al. (27), in which Classes I, II, and III are spontaneously foldable, chaperone-dependent (but partially GroEL-dependent) and obligate GroEL/ES-dependent substrates, respectively.
In the above analyses, we noticed that the low-solubility folds (c1 and a4) were known to be enriched in the obligate chaperonin GroEL substrates (the so-called Class III substrates) (27). Kerner et al. (27) have identified ≈250 GroEL interactors and categorized them into 3 classes (I, II, and III), based on a quantitative proteomic analysis. The Class I and II substrates are only partially chaperonin dependent, whereas ≈85 Class III substrates are considered as obligate substrates that engage >75% of the GroEL capacity. The solubilities of the GroEL substrates are shown in the histograms (Fig. 4C). Notably, ≈60% of the Class III substrates were in the Agg group (44 of 74), indicating that the Class III substrates are extremely aggregation-prone. In contrast, the Class I substrates tended to be soluble. This analysis suggests that GroEL preferentially binds the aggregation-prone proteins in vivo.
Attempts to Predict the Aggregation Propensity.
Finally, we tested whether our data can be applicable to several recently developed web tools to predict protein aggregation. We chose the TANGO (10), AGGRESCAN (12), and PASTA programs (13). However, none of the tools tested extracted a notable positive correlation between our datasets and the predicted results (Fig. S8), probably because the algorithms used in those programs basically relied on data from amyloid aggregates in eukaryotes. Our attempt to predict the solubilities by using a support vector machine (SVM) algorithm (28), with the parameters including molecular mass, pI, and amino acid content, resulted in ≈80% accuracy. The algorithm provides a reasonable prediction but is not completely satisfactory. For more accurate prediction, we should incorporate information about the tertiary structure, because the solubilities depended strongly on the SCOP folds. A combination of 3-dimensional structure prediction with other physicochemical properties might improve the solubility prediction.
Discussion
In this article, we conducted a global aggregation analysis of whole E. coli proteins, coupled with a reconstituted cell-free translation system [the PURE system (17)]. The aggregation propensities of >3 thousands of proteins, which were evaluated under the chaperone-free condition, showed that the proteins were categorized into 2 groups, soluble and aggregation-prone. In addition, statistical analysis revealed that some structural classes of proteins were strongly biased to the aggregation propensities.
Several caveats should be stated regarding the interpretation of our data. First, because our aggregation analysis completely depends on the centrifugation, other conditions like a higher-speed centrifugation might cause a change in the shape of the histogram. Thus, there is a possibility that soluble fractions might include oligomeric assemblies that are aggregation precursors. This is of particular interest because recent advances on amyloid-forming proteins have revealed that soluble oligomeric species of some amyloid proteins are toxic to the cell (29). Second, even a soluble protein does not always have the native structure. Some might be soluble in an unstructured state. Indeed, we have previously shown that a fraction of the proteins produced in the chaperone-free PURE system are soluble but not functional (14, 15). The addition of chaperones, such as the DnaK system or GroEL/ES, helps the proteins to reach their functional native states (14, 15). Third, the centrifugation assay cannot discriminate amorphous aggregates from structured aggregates, such as amyloid fibrils. Recent study by Wang et al. (30), which showed that bacterial inclusion bodies can contain amyloid-like structures, raises the possibility of the amyloid-like structures in insoluble aggregates in our assay, although it has been assumed that bacteria have few amyloid-forming proteins (e.g., ref. 22).
Nevertheless, the data presented here provide a unique viewpoint for protein science. The most important finding in our analyses is that, in terms of their solubility, proteins belong to 2 subgroups. Because the proteins tested are basically soluble in the cell, mainly because of the assistance of chaperones, the bimodal property was revealed by the use of the PURE system, a reconstituted cell-free translation system that lacks chaperones. This hidden bimodal solubility of the proteins prompted us to imagine the evolution of protein folding in the cell: The aggregation-prone groups might have evolved to fold correctly only with the aid of chaperones. In support of this concept, we found that the obligate GroEL substrates are aggregation-prone. In this context, the presence of the aggregation-prone group might guarantee a hypothetical buffering capacity of chaperones during evolution, by releasing the genetic variation under certain conditions, as has been suggested in the case of Hsp90 in eukaryotic organisms (31, 32).
