Abstract
The lack of a primate model that utilizes HIV-1 as the challenge virus is an impediment to AIDS research; existing models generally employ simian viruses that are divergent from HIV-1, reducing their usefulness in preclinical investigations. Based on an understanding of species-specific variation in primate TRIM5 and APOBEC3 antiretroviral genes, we constructed simian-tropic (st)HIV-1 strains that differ from HIV-1 only in the vif gene. We demonstrate that such minimally modified stHIV-1 strains are capable of high levels of replication in vitro in pig-tailed macaque (Macaca nemestrina) lymphocytes. Importantly, infection of pig-tailed macaques with stHIV-1 results in acute viremia, approaching the levels observed in HIV-1-infected humans, and an ensuing persistent infection for several months. stHIV-1 replication was controlled thereafter, at least in part, by CD8+ T cells. We demonstrate the potential utility of this HIV-1-based animal model in a chemoprophylaxis experiment, by showing that a commonly used HIV-1 therapeutic regimen can provide apparently sterilizing protection from infection following a rigorous high-dose stHIV-1 challenge.
Keywords: AIDS, PrEP, SIV, stHIV-1
AIDS research has been hampered by the lack of an animal model that utilizes HIV-1 as the challenge virus, the central problem being the absence of a practical host species in which HIV-1 replicates efficiently (1). While chimpanzees can be productively infected, they are endangered, expensive, do not typically develop AIDS after HIV-1 infection, and their use in research engenders ethical concerns (2). Consequently, the most widely used animal models of human HIV-1 infection and AIDS comprise infection of rhesus or pig-tailed macaques with simian immunodeficiency viruses (SIV) or chimeras encoding the HIV-1 envelope or reverse transcriptase (SHIV, or RT-SHIV), which can cause simian AIDS (3–7). These models have been extremely informative, particularly in studies of SIV and SHIV pathogenesis (1, 8–10). However, they have shortcomings, mostly because of the large genetic distance between SIVs and HIV-1. The divergence between human and simian viruses necessitates a 2-stage process for evaluation of candidate vaccines in macaques, with proof of concept-challenge studies using SIV followed by immunogenicity studies using the corresponding HIV-1 immunogens, which often cannot be directly evaluated for indications of efficacy before human trials. Additionally, differences between HIV-1 and SIV enzymes preclude preclinical evaluation of the efficacy of some drugs, such as non-nucleoside RT inhibitors that are inactive against SIVs. Thus, the development of animal models that use HIV-1 as the challenge virus would undoubtedly facilitate the evaluation of candidate prevention and treatment strategies.
A major reason underlying the inability of HIV-1 to replicate in nonhuman primate cells is the existence therein of gene products that have evolved to inhibit retroviral replication. The TRIM5α protein blocks retroviral infection by inactivating incoming capsids (11), and species-specific variation in the TRIM5α C-terminal domain that is responsible for capsid recognition (12, 13) is a major determinant of host susceptibility to various retroviruses (14–16). Additionally, APOBEC3 proteins infiltrate virions during their assembly and inactivate viral genomes during subsequent reverse transcription, largely by catalyzing the deamination of cytidines in nascent retroviral DNA (17–20). While primate lentiviruses encode Vif proteins that antagonize APOBEC3 proteins by inducing their degradation (21–23), resistance to Vif-induced degradation often occurs as a result of species-specific variation in APOBEC3 proteins (24–28).
By virtue of its particular capsid and Vif protein sequences, HIV-1 avoids and antagonizes the human forms of TRIM5α and APOBEC3 proteins. However, HIV-1 fails to avoid or antagonize the rhesus macaque TRIM5α and APOBEC3 proteins, and cannot replicate in rhesus macaque T cells (29). Previously, we and others demonstrated that the inability of HIV-1 to replicate in rhesus macaque cells in vitro can be overcome by engineering resistance to TRIM5α and APOBEC3 proteins (30, 31). Notably Macacca nemestrina (pig-tailed macaques) lack a TRIM5α protein, and the dominant form of TRIM5 expressed in this species is a TRIMCyp fusion protein that arose through retrotransposition of a CypA cDNA into the TRIM5 locus of an ancestral macaque (32–36). The resulting protein is similar to the TRIMCyp protein previously identified in owl monkeys (37, 38) but, crucially, differs from the owl monkey TRIMCyp in that it does not inhibit HIV-1 infection (32–36). Thus, pig-tailed macaques appear to present fewer impediments to HIV-1 replication and could potentially be more amenable to infection by stHIVs that are derived by minimal modifications of HIV-1.
