Abstract
Most bacteriophages possess long tails, which serve as the conduit for genome delivery. We report the solution structure of the N-terminal domain of gpV, the protein comprising the major portion of the noncontractile phage λ tail tube. This structure is very similar to a previously solved tail tube protein from a contractile-tailed phage, providing the first direct evidence of an evolutionary connection between these 2 distinct types of phage tails. A remarkable structural similarity is also seen to Hcp1, a component of the bacterial type VI secretion system. The hexameric structure of Hcp1 and its ability to form long tubes are strikingly reminiscent of gpV when it is polymerized into a tail tube. These data coupled with other similarities between phage and type VI secretion proteins support an evolutionary relationship between these systems. Using Hcp1 as a model, we propose a polymerization mechanism for gpV involving several disorder-to-order transitions.
Keywords: NMR structure, disordered regions, macromolecular assembly
Bacteriophages are likely the most abundant biological entities on the planet, with a global population estimated at 1031 (1). Of these phages, >80% possess dsDNA genomes packaged within an icosahedral head connected to a long (≈50–200 nm) tail (2). The tail serves as the conduit for genome injection. Long phage tails may be contractile, as found in the Myoviridae (e.g., phage T4), or noncontractile, as found in the Siphoviridae (e.g., phage λ). The tube of noncontractile tails is composed primarily of multiple copies of one protein, known as the major tail protein (MTP). During infection, the overall shape of the noncontractile tail is maintained. In contrast, contractile tails include a central tail tube protein (TTP) that is surrounded by a tail sheath protein (TSP). On infection, the tail sheath contracts and the TTP penetrates the outer cell membrane and cell wall (3). The marked structural and functional differences between contractile and noncontractile tails [supporting information (SI) Fig. S1A] raises an intriguing question as to whether any evolutionary relationship exists between them. This issue is difficult to address, because the extensive sequence divergence among phage proteins results in a lack of significant sequence similarity even among proteins that are clearly evolutionarily related. For example, sequence similarities between experimentally verified MTPs from different Siphoviridae are often undetectable, despite the high probability of a common origin for these proteins (4). In the absence of sequence similarity, an evolutionary connection between different proteins with similar functions can be inferred if they possess similar 3D structures (5). Structural similarities between phage and viral capsid proteins point to a common evolutionary origin for the heads of tailed dsDNA phage in bacteria and archea (6, 7), and eukaryotic viruses of the Herpes family (8). Similar comparative studies for phage tails have not yet been possible, because no structure of any tail tube or TSP has been reported.
One indication that contractile and noncontractile tails may share a common ancestor and assembly mechanism is provided by comparative analyses of phage genomes. All long-tailed phages possess a large gene (usually, >2 kbp) encoding a tape measure protein (TMP), which is responsible for precisely determining tail length. Genes encoding the TTPs are generally upstream of the TMP gene, and are separated from it by a gene encoding a tail assembly chaperone. This gene is distinguished by a highly conserved programmed translational frameshift that is observed in the genomes of both Siphoviridae and Myoviridae (9). The high conservation of this distinctive gene module among diverse phages is most easily explained by hypothesizing a common ancestor for these phages. However, a tape measure-like protein has also been identified as having a crucial role in assembly of the type III secretion system injectisome (10), and this type of protein may be a requisite for the formation of various unrelated needle-like structures where length must be regulated. In the case of phage tails and these other structures, the mechanisms by which polymerization of the tube subunits is initiated and controlled remain poorly understood.
