Abstract
Phytohormones have essential roles in coordinately regulating a large array of developmental processes. Studies have revealed that brassinosteroids (BRs) and abscisic acid (ABA) interact to regulate hundreds of expression in genes, governing many biological processes. However, whether their interaction is through modification or intersection of their primary signaling cascades, or by independent or parallel pathways remains a big mystery. Using biochemical and molecular markers of BR signaling and ABA biosynthetic mutants, we demonstrated that exogenous ABA rapidly inhibits BR signaling outputs as indicated by the phosphorylation status of BES1 and BR-responsive gene expression. Experiments using a bri1 null-allele, bri1-116, and analysis of subcellular localization of BKI1-YFP further revealed that the BR receptor complex is not required for ABA to act on BR signaling outputs. However, when the BR downstream signaling component BIN2 is inhibited by LiCl, ABA failed to inhibit BR signaling outputs. Also, using a set of ABA insensitive mutants, we found that regulation of ABA on the BR primary signaling pathway depends on the ABA early signaling components, ABI1 and ABI2. We propose that the signaling cascades of ABA and BR primarily cross-talk after BR perception, but before their transcriptional activation. This model provides a reasonable explanation for why a large proportion of BR-responsive genes are also regulated by ABA, and provides an insight into the molecular mechanisms by which BRs could interact with ABA.
Keywords: cross-talk, gene expression, phosphorylation, seed germination, signal transduction
Unlike animals, plants are sessile and need to constantly regulate their developmental and physiological processes to respond to various internal and external stimuli. Studies have revealed that many biological processes result from integrating multiple hormonal signals, and extensive cross-talk among different hormonal signaling pathways is present in plants (1, 2). Recently, a large set of microarray data verified that many genes are coregulated by multiple hormones, suggesting the importance of hormones to coordinately regulate biological processes in plants (3, 4). A few studies have also elucidated specific molecular mechanisms of hormonal cross-talk. They include the role of auxin and ethylene in regulating root meristem development (1), the antagonistic relationship between abscisic acid (ABA) and gibberellins (GAs) on seed dormancy and germination (5, 6), and integration of the primary signaling pathways of auxin and brassinosteroids (BRs) by auxin response factor 2 (ARF2) (7).
Studies have indicated that BRs and ABA can coregulate the expression of hundreds of genes (4), and they interact physiologically in controlling many developmental processes (5, 8–10). It is also well known that ABA is required to establish seed dormancy during embryo maturation and to inhibit seed germination (5), whereas BRs promote seed germination, likely through enhancing the embryo growth potential to antagonize the effect of ABA (5, 10, 11). In Arabidopsis, the BR biosynthetic mutant, det2-1, and the BR responsive mutant, bri1-1, are more sensitive to inhibition of ABA on seed germination than wild-type, although these mutants germinate well in normal growth condition (10). Also, microarray data showed that BRs and ABA regulate the expression of a greater number of genes than expected at random (3, 4). However, the underlying molecular mechanism of this interaction, and whether the cross-talk between BRs and ABA is through their primary signaling pathways, are unknown.
Genetic and biochemical studies have identified several key components in the signaling pathway of BRs. BRs are perceived by a cell surface receptor, BRI1, a leucine-rich-repeat receptor-like kinase (12). Direct binding of BR to a small region of the extracellular domain of BRI1 (12–15) induces phosphorylation of multiple sites and conformational change of its cytosolic domain (16, 17), and dissociation of BKI1, a negative regulator of BR signaling, from the plasma membrane (18). These events result in the activation of BRI1 and its association with another receptor kinase, BAK1 (19), or other substrates (20), which subsequently transduces the BR signal to downstream components through a yet unknown mechanism. Two downstream components, a GSK3-like protein kinase, BIN2 (21, 22), and a protein phosphatase, BSU1 (23), control the phosphorylation states of a family of plant-specific transcription factors, including BES1 (24) and BZR1 (25). Phosphorylated BES1 and BZR1 are less stable (24, 26), more cytoplasmically retained by 14-3-3 proteins (27), and weaker at DNA binding (28) than dephosphorylated forms. Thus, the phosphorylation status of BES1 and/or BZR1 has been used as a direct biochemical marker for BR signaling outputs (18). BZR1 and BES1 can bind directly to the promoter regions of the BR biosynthetic genes, CPD and DWF4, and inhibit their expression, forming a negative feedback loop (25, 29). Therefore, the transcript levels of CPD and DWF4 are excellent molecular markers, and have been widely used for determining the strength of BR signaling outputs (18).