Here, we constructed stHIV-1 strains that differ from HIV-1 only in the vif gene, and show that such viruses replicate robustly in pig-tailed macaque lymphocytes in vitro. Additionally, we show that infection of pig-tailed macaques with stHIV-1 results in acute viremia at a level approaching that observed in HIV-1-infected humans. Moreover, stHIV-1 can establish infection in pig-tailed macaques that persists for months but is controlled thereafter, at least in part, by CD8+ T cells. Finally, we demonstrate the potential utility of this HIV-1-based animal model by showing that a commonly used HIV-1 therapeutic regimen, used as chemoprophylaxis, can protect pig-tailed macaques from infection by stHIV-1 after a rigorous high-dose challenge.
Results
Vif-Substituted HIV-1 Replicates in Macacca nemestrina Lymphocytes.
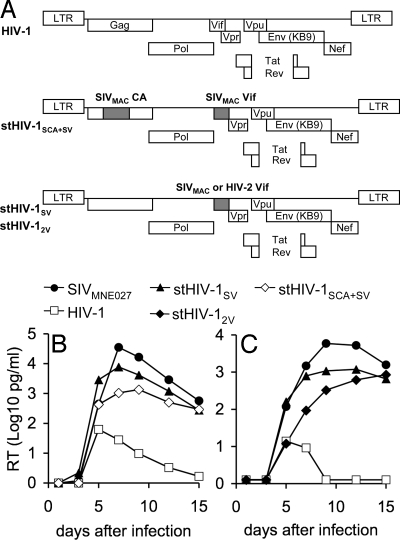
The absence of a TRIM5 protein capable of inhibiting HIV-1 infection raised the possibility that pig-tailed macaque lymphocytes might support the replication of HIV-1. However, rhesus macaques express several APOBEC3 proteins that potently inhibit HIV-1 infectivity (28), and the same was expected to be true of pig-tailed macaques. Therefore, we generated 2 HIV-1-derived constructs, termed stHIV-1SV and stHIV-12V, encoding a macaque-adapted HIV-1 envelope protein (from SHIVKB9), in which the HIV-1 vif gene was replaced by the vif genes from SIVMAC239 and HIV-2ROD, respectively (Fig. 1A). These vif genes were selected because we previously found that Vif proteins from SIVMAC and HIV-2ROD efficiently antagonized the antiretroviral activity of several APOBEC3 proteins from rhesus and pig-tailed macaques (ref. 28 and data not shown). Spreading infection assays in pig-tailed macaque lymphocytes revealed that the replication of the parental HIV-1 construct was transient and low, whereas SIVMNE027, an SIV strain that is highly pathogenic in pig-tailed macaques (10), replicated robustly, with reverse transcriptase (RT) activity reaching a peak at approximately day 6 to 8 after infection (Fig. 1B). Importantly, stHIV-1SV also replicated very efficiently in pig-tailed macaque lymphocytes, and RT levels reached a peak at the same time and at only marginally lower levels than SIVMNE027, (see Fig. 1B). Notably, the presence of SIVMAC CA provided no advantage over HIV-1 CA (compare replication of stHIV-1SCA+SV with stHIV-1SV in Fig. 1B). Further experiments in pig-tailed macaque lymphocytes confirmed that stHIV-1SV replicated with an efficiency comparable to that of SIVMNE027, while stHIV-12V was marginally impaired as compared to stHIV-1SV (Fig. 1C). Importantly, however, both stHIV-1SV and stHIV-12V replicated with far greater efficiency than a matched HIV-1 construct in which vif was unaltered.