To address the evolution and assembly mechanism of phage tails, we have carried out structural and functional studies on gpV, the MTP of phage λ. The noncontractile tail of λ is comprised of 32 hexameric rings of gpV, which are stacked like tires onto an initiator complex composed of 6 phage proteins including gpH, the TMP (Fig. S1B) (11); gpV does not assemble on its own, but readily polymerizes into a tube when the initiator complex is encountered. The extent of gpV polymerization is regulated by the presence of the TMP, and another protein, gpU, which serves to cap the finished tail when it reaches its proper length (12). The mechanism by which gpV is maintained in a mostly monomeric state in the absence of the initiator complex, and the locations of the interfaces involved in gpV hexamerization and polymerization have not been identified. In this work, we describe the NMR solution structure of the domain of gpV required for tail tube formation, the N-terminal domain of gpV (gpVN). The structure of gpVN displays a striking similarity to a previously solved but unpublished structure of a Myoviridae TTP, and to a hexameric ring-shaped component of a bacterial type VI secretion system. These structural similarities provide strong evidence for the common evolution of all long-tailed phages, and the type VI secretion system. Also, we present a model for the mechanism of gpV polymerization.
Results and Discussion
Solution Structure and Dynamics of gpVN.
The gpV is comprised of 2 domains, and only the N-terminal domain, gpVN, is required for assembly of functional phage particles (13). Here, we have used NMR spectroscopy to solve the solution structure of a gpVN construct comprising gpV residues 1–153, boundaries that we determined through bioinformatic analysis (14). An ensemble of the 10 lowest energy structures for gpVN are shown in Fig. S2A. The structured regions of the protein are well defined, having a backbone rmsd of 0.69 Å (Table S1). However, 3 regions, spanning residues 1–14, 50–78, and 149–153, displayed no long-range NOEs, and could be assigned no fixed structure. The disordered nature of these regions was confirmed through an assessment of {1H}-15N heteronuclear NOEs, which indicated that they are dynamic on the ns-ps timescale (Fig. S2B), similar to the behavior of an unfolded protein. Strikingly, these 3 regions account for ≈30% of the entire domain.
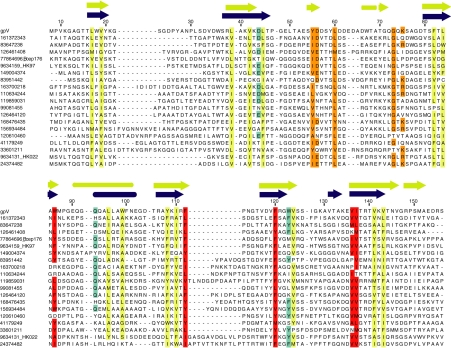
The structured part of gpVN is composed of 7 β-strands and 1 α-helix. The secondary structure assignments were confirmed by examination of short-range NOE data and NMR chemical shift information. The 7 strands are arranged into 2 antiparallel sheets, which fold into a twisted β-sandwich conformation. Sheet 1 is formed from strands 1, 2, 4, and 5, whereas sheet 2 is formed from strand 2, 3, 6, and 7. The α-helix is located between strands 3 and 4 (Fig. 1A). The high quality of this structure is reflected by its well-packed core consisting of 16 hydrophobic residues that display <10% solvent accessibility (Fig. 1B). In an alignment of a very diverse set of gpVN homologues collected through multiple iterations of PSI-BLAST (Fig. 2), an almost complete conservation of hydrophobicity is seen at 14 of the 16 core positions, which is remarkable in light of the low sequence identity among these proteins. Also, secondary structure prediction performed on the whole alignment (15) showed concordance at every secondary structure element in gpVN, except for the short β6-strand. The agreement between the gpVN structure and the alignment data strongly suggests that all of these homologues adopt a tertiary structure similar to gpVN.
Fig. 1.
Solution structure of gpVN. (A) A ribbon representation of the lowest energy structure of gpVN. (B) The 16 hydrophobic core side-chains are represented as yellow bonds. Strands are colored in dark blue, the helix in red, and loops in gray. A and B were generated by using PyMOL (http://pymol.sourceforge.net/).
Fig. 2.