The ABA signaling pathway appears to be more complicated than that of BRs, but several key components have been identified and studied. A number of PP2C family serine/threonine phophatases, including ABI1, ABI2, HGA1, and HAB1, apparently have essential roles in ABA signaling, and their gain-of-function mutations show significant ABA insensitive phenotypes in seed germination (30–33). Recently, some putative ABA receptors (34, 35), some protein kinase (36), and several transcription factors (37–39) involved in the ABA signaling pathway have been reported, but their targets are unknown, and their position in the ABA signaling pathway remains to be determined.
In this study, we used genetic and biochemical approaches, and systematically analyzed BR and ABA signaling mutants to investigate whether and how ABA regulates BR signaling outputs. The results demonstrate that ABA can rapidly inhibit BR signaling, as indicated by the enhanced phosphorylation of BES1 and regulation of BR-responsive gene expression. Using BR signaling mutants, we found that the effect of ABA on BR signaling does not depend on BR perception, but it relies on BIN2, a more downstream component of BR signaling. Using ABA signaling mutants, we found that the regulatory effect of ABA on BR signaling largely depends on ABI2 and slightly on ABI1. These findings confirm that ABA signaling acts downstream of the BR receptor complex, but upstream of BES1 to regulate the signaling outputs of BRs.
Results
Newly Identified BR-Related Mutants and Overexpression Lines Confirm that BRs Have an Important Role in Antagonizing the Inhibitory Effect of ABA on Seed Germination.
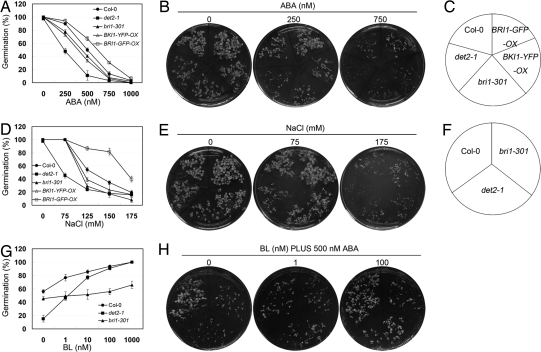
Previous studies imply that BR-deficient and BR-perceptional mutants are more sensitive to ABA during seed germination (10). With more BR-related mutants and some overexpression lines identified or created recently, we were interested in testing the responses of these genetic materials to ABA during seed germination. These genetic materials include the BR-deficient mutant, det2-1, a weak bri1 allele, bri1-301 (40), BKI1-YFP-OX (18), and BRI1-GFP-OX (Table S1). As shown in Fig. 1A, seed germination was significantly inhibited by 500 nM ABA in BR-deficient or signaling mutants (det2-1, 11.01%; bri1-301, 40.46%; and BKI1-YFP-OX, 33.5%), whereas the germination rate of Col-0 was just reduced to 50%. In contrast, overexpression of BRI1 (BRI1-GFP-OX) significantly enhanced resistance to ABA, resulting in a seed germination rate of ≈67% in 500 nM ABA (Fig. 1A). Similarly, BRI1-GFP-OX grew better than the others in 750 nM ABA, but the growth of det2-1 was already largely retarded by 250 nM ABA (Fig. 1B).
Fig. 1.
The antagonizing effect of BR and ABA on seed germination. (A) Germination rates of BR-related mutants under ABA condition. (B) Nine-day old seedlings on 1/2 MS medium containing 0, 250 and 750 nM ABA. (C) Arrangements for genotypes in B and E. (D) Germination rates of BR biosynthetic and signaling mutants under NaCl condition. (E) Nine-day old seedlings on 1/2 MS medium containing 0, 75 and 175 mM NaCl. (F) Arrangements for genotypes in H. (G) The inhibitory effect of ABA on seed germination can be rescued by applied BL. (H) Nine-day old seedlings on 1/2 MS medium containing 500 nM ABA with 0, 1, or 100 nM BL. Germination rates were recorded at 5 days after plates were transferred to growth room. The experiments were repeated 3 times, and data are the average value (n ≈ 50) for each independent experiment. The error bars represent SE.