Fig. 1.
Replication of stHIV-1 variants in pig-tailed macaque lymphocytes in vitro. (A) Schematic representation of the viral constructs used in this study, shaded regions indicate sequences from HIV-2ROD or SIVMAC239, as indicated. All HIV-1-derived constructs used the macaque-adapted HIV-1 envelope from SHIVKB9. (B, C) RT activity in culture supernatants (expressed as log10 pg/ml) was measured every 2 or 3 days for 15 days following inoculation of pig-tailed macaque lymphocytes with the indicated viruses. The 2 graphs shown in (B) and (C) represent independent experiments employing 2 different pig-tailed macaque donors.
Acute Viremia and Persistent Replication of stHIV-1 in Pig-Tailed Macaques.
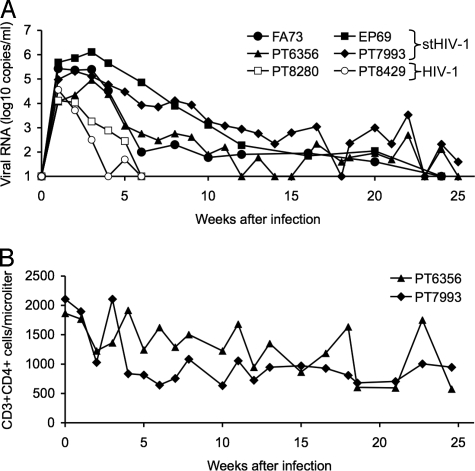
The ability of minimally modified stHIV-1 strains to replicate robustly in pig-tailed macaque lymphocytes suggested that most or all of the intrinsic barriers to HIV-1 replication in that species had been overcome simply by replacement of the vif gene. Therefore, 4 animals were inoculated intravenously with an admixture of stHIV-1SV and stHIV-12V. Infection was established in all 4 animals, with peak plasma viral load reaching 105 to 106 copies of viral RNA (vRNA) per ml at 2 to 4 weeks after inoculation (Fig. 2A). Viremia declined gradually thereafter, but persisted at detectable levels for >20 weeks after inoculation. A modest decline in CD4+ T cells was observed during the first 5 weeks after inoculation, but this parameter subsequently remained stable throughout the course of infection without clear evidence of CD4+ T-cell depletion [Fig. 2B, supporting information (SI) Fig. S1]. Infection of 2 pig-tailed macaques with the same dose of a matched HIV-1 stock resulted in a peak viral load of ≈104 RNA copies per ml at 1-week after inoculation, but plasma viremia declined rapidly thereafter, and was cleared by 6 weeks following infection in both animals (see Fig. 2A). Thus, both HIV-1 and stHIV-1SV/2V could clearly establish infection in pig-tailed macaques, but the differential ability of HIV-1 versus stHIV-1SV/2V to replicate in pig-tailed macaque lymphocytes in vitro quite accurately predicted their different properties in vivo. In particular, the presence of the vif genes from SIVMAC and HIV-2 gave a substantial advantage to stHIV-1 in terms of the level of acute viremia observed during primary infection, and in the degree to which infection and measurable plasma viremia persisted in pig-tailed macaques (see Fig. 2A).
Fig. 2.
Infection of pig-tailed macaques with HIV-1 and stHIV-1. (A) Four macaques were infected with a 1:1 mixture of stHIV-1SV and stHIV-12V (filled symbols) and 2 macaques were infected with the same dose (2 × 106 i.u.) of HIV-1 (open symbols). Plasma viremia was measured by RT-PCR and is given as the log10 viral RNA copies per milliliter of plasma. (B) CD4+ T cells, enumerated using FACS analyses in 2 of the 4 stHIV-1-infected macaques over the course of infection. CD4 counts in the 2 other macaques were monitored on a different schedule (see SI Text, Fig. S1).
Immune Responses Contribute to the Control of stHIV-1 Replication in Vivo.