Alignment of diverse homologues of gpV. The gpVN homologues shown here are representative of many sequences collected through many iterations of PSI-BLAST (for details, see Methods). These sequences were chosen to maximize diversity, and are derived mostly from prophages in various bacteria (sequences are designated by National Center for Biotechnology Information GI nos.). Sequences taken from characterized phages are indicated. The average pairwise identity of these sequences to gpVN is 17%, with no sequence being >23% identical. The average pairwise identity among all of the sequences in the alignment is 18%. No 2 sequences in the alignment are >37% identical. Nonconserved sequences at the N termini of these proteins were truncated (gpV starts at Met 8). In the last 2 sequences, a large loop after β-strand 4 is truncated. The 14 highly conserved hydrophobic core positions are indicated in yellow. Conserved nonpolar positions in the structured regions of gpVN that are >20% exposed in the gpVN monomeric structure are indicated in red. Conserved residues within the middle unstructured loop are indicated in orange, and additional residues that are conserved and found at hexamer interface positions are indicated in cyan. Experimentally derived secondary structure elements are indicated on top of the alignment in blue with the computationally predicted secondary structure shown in yellow (15).
Central Disordered Region of gpVN Is Functionally Important.
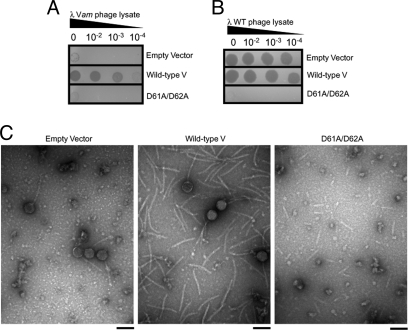
Disordered regions are a hallmark of many unassembled bacteriophage morphogenetic proteins, and the functional importance of these regions has been demonstrated in several cases (16, 17). A potentially important feature of the large unstructured region in the middle of gpVN (residues 50–78) is its high concentration of negatively charged residues, given that electrostatic interactions were previously implicated in the polymerization of gpV hexamers (18). To investigate the role of this region and the charged residues within it, we substituted a tandem pair of Asp residues at positions 61 and 62 with Ala. The biological activity of this mutant was assessed by using an in vivo complementation assay whereby a λ phage that is unable to make gpV (Vam mutant) is complemented by gpV expressed from a plasmid. It can be seen that the D61A/D62A mutant is completely devoid of biological activity (Fig. 3A), proving that this region of gpV is critical for its function. Also, this mutant also displays a dominant negative phenotype in that it inhibits the growth of wild-type λ phage (Fig. 3B). This effect implies that the D61A/D62A mutant incorporates into tail assembly intermediates, but that its presence inhibits further tail formation. Because gpV does not hexamerize until it is incorporated into the tail, this behavior is most simply explained if the mutant is still able to form hexamers on interacting with the tail initiation complex, but is debilitated in the hexamer-hexamer interaction required for the formation of a tail tube. To test this hypothesis, we overexpressed either wild-type or mutant gpV, while simultaneously inducing a wild-type prophage. The resulting phage particles were examined by using transmission electron microscopy. Fully assembled phages were observed in the empty vector negative control indicating that phage induction and assembly was successful (Fig. 3C). Overexpression of wild-type gpV resulted in the presence of fully assembled phage and an abundance of free λ tails of wild-type length (Fig. 3C). Remarkably, the overexpression of the D61A/D62A mutant resulted in the formation of short tails (Fig. 3C). Although fully assembled phage and tails of normal length were also observed, the most abundant specimen found on the grid was short tails. This observation supports the idea that the D61A/D62A mutant inhibits the hexamer-hexamer interactions involved in tail polymerization.
Fig. 3.
Role of the central disordered loop in λ tail assembly. (A) The ability of the D61A/D62A gpV mutant expressed from a plasmid to complement a λ Vam lysate in vivo was assessed by observing plaque formation. The “empty vector” experiment constitutes the negative control. As indicated, 3 different dilutions of the Vam lysate were spotted onto the plates. (B) The effect of plasmid-expressed WT gpV and the D61A/D62A mutant on the growth of WT λ phage was assayed. A WT phage lysate was spotted at the indicated dilutions on cells carrying these plasmids. (C) Representative electron micrographs of phage particles produced when a WT λ prophage was induced in the presence of empty vector, overexpressed wild-type gpV, or overexpressed D61A/D62A gpV. (Scale bar, 100 nm.)