Because mutants with altered ABA sensitivity usually have different germination rates under NaCl treatment, we also checked whether these BR-related mutants have different responses to NaCl. As shown in Fig. 1D, NaCl significantly inhibited the germination of BR-deficient or BR signaling-defective mutants (det2-1, 24.21%; bri1-301, 29.16%; and BKI1-YFP-OX, 39.22%; Col-0, 54.2%). In contrast, BRI1-GFP-OX was more resistant to NaCl, with a germination rate of ≈86.7% in 150 mM NaCl (Fig. 1D). Also, the inhibitory effect of NaCl on seedling growth of these mutants was similar to the effect of ABA (Fig. 1E).
To further assess how BR and ABA interact to influence seed germination, we examined whether the inhibitory effect of ABA on seed germination can be reversed by applying brassinolide (BL), the most active form of BRs. With 500 nM ABA in the medium, we found that seed germination and growth of Col-0 and det2-1 was rescued by BL in a dosage-dependent manner (Fig. 1 G and H). In contrast, seed germination of bri1-301 was almost unchanged by applied BL (Fig. 1G), indicating that stronger BR signaling could overcome the inhibitory effect of ABA on seed germination, and they may regulate common components with opposite direction to control seed germination.
ABA Inhibits the BR Signaling Outputs.
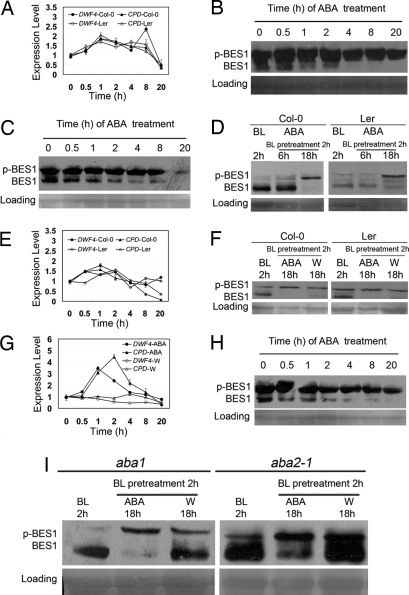
To explore whether ABA is directly involved in the BR signaling pathway, we used BR-regulated genes as molecular markers and the phosphorylation status of BES1 as a biochemical marker to evaluate BR signaling outputs after ABA treatment. First, we measured the transcript levels of 2 BR down-regulated genes, DWF4 and CPD, via real-time qRT-PCR in wild-type Col-0. As shown in Fig. 2A, treatment of 1 μM ABA for 30 min enhanced the expression of DWF4 and CPD, suggesting that BR signaling was rapidly inhibited by ABA. Then, we checked the phosphorylation status of a biochemical marker, BES1, by immunoblot analysis in wild-type Col-0. With the treatment of 1 μM ABA, the amount of dephosphorylated BES1 was rapidly reduced (Fig. 2B). Because under normal growth conditions BES1 is mainly present in the phosphorylated state, we pretreated seedlings with 1 μM BL for 2 h to allow dephosphorylation of BES1. Then, we treated seedlings with 1 μM ABA to determine whether ABA induces the phosphorylation of BES1. We observed that BL greatly stimulated the dephosphorylation of BES1 in Col-0, and the subsequent treatment of ABA for 6 h slightly but significantly induced the phosphorylation of BES1. A longer (18 h) treatment with ABA further enhanced the phosphorylation of BES1, and the dephosphorylated BES1 was almost undetectable (Fig. 2D). Similar results were also observed in seedlings of Ler (Fig. 2 A, C, and D).
Fig. 2.
ABA inhibits the BR signaling outputs. (A) ABA promotes the BR-suppressed gene expression. Seedlings of Col-0 and Ler were treated with 1 μM ABA. (B) ABA induces the phosphorylation of BES1 in wild-type Col-0. Seedlings were treated with 1 μM ABA. (C) ABA induces the phosphorylation of BES1 in wild-type Ler. Seedlings were treated with 1 μM ABA. (D) ABA induces the phosphorylation of BES1 in BR-treated seedlings. Seedlings of Col-0 and Ler were pretreated with 1 μM BL for 2 h; then, with 1 μM ABA for another 6 h and 18 h. (E) Aqueous solution treatment promotes the expression of BR-suppressed genes in wild-type Col-0 and Ler. (F) Aqueous solution treatment induces the phosphorylation of BES1 in wild-type Col-0 and Ler. Seedlings were pretreated with 1 μM BL for 2 h; then, with 1 μM ABA or aqueous solution (W) as control for 18 h. (G) ABA significantly alters the expression of BR-regulated genes, but aqueous solution does not in the ABA-deficient mutant aba1. (H) ABA induces the phosphorylation of BES1 in aba1. Seedlings were treated with 1 μM ABA. (I) ABA significantly decreases the accumulation of dephosphorylated BES1 in ABA-deficient mutants, aba1 and aba2-1. Seedlings were pretreated with 1 μM BL for 2 h; then, with 1 μM ABA or water as control for 18 h. In B, C, D, F, H, and I, immunoblot with anti-BES1 shows levels of phosphorylated BES1 (p-BES1) and dephosphorylated BES1 (BES1); Ponceau-S stained Rubisco large subunit serves as a loading control. Nine-day old seedlings were used.