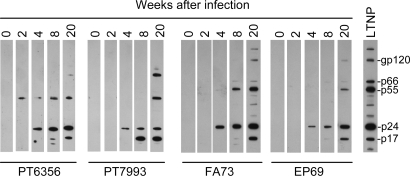
To determine whether animals challenged with stHIV-1 mounted a specific viral antibody response, plasma was obtained at 0, 2, 4, 8, and 20 weeks after inoculation. All infected animals generated antibodies against HIV-1 Gag proteins that became detectable from weeks 2 to 4 after inoculation in Western blot assays; antibody levels increased over time thereafter (Fig. 3). Antibodies that were capable of neutralizing stHIV-1SV were detectable as early as 2 weeks after inoculation in some animals, while up to 8 weeks of infection elapsed before neutralizing antibodies were detected in other animals (Table 1). Overall, a robust serological response to stHIV-1 infection was observed in each animal. Additionally, modest but measurable responses to HIV-1 antigens by both CD4+ and CD8+ T cells were observed using intracellular cytokine staining assays in which cells were stimulated by overlapping peptides from HIV-1 Gag, Pol, or accessory proteins (Fig. S2).
Fig. 3.
Antibody responses to stHIV-1 infection in pig-tailed macaques. Western blot analysis using plasma recovered from the 4 stHIV-1-infected macaques at the indicated time points following stHIV-1 infection. Serum from an HIV-1-infected human long-term nonprogressor was used as a positive control.
Table 1.
Neutralization of stHIV-1SV by pig-tailed macaque plasma
| Animal | Weeks after infection |
|||||
|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 8 | 20 | ||
| PT6356 | 50% | <20a | <20 | <20 | 160 | 160 |
| 90% | <20 | <20 | <20 | <20 | 28 | |
| PT7993 | 50% | <20 | <20 | 90 | 1,000 | 1,200 |
| 90% | <20 | <20 | <20 | 90 | 160 | |
| EP69 | 50% | <20 | 80 | 160 | 1,600 | 1,800 |
| 90% | <20 | <20 | 20 | 320 | 320 | |
| FA73 | 50% | 30 | 40 | 320 | 10,240 | 7,000 |
| 90% | <20 | <20 | 80 | 120 | 600 | |
aPlasma neutralization of stHIV-1SV was measured using TZM target cells as described in the SI Text and is given as the reciprocal of the dilution at which 50% or 90% (as indicated) reduction of virus infectivity was achieved.
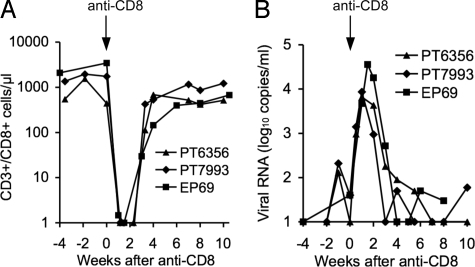
In all 4 stHIV-1-infected animals, plasma virus levels gradually declined from weeks 4 to 25 following infection, to <102 vRNA copies per milliliter at around 25 weeks after infection (see Fig. 2A). To assess the contribution of CD8+ lymphocytes to control of viral replication, 3 animals were treated with the humanized anti-CD8 antibody cM-T807 (39) to deplete CD8+ T cells. This treatment removed >99% of the CD8+ T cells from peripheral blood in each animal (Fig. 4A), and a rapid, ≈100-fold increase in vRNA levels ensued in all 3 CD8+ T-cell-depleted animals (Fig. 4B). Indeed, plasma virus levels peaked at between ≈104 and 105 vRNA copies per milliliter at 1 to 2 weeks after anti-CD8 antibody administration, but declined rapidly thereafter, coincident with the reappearance of CD8+ T cells in peripheral blood (see Fig. 4). Viremia re-established its low pre-CD8+ T-cell-depletion levels within 3 to 5 weeks of the CD8-antibody infusion in all 3 animals.
Fig. 4.
Effect of depletion of CD8+ cells on stHIV-1 replication in vivo. (A) Effect of anti-CD8-antibody infusion on the numbers of CD8+ T cells in peripheral blood, as determined by FACS analyses. Note that analysis of lymph node biopsies at 1 week after antibody infusion revealed that 0%, 8%, and 0.1% of the CD3+ cells were CD8+ for animals PT6356, PT7993, and EP69, respectively. (B) Plasma viremia, measured by RT-PCR in the weeks preceding, during, and following CD8+ T-cell depletion in 3 animals.