Another main finding is that some of the SCOP folds are strongly biased to the aggregation propensity. In particular, the presence of aggregation-prone folds is apparently paradoxical because aggregates formation should occur before the completion of folding. The apparent correlation between some SCOP folds and the aggregation tendency suggests that folding intermediates have 2 classes, aggregation-prone and soluble. Then, what is the difference between the aggregation-prone and soluble intermediates? Regarding this point, the competition between the correct folding and the aggregates formation, known as kinetic partitioning in the protein folding (33, 34), should be considered. Because the kinetic partitioning is closely related to folding kinetics itself, understanding the mechanism of aggregate formation would require a detailed mechanism of protein folding. Our approach using the PURE system will have a potential to investigate a global analysis of the folding kinetics, providing a unique insight into the kinetic partitioning.
Finally, our approach using the PURE system provides an invaluable resource for a broad range of protein sciences, including protein folding prediction, protein design, folding coupled with translation, and the role of chaperones with nascent proteins. In addition, the comprehensive cell-free synthesis of all proteins encoded in a genome, termed a reconstituted proteome, paves the way for the construction of an on-demand protein bank system, which would useful for a variety of protein research, including emerging synthetic biology (35) in the future.
Materials and Methods
E. coli ORF Library.
The ASKA library (19, 36) was originally provided by Hirotada Mori (Nara Institute of Science and Technology, Nara, Japan), and the purified ASKA library (37) plasmid set was kindly provided by Tomoaki Matsuura (Osaka University, Osaka, Japan). A total of 4,132 ORFs were individually amplified by PCR using the ASKA library plasmids as templates. The sequences of the common primers were as follows: primer1, 5′-GGCCTAATACGACTCACTATAGGAGAAATCATAAAAAATTTATTTGCTTTGTGAGCGG-3′, and primer2, 5′-GTTATTGCTCAGCGGTTAGCGGCCGCATAGGCC-3′. Primer1 contains the T7 promoter (italicized) for expression by the PURE system, and primer2 contains the UAA stop codon (italicized).
Cell-Free Protein Synthesis and Protein Aggregation Assay.
The method for the evaluation of the protein aggregation propensity was based on a previously reported method (14–16), with several modifications. Each ORF was translated with N- and C-flanking regions, with the following amino acid sequences: N-, MRGSHHHHHHTDPALRA and C-, GLCGR. The transcription–translation-coupled PURE system (17, 18) reaction, including [35S]methionine, was performed at 37 °C for 1 h. After the protein synthesis, an aliquot was withdrawn as the total fraction, and the remainder was centrifuged at 21,600 × g for 30 min. Both the total and supernatant fractions were separated by SDS/PAGE, and the band intensities were quantified by autoradiography. The ratio of the supernatant to the total protein was defined as the solubility, the index of protein aggregation tendency.
Data Analyses.
All analyses, except for those described in Fig. 2 A, C, and D and Figs. S1 and S2, were performed with quantified cytoplasmic proteins. The amino acid sequence, subcellular location, type of gene product, and SCOP fold information was obtained from GenoBase (http://ecoli.naist.jp/GB6/search.jsp). The SCOP fold annotation in GenoBase was based on the SUPERFAMILY database (36, 38). The information on essential genes was obtained from the PEC database (39). The information on secondary structure prediction [PSIPRED (24, 25)] was obtained from the GTOP database (40). Molecular masses were calculated from the deduced amino acid sequences. Estimation of pI values was accomplished by using a web tool (http://isoelectric.ovh.org/) (41). For the prediction of protein aggregation from the amino acid sequence, 3 programs [TANGO (10), PASTA (13), and AGGRESCAN (12)] were obtained from their web sites.
Prediction by SVM Algorithm.