Waterlogging can induce ABA production in plants (41). In our experiments, including the mock treatments, seedlings were submerged in aqueous solution, which might result in an increase of ABA content in the wild-type background. This procedure may have masked the effect of applied ABA on BR signaling outputs. Therefore, we checked whether water alone can affect BR signaling outputs. We found that aqueous solution treatment without ABA also induced the expression of DWF4 and CPD (Fig. 2E), and stimulated the phosphorylation of BES1 in wild-type Col-0 (Fig. 2F). Thus, to avoid the effect of an increased endogenous ABA level induced by aqueous solution, we performed the ABA treatment using seedlings of the ABA-deficient mutants, aba1 (42) and/or aba2-1 (43). As predicted, the expression levels of both DWF4 and CPD were almost unchanged after water treatment in the aba1 mutant (Fig. 2G), but treatment with 1 μM ABA led to a greater increase in the expression levels of DWF4 and CPD in aba1 than that in wild type (Fig. 2 A and G). Also, ABA treatment caused a significant decrease in the accumulation of dephosphorylated BES1in the aba1 mutant (Fig. 2H). We further tested the phosphorylation status of BES1 in aba1 and aba2-1, and found that pretreatment with 1 μM BL for 2 h, and after treatment with ABA for 18 h, significantly induced the phosphorylation of BES1, and the dephosphorylated BES1 was almost undetectable. In contrast, a significant amount of dephosphorylated BES1 was still present after treatment with aqueous solution for an additional 18 h (Fig. 2I). Given that aba1 and aba2-1 have different genetic backgrounds (Table S1), we did not find an obvious difference in response to ABA between the 2 ecotypes, Col-0 and Ler (Fig. 2F). In deed, because ABA promoted rather than inhibited the expression of BR biosynthetic genes, the inhibitory effect of ABA on BR signaling is unlikely through changing the level of endogenous BR. Together, these results strongly suggest that ABA can rapidly and directly inhibit the BR signaling.
Effect of ABA on BR Signaling Does Not Depend on the Perception of BR.
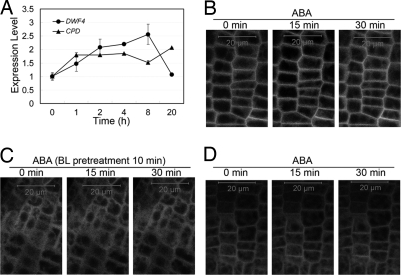
To investigate whether the effect of ABA on BR signaling is through the BR receptor complex, we first examined whether ABA regulates the expression of the BR marker genes DWF4 and CPD in the bri1-116 mutant, which is a null allele of bri1 and contains a stop codon in the extracellular domain (12). As shown in Fig. 3A, treatment with 1 μM ABA led to a significant increase of the expression of CPD and DWF4 in bri1-116, which is similar to that in wild type (Fig. 2A), indicating that the regulation of ABA on BR signaling may not rely on BRI1, and the cross-talk between BR and ABA signaling is likely to occur downstream of BRI1. Because BKI1 is an early BR signaling component, and its plasma membrane dissociation is induced by BR (18), we tested whether ABA treatment alters the subcellular localization of BKI1-YFP in BKI1-YFP-OX and BKI1-YFP-OX/bri1-116. As shown in Fig. 3 B and D, 15 or 30 min treatment with ABA did not change the subcellular localization of BKI1-YFP in both the wild-type and bri1-116 background. To further test whether the inhibitory effect of ABA on BR signaling is through the enhanced plasma membrane association of BKI1-YFP, we pretreated seedlings of BKI1-YFP-OX with 1 μM BL for 10 min to abolish the plasma membrane association of BKI1-YFP. Subsequent treatment with ABA did not enhance the plasma membrane association of BKI1-YFP (Fig. 3C). These results imply that the primary role of ABA in BR signaling does not depend on BRI1 and BKI1, and it is not through the change in endogenous levels of BRs.