Prophylaxis of stHIV-1 Infection.
The profound difficulties in developing effective HIV-1 vaccines (40) has highlighted the need for alternative infection-prevention strategies, including pre-exposure prophylaxis (PrEP) using antiretroviral drugs (41). Previous nonhuman primate studies of PrEP have been restricted to the use of SIV or SHIVs, with the aforementioned attendant limitations (42).
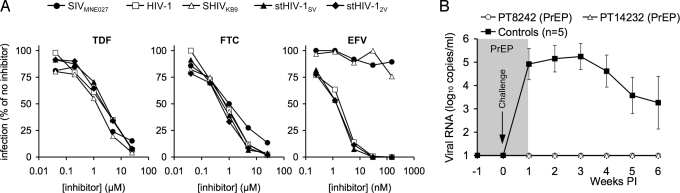
To explore the use of stHIV-1 in antiretroviral drug prophylaxis experiments, we first determined sensitivity of several potential challenge viruses to a panel of antiretroviral drugs that are used clinically in HIV-1 infection (Fig. 5A, Table S1). Both tenofovir disoproxil fumarate (TDF) and emtricitabine (FTC), which are nucleoside RT inhibitors, were effective inhibitors of HIV-1, stHIV-1SV, stHIV-12V, SHIVKB9, and SIVMNE027. Conversely, efavirenz (EFV), a non-nucleoside RT inhibitor, was effective against HIV-1, stHIV-1SV, and stHIV-12V but, as expected, failed to inhibit SHIVKB9 or SIVMNE027 (see Fig. 5A). Moreover, 2 protease inhibitors, amprenavir and atazanavir, exhibited more than 10-fold greater potency against HIV-1, stHIV-1SV, and stHIV-12V than against SIVMNE027 (see Table S1). Thus, stHIV-1 is better suited for use in prophylaxis experiments using important classes of anti-HIV drugs that are ineffective against SIV or SHIV strains.
Fig. 5.
Antiretroviral drug prophylaxis of stHIV-1 infection. (A) Sensitivity of SIVMNE027, SHIVKB9, HIV-1, and stHIV-1 strains to inhibition by the nucleoside (TDF and FTC) and non-nucleoside (EFV) RT inhibitors, that comprise a mixture commonly used for treatment of HIV-1 infection. TZM cells were infected with each virus in the presence of the indicated concentrations of each drug, and the level of infection is plotted as the percentage of that in the absence of drug. (B) Two macaques received chemoprophylaxis with a TDF/FTC/EFV combination (PrEP) and were challenged intravenously, at 1 week after prophylaxis initiation, with the same dose of stHIV-1SV/2V as was used in Fig. 2. Treatment was continued for 1 week following the challenge. A contemporaneous control macaque was challenged but untreated. Plasma viremia was measured for the ensuing 5 weeks, using RT-PCR as in Fig. 2, and compared to the 4 previously inoculated and the contemporaneous control for which the mean and standard deviation of the plasma viremia over the first 6 weeks of infection is plotted.
A commonly used therapy for HIV-1 infection consists of a TDF/FTC/EFV combination and is available formulated as a single tablet (Atripla) for once-per-day dosing, and thus might be considered an ideal candidate for use in PrEP approaches. To simulate such a regimen, the TDF/FTC/EFV combination was administered daily to 2 pig-tailed macaques for 1 week before challenge with stHIV-1SV/2V. These animals and a contemporaneous untreated control macaque received the identical dose of stHIV-1SV/2V as was used previously (see Fig. 2A). The 2 treated animals received treatment for 1 more week after challenge. As expected, the untreated animal, like the previous 4 untreated stHIV-1SV/2V-infected animals, became viremic within 1 week of challenge (Fig. 5B). In contrast, vRNA remained undetectable in the plasma of the TDF/FTC/EFV-treated animals. Even though drug treatment ceased 1 week after challenge, plasma vRNA remained undetectable for the remaining 5 weeks of the experiment (see Fig. 5B). Additionally, viral DNA or RNA could not be detected in the peripheral blood or lymph nodes of the PrEP-treated macaques, while cell-associated viral DNA and RNA were detected in the untreated contemporaneous control animal (Table S2). Thus, within the limits of assay sensitivity, and with the caveat that only two PrEP-treated animals were challenged, this regimen gave apparently sterilizing protection against a high-dose i.v. challenge by stHIV-1.