SVM (28) performance was analyzed with the Agg and Sol proteins (1,599 samples). The SVM classifier was trained with 1,000 randomly chosen samples with molecular mass, pI values, and ratios of each amino acid content. The prediction accuracy was calculated by the other 599 samples. The calculation was performed by using the KSVM library in the kernlab package with R software.
Supplementary Material
Acknowledgments.
We thank Hirotada Mori and Tomoaki Matsuura for the gift of the ASKA library plasmid set and Yoshihiro Shimizu and Takashi Kanamori for technical advice and useful suggestions. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Area 17049009, 19037007, and 19058002 (to H.T.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811922106/DCSupplemental.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: Fom nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 3.Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006;24:179–185. doi: 10.1016/j.tibtech.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, et al. Kinetic partitioning of protein folding and aggregation. Nat Struct Biol. 2002;9:137–143. doi: 10.1038/nsb752. [DOI] [PubMed] [Google Scholar]
- 7.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 8.Williams AD, et al. Mapping abeta amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 9.de Groot NS, Aviles FX, Vendrell J, Ventura S. Mutagenesis of the central hydrophobic cluster in Abeta42 Alzheimer's peptide. Side-chain properties correlate with aggregation propensities. FEBS J. 2006;273:658–668. doi: 10.1111/j.1742-4658.2005.05102.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 11.Tartaglia GG, Cavalli A, Pellarin R, Caflisch A. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Protein Sci. 2005;14:2723–2734. doi: 10.1110/ps.051471205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conchillo-Sole O, et al. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trovato A, Seno F, Tosatto SC. The PASTA server for protein aggregation prediction. Protein Eng Des Sel. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 14.Ying BW, Taguchi H, Ueda H, Ueda T. Chaperone-assisted folding of a single-chain antibody in a reconstituted translation system. Biochem Biophys Res Commun. 2004;320:1359–1364. doi: 10.1016/j.bbrc.2004.06.095. [DOI] [PubMed] [Google Scholar]
- 15.Ying BW, Taguchi H, Kondo M, Ueda T. Co-translational involvement of the chaperonin GroEL in the folding of newly translated polypeptides. J Biol Chem. 2005;280:12035–12040. doi: 10.1074/jbc.M500364200. [DOI] [PubMed] [Google Scholar]
- 16.Ying BW, Taguchi H, Ueda T. Co-translational binding of GroEL to nascent polypeptides is followed by post-translational encapsulation by GroES to mediate protein folding. J Biol Chem. 2006;281:21813–21819. doi: 10.1074/jbc.M603091200. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 20.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 24.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 25.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 27.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 29.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6:e195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 32.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 33.Jaenicke R. Folding and association versus misfolding and aggregation of proteins. Philos Trans R Soc London Ser B. 1995;348:97–105. doi: 10.1098/rstb.1995.0050. [DOI] [PubMed] [Google Scholar]
- 34.King J, Haase-Pettingell C, Robinson AS, Speed M, Mitraki A. Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 1996;10:57–66. doi: 10.1096/fasebj.10.1.8566549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Channon K, Bromley EH, Woolfson DN. Synthetic biology through biomolecular design and engineering. Curr Opin Struct Biol. 2008;18:491–498. doi: 10.1016/j.sbi.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Riley M, et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazuta Y, et al. Comprehensive analysis of the effects of Escherichia coli ORFs on protein translation reaction. Mol Cell Proteomics. 2008;7:1530–1540. doi: 10.1074/mcp.M800051-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: Additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato J, Hashimoto M. Construction of consecutive deletions of the Escherichia coli chromosome. Mol Syst Biol. 2007;3:132. doi: 10.1038/msb4100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawabata T, et al. GTOP: A database of protein structures predicted from genome sequences. Nucleic Acids Res. 2002;30:294–298. doi: 10.1093/nar/30.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sillero A, Maldonado A. Isoelectric point determination of proteins and other macromolecules: Oscillating method. Comput Biol Med. 2006;36:157–166. doi: 10.1016/j.compbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.