Fig. 3.
The effect of ABA on BR signaling outputs does not depend on the BR receptor complex. (A) The effect of ABA on the expression of BR-responsive genes in bri1-116; bri1-116 is a null allele of bri1 and contains a stop codon in the extracellular domain. Were treated 20-day old seedlings with 1 μM ABA. (B) The subcellular localization of BKI1-YFP after ABA treatment. Were treated 4-day old seedlings of BKI1-YFP-OX with 1 μM ABA for 15 min or 30 min. (C) ABA does not alter the subcellular localization of BKI1-YFP after BL treatment. Were pretreated 4-day old Seedlings of BKI1-YFP-OX with 1 μM BL for 10 min; then, they were treated with 1 μM ABA for 15 min or 30 min. (D) ABA does not alter the subcellular localization of BKI1-YFP in bri1-116 background. Were treated 4-day old seedlings of BKI1-YFP-OX/bri1-116 with 1 μM ABA for 15 or 30 min.
Effect of ABA on BR Signaling Is Through BIN2.
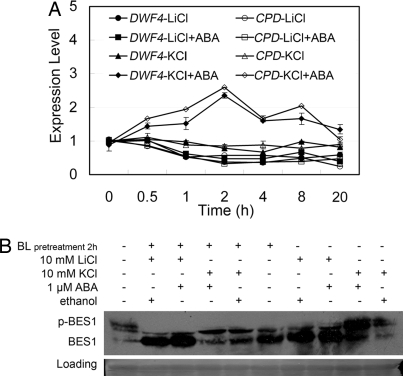
To understand at which point ABA becomes involved in the BR signaling cascade, we examined whether the downstream BR signaling component, BIN2, a negative regulator of BR signaling, is required for their cross-talk. Because BIN2 has several homologues, we used a specific GSK3 kinase inhibitor, lithium chloride (LiCl), to block the activity of BIN2 kinase (44). First, we treated the seedlings of aba1 with 10 mM LiCl, 10 mM KCl, and 10 mM LiCl plus 1 μM ABA or 10 mM KCl plus 1 μM ABA, and measured the transcript levels of DWF4 and CPD. As shown in Fig. 4A, both treatments of LiCl and LiCl plus ABA down-regulated the expression of DWF4 and CPD, whereas the mock treatment of KCl plus ABA promoted the expression of DWF4 and CPD, indicating that, once the BIN2 activity was inhibited by LiCl, ABA failed to induce the expression of DWF4 and CPD. Then, we treated seedlings of aba1 with the above 4 types of solutions, or pretreated the seedlings of aba1 with 1 μM BL to allow a complete dephosphorylation of BES1 following treatment of the above 4 types of solutions to test the phosphorylation status of BES1. As shown in Fig. 4B, when BIN2 kinase activity was inhibited by LiCl, ABA failed to induce the phosphorylation of BES1, whereas in the mock treatment with KCl, ABA still stimulated the phosphorylation of BES1 both in BL-treated and BL-untreated seedlings. These results indicate that ABA most likely inhibits BR signaling by regulating BIN2 or unknown components upstream of BIN2.
Fig. 4.
LiCl suppresses ABA-inhibited BR signaling outputs. (A) LiCl suppresses ABA-promoted expression of BR down-regulated genes. Were treated 9-day old seedlings of aba1 with the following solutions, respectively: 10 mM LiCl, 10 mM KCl, 10 mM LiCl plus 1 μM ABA, and 10 mM KCl plus 1 μM ABA. (B) LiCl suppresses ABA-stimulated phosphorylation of BES1. With solutions used in A, 9-day old seedlings of aba1 were treated for 20 h, or were pretreated with 1 μM BL for 2 h and then treated with solutions used in A for additional 18 h. Immunoblot with anti-BES1 shows levels of p-BES1 and BES1. Ponceau-S stained Rubisco large subunit serves as a loading control.
Effect of ABA on BR Signaling Depends on ABA Early Signaling.