Discussion
Here, we show that stHIV-1 strains encoding Vif proteins from the HIV-2/SIVSM/SIVMAC lineage of lentiviruses can replicate efficiently in pig-tailed macaque cells in vitro and in vivo. Both HIV-1 and stHIV-1 were able to infect pig-tailed macaques, but HIV-1 replication was rapidly curtailed. In contrast, stHIV-1 plasma viremia peaked at a level approaching that observed in HIV-1-infected humans, and persisted at detectable levels for several months. Clearly, the replacement of the vif gene conferred a major advantage to stHIV-1 in pig-tailed macaques, and this result underscores the effectiveness of the APOBEC3 in blocking the replication of retroviruses that are unable to antagonize this intrinsic immune-defense mechanism.
Early reports indicated that pig-tailed macaques might be partly permissive for HIV-1 replication (43–45), and a more recent study using HIV-1 strains expressing modified Gag and Vif proteins achieved transient replication in pig-tailed macaques (46). However the levels and persistence of stHIV-1 replication documented herein are far greater than previously observed using HIV-1-derived viruses in monkeys. The relative success of stHIV-1 as compared to previous attempts to establish HIV-1-based nonhuman primate models is likely a consequence of the fact that only minimal, but critical, modifications to the HIV-1 genome were made to antagonize intrinsic host defenses. The fact that a macaque-adapted HIV-1 envelope was used may also have contributed to the comparatively robust replication of stHIV-1 in vivo. Nonetheless, stHIV-1 replication was eventually controlled, at least in part by CD8+ T cells, and the overall course of stHIV-1 infection resembled that expected of human long-term nonprogressors who harbor low levels of replicating HIV-1 for many years, and in whom viral replication is thought to be controlled largely by effective immune responses.
The development of an HIV-1 vaccine will likely be extremely difficult, and may not be possible in a reasonable period (40). Consequently, alternative strategies, such as PrEP and topical microbicides, may be the most viable approaches for preventing HIV-1 infection, at least in the short to medium term. However, in considering alternative PrEP approaches, critical parameters, such as the optimal agents, dose, and timing of drug administration relative to exposure to HIV-1 cannot readily be tested in human clinical trials. Because stHIV-1 encodes all of the targets for licensed HIV-1 therapeutics, the model described herein and refinements of it could be useful in exploring these parameters. Notably, a drug combination that simulates a commonly used regimen, which is administered to humans as a single tablet per day, gave apparently sterilizing protection against a very high-dose i.v. stHIV-1 challenge.
While the stHIV-1 may be useful in its current form for certain experiments, such as those documented herein, further developments in the model will be required to improve its utility. For example, it will be important to determine whether stHIV-1 infection can be initiated via mucosal challenge. Moreover, we caution against the misuse of the stHIV-1 model in its current form as testing platform for HIV-1 vaccines. The current generation of stHIV-1 employs the KB9 (HIV-189.6-derived) dual-tropic envelope protein. This envelope was selected for our studies because, although it is HIV-1-derived, it has previously been adapted and demonstrated to support efficient SHIV replication in macaques. We note that SHIV89.6 appears significantly more sensitive to inhibition by vaccine-induced immune responses than does, for example, SIVMAC239, perhaps because of the particular T-cell subsets targeted by X4 SHIVs (9), or its greater sensitivity to antibody neutralization (47). At present, the behavior of stHIV-1 in vaccination and challenge experiments is unknown, but it is reasonable to expect that it would exhibit the comparatively high sensitivity to vaccine-induced immune responses observed with SHIV89.6. Thus, further development of the stHIV-1 model will include the derivation of strains that replicate at high levels throughout the course of infection, and cause HIV-1-like CD4+ T-cell depletion and disease, as well as the construction of stHIV-1 variants that use the CCR5 coreceptor.
Materials and Methods
Viral Constructs, Viral Stock Generation, and in Vitro Replication Assays.
To generate the HIV-1 and stHIV-1 constructs used throughout these studies, the env gene in an HIV-1NL4–3 proviral plasmid was replaced by the rhesus macaque-adapted KB9 envelope (obtained from pSHIV-KB9). In stHIV-1SV and stHIV-12V proviral plasmids, the HIV-1 vif gene was also replaced with SIVMAC239 vif and HIV-2ROD vif, respectively (see Fig. 1A and SI Text for detailed methodology). Viral stocks were generated by transfection in 293 T cells and were titered using GHOST-X4/R5 cells. Replication in pig-tailed macaque lymphocytes was monitored using of ELISA-based RT assays (see SI Text).
Animal Inoculation and Monitoring.
Pig-tailed macaques (Macaca nemestrina) were housed in accordance with American Association for Accreditation of Laboratory Animal Care standards and all animal procedures were according to protocols approved by local Institutional Animal Care and Use Committees (see SI Text). Infection was initiated by i.v. infusion of 1 × 106 i.u. each of stHIV-1SV and stHIV-12V, or 2 × 106 i.u of HIV-1. Plasma was separated for analysis of the presence of vRNA by quantitative PCR, or antibodies by Western blotting or neutralization assays. Whole blood was used for monitoring lymphocyte subsets via FACS assays. For intracellular cytokine assays, cryopreserved peripheral blood mononuclear cells were used. Details of these monitoring assays are described in the SI Text.
CD8+ T-Cell Depletion.
Three stHIV-1 infected animals were transiently depleted of CD8+ lymphocytes at ≈26 weeks after infection by the administration of the chimeric antihuman CD8 monoclonal antibody (MAb), cM-T807, given intravenously at 50 mg/kg of body weight. The extent of CD8 T-cell depletion in peripheral blood was monitored by FACS, as described in the SI Text.
Antiretroviral Drug Sensitivity and Prophylaxis Experiments.
The sensitivity of viruses to inhibition by RT and protease inhibitors was determined using in vitro assays described in the SI Text. Two macaques were treated with once-daily doses of TDF (20 mg/kg) and FTC (50 mg/kg), in a combined s.c. injection, and EFV (200 mg), given orally in various food treats. One week after the initiation of drug administration, the 2 animals and an untreated control animal were challenged intravenously with stHIV-1SV and stHIV-12V, as described above. Drug administration was maintained for 1 further week and discontinued thereafter. Plasma viremia and cell associated viral DNA and RNA were measured as described in the SI Text.
Supplementary Material
Acknowledgments.
We thank Zerina Kratovac, Cesar Virgen, Bernice Kaack, Lara Doyle, Linda Green, Kelsi Rasmussen, Janell LeBlanc, and Adam Wiles for assistance, Sunil Ahuja for the pR1-D plasmid, Norbert Bischofberger and Gilead Sciences for providing TDF and FTC, and Ronald Desrosiers and Joseph Sodroski for the SIVMAC239 and SHIVKB9 clones, which were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH, as were the overlapping 15-mer SIVMAC239 and HIV-1 peptides. The cM-T807 antibody was provided by the NIH Nonhuman Primate Reagent Resource (R24 RR016001, N01 AI040101). This work was supported by Grants from the NIH, R01AI078788 (to T.H.) and R01AI64003 (to P.D.B.), American Foundation for AIDS Research (amFAR) 106404–33-RFMC and 107149–44-RGRL (to Z.A.), and with federal funds from the National Cancer Institute, NIH, under contracts N01-CO-12400 and HHSN266200400088C (to J.D.L.) and through the Intramural Center for Cancer Research, which supports the HIV Drug Resistance Program (V.N.K.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812587106/DCSupplemental.