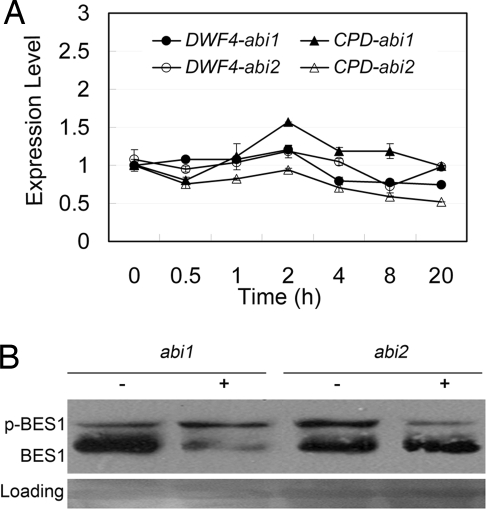
To further determine how ABA alters BR signaling outputs, we tested the transcript level of DWF4 and CPD and the phosphorylation status of BES1 in response to ABA in the likely early signaling mutants abi1 and abi2 (33). Compared with wild-type and aba1 (Fig. 2 A and G), the expression levels of both DWF4 and CPD were slightly changed after treatment with 1 μM ABA in the abi1 and abi2 mutants (Fig. 5A). Also, as shown in Fig. 5B, treatment with 1 μM ABA for the seedlings pretreated with 1 μM BL led to different levels of BES1 phosphorylation between abi1 and abi2. Interestingly, a significant amount of dephosphorylated BES1 was present in abi1 and abi2, with the abi2 mutant having a larger amount of dephosphorylated BES1 after ABA treatment, which is different from the wild-type (Fig. 2F), aba1, and aba2-1 (Fig. 2I). This result suggests that the inhibitory role of ABA on BR signaling depends largely on ABI2 and slightly on ABI1.
Fig. 5.
The role of ABA on BR signaling is through ABI2 and ABI1. (A) The slight effect of ABA on the expression of BR-responsive genes in ABA signaling mutants. Nine-day old seedlings of ABA signaling mutants (abi1 and abi2) were treated with 1 μM ABA. (B) The alteration of the effect of ABA on phosphorylation status of BES1 in ABA signaling mutants. Nine-day old seedlings of ABA signaling mutants abi1 and abi2 were pretreated with 1 μM BL for 2 h; then, with 1 μM ABA for 18 h. Seedlings pretreated with BL for 2 h as “−,” and pretreated with BL for 2 h then pretreated with ABA for 18 h as “+.” Immunoblot with anti-BES1 shows the levels of p-BES1 and BES. Ponceau-S stained Rubisco large subunit serves as a loading control.
Discussion
This study provides several lines of strong evidence demonstrating that ABA inhibits the primary signaling outputs of BR. First, in both wild-type and ABA-deficient mutants, ABA can rapidly induce the expression of the BR-suppressed genes, DWF4 and CPD, and enhance the phosphorylation of BES1. In ABA-deficient mutants, the effect of ABA on BR signaling outputs is more significant. Second, the regulatory role of ABA on BR signaling outputs does not depend on the BR receptor complex, indicating that the effect of ABA is not through an alteration of the BR level. Third, the inhibitory effect of ABA on BR signaling is blocked in the ABA signaling mutants, abi1 and abi2. This finding further suggests that its role in BR signaling is likely through the primary signaling cascade, rather than a secondary effect through the early responsive genes of ABA.
Our studies also narrow down where the ABA and BR signaling cascades cross-talk. On the side of BR signaling, ABA apparently acts downstream of BR receptor complex, because the bri1 mutant, which lacks the BR receptor, displayed a similar response to ABA as wild-type. Also, the ABA signal affects BR signaling likely by regulating BIN2 or components upstream of BIN2, as indicated by the observation that, once the activity of BIN2 is inhibited by LiCl, the response of marker genes of DWF4 and CPD, and the biochemical maker BES1 to ABA, was also abolished. On the side of ABA signaling, abi1 and abi2 mutants showed different responses to ABA in BR signaling from wild-type and ABA deficient mutants, indicating that ABA directly affects BR signaling outputs through ABI2 and ABI1. However, using a yeast 2-hybrid assay, we did not find a physical interaction of ABI2 with any known components of BR signaling, including BRI1, BAK1, TTL, BKI1, BIN2, and BES1 (Fig. S1). Because the components between BRI1 and BIN2 in the BR signaling pathway, and the components directly downstream of ABI1 and ABI2 in the ABA signaling pathway, remain to be identified, these unknown components could participate in their direct interaction. A more significant phenotype exhibited in abi2 than in abi1 suggests that ABI1 and ABI2 may have different roles in the cross-talk between ABA and BR signaling, which is consistent with previous studies that suggested ABI1 and ABI2 having distinct physiological functions (45, 46).