References
- 1.Ambrose Z, KewalRamani VN, Bieniasz PD, Hatziioannou T. HIV/AIDS: in search of an animal model. Trends Biotechnol. 2007;25:333–337. doi: 10.1016/j.tibtech.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Fultz PN. Nonhuman primate models for AIDS. Clin Infect Dis. 1993;17(Suppl 1):S230–S235. doi: 10.1093/clinids/17.supplement_1.s230. [DOI] [PubMed] [Google Scholar]
- 3.Daniel MD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 4.Reimann KA, et al. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harouse JM, Gettie A, Tan RC, Blanchard J, Cheng-Mayer C. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 6.Uberla K, et al. Animal model for the therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc Natl Acad Sci USA. 1995;92:8210–8214. doi: 10.1073/pnas.92.18.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambrose Z, et al. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J Virol. 2007;81:12145–12155. doi: 10.1128/JVI.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lackner AA, Veazey RS. Current concepts in AIDS pathogenesis: insights from the SIV/macaque model. Annu Rev Med. 2007;58:461–476. doi: 10.1146/annurev.med.58.082405.094316. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura Y, et al. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc Natl Acad Sci USA. 2004;101:12324–12329. doi: 10.1073/pnas.0404620101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimata JT, Kuller L, Anderson DB, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 11.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 12.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 18.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 19.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 22.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 23.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 24.Mariani R, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci USA. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 27.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;81:13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. Early replication block of human immunodeficiency virus type 1 in monkey cells. J Gen Virol. 1995;76(Pt 11):2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 30.Hatziioannou T, et al. Generation of simian-tropic HIV-1 by restriction factor evasion. Science. 2006;314:95. doi: 10.1126/science.1130994. [DOI] [PubMed] [Google Scholar]
- 31.Kamada K, et al. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc Natl Acad Sci USA. 2006;103:16959–16964. doi: 10.1073/pnas.0608289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman RM, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson SJ, et al. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao CH, Kuang YQ, Liu HL, Zheng YT, Su B. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. Aids. 2007;21(Suppl 8):S19–S26. doi: 10.1097/01.aids.0000304692.09143.1b. [DOI] [PubMed] [Google Scholar]
- 37.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 38.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz JE, et al. A nonhuman primate model for the selective elimination of CD8+ lymphocytes using a mouse-human chimeric monoclonal antibody. Am J Pathol. 1999;154:1923–1932. doi: 10.1016/S0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desrosiers RC. Prospects for an AIDS vaccine. Nat Med. 2004;10:221–223. doi: 10.1038/nm0304-221. [DOI] [PubMed] [Google Scholar]
- 41.Grant RM, et al. AIDS Promote HIV chemoprophylaxis research, don't prevent it. Science. 2005;309:2170–2171. doi: 10.1126/science.1116204. [DOI] [PubMed] [Google Scholar]
- 42.Tsai CC, et al. Prevention of SIV infection in macaques by (R)-9-(2-phosphonylmethoxypropyl)adenine. Science. 1995;270:1197–1199. doi: 10.1126/science.270.5239.1197. [DOI] [PubMed] [Google Scholar]
- 43.Agy MB, et al. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science. 1992;257:103–106. doi: 10.1126/science.1621083. [DOI] [PubMed] [Google Scholar]
- 44.Gartner S, et al. HIV-1 infection in pigtailed macaques. AIDS Res Hum Retroviruses. 1994;10(Suppl 2):S129–S133. [PubMed] [Google Scholar]
- 45.Kent SJ, et al. Cytotoxic and proliferative T cell responses in HIV-1-infected Macaca nemestrina. J Clin Invest. 1995;95:248–256. doi: 10.1172/JCI117647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igarashi T, et al. Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J Virol. 2007;81:11549–11552. doi: 10.1128/JVI.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato S, Johnson W. Antibody-mediated neutralization and simian immunodeficiency virus models of HIV/AIDS. Curr HIV Res. 2007;5:594–607. doi: 10.2174/157016207782418515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.