The cross-talk between BR and ABA responses suggests that transcription factors (such as BES1 and BZR1) regulated by BRs could also serve as major modulators to integrate multiple cues. Consistent with the previous studies (10), seed germination experiments suggest that the signaling pathways of BRs and ABA may work oppositely to regulate seed germination. Microarray data have shown that a large portion of BR-responsive genes are also regulated by ABA (3, 4), suggesting that ABA is significantly involved in the BR signaling pathway. Previous studies also show that central repressors, such as DELLAs, could act to modulate diverse hormone effects (47, 48), integrate responses to adverse conditions, and regulate plant growth. Therefore, it is likely that ABA could be significantly involved in many hormone signaling pathways. Many stress signals promote the production of ABA, so stress signals could use ABA to cross-talk with BR signaling and regulate the activity of BES1 or its homologues to coordinately control biological processes.
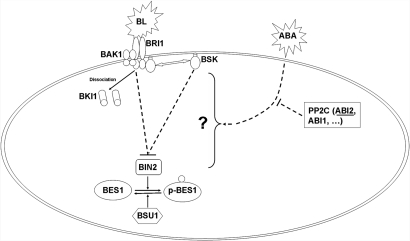
These findings and previous information suggest a model to explain the interaction between BR and ABA signaling pathways (Fig. 6). ABA regulates BR signaling via the PP2C family, including ABI1 and ABI2. This involvement is downstream of BR perceptional complex (at least independent of BRI1 and BKI1), but through BIN2 or unknown upstream components of BIN2. Also, the inhibitory role of ABA on BR signaling is primarily through ABI2 and partially through ABI1. This study provides insight into the mechanism of the interaction between ABA and BR, and defines a specific region where the 2 signals may interact. A recent study demonstrates that ARF2, a transcription factor that responds to auxin, can be regulated by BIN2, a negative regulator of BR signaling (7). Our study found that ABA signaling is involved in the primary signaling of BRs through regulating BIN2 or its upstream components. When more components in the BR and ABA signaling pathways are discovered, the specific components linking the 2 pathways will surely be identified.
Fig. 6.
A proposed model for the effect of ABA on BR signaling outputs. For BR signaling pathway, BR binding to BRI1 activates BRI1 complex via releasing plasma membrane association of BKI1, association with BAK1, and phosphorylation of BSKs. Activated BRI1 complex likely inhibits BIN2 kinase through yet an unknown mechanism, leading to the dephosphorylation of BES1, which actively regulates BR-responsive gene expression. ABA could regulates unknown components downstream of BRI1, but upstream of BIN2 in the BR signaling, which then lead to the activation of BIN2 through an undiscovered mechanism. Gain-of-function mutation of ABI1 and ABI2 in the ABA early signaling can inhibit the effect of ABA on BR signaling.
Materials and Methods
Plant Materials and Growth Condition.
Arabidopsis ecotypes Columbia (Col-0) and Landsberg erecta (Ler), the BR-related and ABA-related mutants and transgenic lines, were listed in Table S1. Phenotypic analyses were conducted on culture plates with 1/2 MS medium (Sigma–Aldrich) and 0.8% phytagel (Sigma–Aldrich). Plants were grown at 22 °C under long-day condition (300 μE m-2 s-1, with 16-hr light/8-hr dark cycles) in growth room.
Germination Experiments.
Seeds were imbibed for 4 days at 4 °C, and then moved to a growth room. Germination rate was determined after 5 days with emerging cotyledons. Stock solutions of ABA (Sigma–Aldrich) were in ethanol; BL (Wako) was dissolved in DMSO, and added to autoclaved media after cooling to ≈55 °C.
Gene Expression Analysis by Real-Time qRT-PCR.
Seedlings were first submerged in aqueous solution for 30 min. Then, they were treated with solutions as indicated in the main text. Total RNA was extracted by using a Tiangen RNAprep plant kit (Tiangen), and the first strand cDNA was synthesized by using Takara PrimeScript First-Strand cDNA Synthesis kit (Takara); cDNAs were combined with SYBR master mix for PCR (Invitrogen). Primers for each gene are shown as follows: CPD (At5g05690): 5′- TTACCGCAAAGCCATCCAA-3′ and 5′- TCATCACCACCACCGTCAAC-3′; and DWF4 (At3g50660): 5′- GTTGGCCATTTCTTGGTGAAA-3′ and 5′-TGGCGGTGTACGGTTTAAGAT-3′. A U-box gene (At5g15400) was used to normalize the data (5′-TGCGCTGCCAGATAATACACTATT-3′ and 5′- TGCTGCCCAACATCAGGTT-3′). Real-time PCRs were performed in triplicate with a Bio-Rad iCycler. Data were collected and analyzed with Bio-Rad MyiQ Single-Color Real-Time PCR Detection System.
Protein Extraction from Plants for Immunoblot Analysis.
Seedlings were first submerged in aqueous solution for 30 min. Then, they were treated with solutions as indicated in the main text. Plant tissue was ground to a fine powder in liquid nitrogen. Total protein was extracted with 2× extraction buffer (100 mM Tris·HCl, pH 7.5/100 mM NaCl/5 mM Na-EDTA/1 mM PMSF/5 mM DTT/10 mM 2-mercaptoethanol), separated on a 12.5% SDS/PAGE gel, and transferred to a nitrocellulose membrane (Amersham Bioscisences). Polyclonal anti-BES1 antibody and goat anti-rabbit HRP-conjugated secondary antibody (Pierce Biotechnology) were used to quantify BES1. Signals were visualized by using the SuperSignal west pico chemiluminescent substrate imaging system (Pierce Biotechnology).
Visualization of Protein Localization.
The subcellular localization of BKI1-YFP (18) was visualized and photographed by using a Carl Zeiss LSM 510 META confocal microscope. Root tips of 4-day old seedlings were treated with 1 μM ABA for 15 or 30 min, or pretreated with 1 μM BL for 10 min to allow a complete dissociation of BKI1-YFP from the plasma membrane; then, treated with 1 μM ABA for 15 or 30 min to photograph.
Supplementary Material
Acknowledgments.
We thank Y. Yin (Iowa State University, Ames, IA) for providing the anti-BES1 antibody, C. Song (Henan University, Kaifeng, Henan, China) for providing aba1 and aba2-1 seeds, Y. Guo (National Institute of Biological Sciences, Beijing) for providing abi1 and abi2 seeds, J. Li (University of Michigan, Ann Arbor, MI) for providing bri1-301 seeds, and Prof. D. Shen for helping to set up our laboratory. X.W. was supported by a Fudan University start-up fund, the National Natural Science Foundation of China Grant 30671118, and the Shanghai Pujiang Project Grant 07pj14014.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900349106/DCSupplemental.
References
- 1.Teale WD, et al. Auxin as a model for the integration of hormonal signal processing and transduction. Mol Plant. 2008;1:229–237. doi: 10.1093/mp/ssn006. [DOI] [PubMed] [Google Scholar]
- 2.Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;144:1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goda H, et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis, and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 4.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 5.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular Aspects of Seed Dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 6.Razem FA, Baron K, Hill RD. Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol. 2006;9:454–459. doi: 10.1016/j.pbi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JG, et al. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Wang S, Asami T, Chen J-G. Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein {alpha} subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol. 2008;49:1013–1024. doi: 10.1093/pcp/pcn078. [DOI] [PubMed] [Google Scholar]
- 10.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leubner-Metzger G. Brassinosteroids and gibberellins promote tobacco seed germination by distinct pathways. Planta. 2001;213:758–763. doi: 10.1007/s004250100542. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 13.He Z, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 15.Wang ZY, et al. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plants Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 25.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He JX, et al. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gampala SS, et al. An essential role for 14–3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, et al. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005;138:1117–1125. doi: 10.1104/pp.104.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn JM, et al. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura N, et al. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 33.Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- 34.Liu X, et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 35.Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 36.Wasilewska A, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 37.Finkelstein R, et al. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- 38.Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. [Google Scholar]
- 40.Xu W, et al. Is kinase activity essential for biological functions of BRI1? Cell Res. 2008;18:472–478. doi: 10.1038/cr.2008.36. [DOI] [PubMed] [Google Scholar]
- 41.Jackson MB, Hall KC. Early stomatal closure in waterlogged pea plants is mediated by abscisic acid in the absence of foliar water deficits. Plant Cell Environ. 1987;10:121–130. [Google Scholar]
- 42.Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L. ) Heynh Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 43.Leon-Kloosterziel KM, et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 44.Peng P, Yan Z, Zhu Y, Li J. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant. 2008;1:338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Qin C, Zhao J, Wang X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2004;101:9508–9513. doi: 10.1073/pnas.0402112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishra G, et al. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- 47.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 48.Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.