Structural Similarity Between gpVN and a Tail Tube Protein from a Contractile-Tailed Phage.
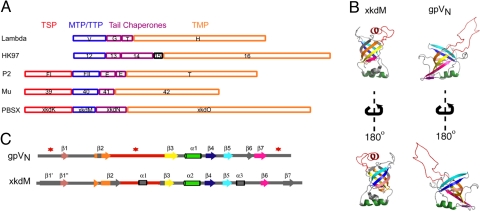
A search for similar structures in the PDB by using VAST (19) yielded many significant hits. Several of the similar structures were members of the PilZ domain, which binds bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) and is involved in bacterial signaling (20). Some other similar structures were members of the pyridoxamine 5′-phosphate oxidase families (21). Although these families of proteins do not appear to be functionally relevant to gpVN, 1 significant hit (PDB ID: 2GUJ, unpublished structure by the Northeast Structural Genomics Consortium) corresponded to XkdM, a protein encoded within the well characterized B. subtilis PBSX prophage, which produces Myoviridae type particles when induced (22). XkdM comprises the tail tube of this particle (23). As would be expected for a tail-tube protein, the gene encoding XkdM is located between the gene encoding the tail sheath and TMP of PBSX (Fig. 4A). Surprisingly, the ORF immediately downstream of XkdM, XkdN, does not appear to encode the conserved frameshift seen in most long-tailed phage, suggesting that this frameshift may not be imperative for all phages.
Fig. 4.
Structural similarity between gpVN and a TTP from a contractile-tailed phage. (A) Gene order conservation among tail proteins from λ (Siphoviridae), HK97 (Siphoviridae), P2 (Myoviridae), Mu (Myoviridae), and PBSX (Myoviridae). In each case, the tail chaperones (gpG and frameshift product, gpGT, in λ), colored purple, are located between the gene encoding the MTP or TTP, colored blue, and the gene encoding the TMP, colored orange. In Myoviridae, the gene encoding the TSP, colored red, is located upstream of the TTP. It should be noted that PBSX was previously reported to possess the conserved frameshift (9), but this was an error. (B) Tertiary and (C) secondary structure comparison between gpVN and XkdM, the TTP from PBSX. Each secondary structure element in XkdM is color-coded based on its tertiary alignment with gpVN (β1, salmon; β2, orange; β3, yellow; β4, blue; α1, green; β5, sky blue; β6, purple; and β7, deep pink). Loops and secondary structure elements that do not align are colored in gray. In C, regions that are disordered in gpVN are denoted with a red star.
gpVN and XkdM possess strikingly similar folds (Fig. 4B), and can be overlaid with an RMSD of 1.9 Å over 57 equivalent Cα positions. The topology of the 2 proteins is essentially the same, except that gpVN displays disordered regions at its N and C termini where strands are present in XkdM (Fig. 4C). The common core tertiary structure of each consists of 2 orthogonally packed sheets. Both proteins have large loops between strands 2 and 3. Although this loop is completely disordered in gpVN, it forms a structured loop with a helix in XkdM that folds back into the top of the structure (Fig. 4 B and C). This conformation could be induced through the crystallization process, and this region may be disordered when XkdM is in solution. One other difference between the structures is the presence of a helix between strands 5 and 6 in XkdM, where gpVN has a short strand.
The strong structural similarity between gpVN, a noncontractile TTP, and XkdM, a contractile TTP, provides direct evidence that these 2 types of tails are structurally related at the molecular level. Also, this structural similarity combined with the gene order conservation seen in most Siphoviridae and Myoviridae genomes (Fig. 4A) provides compelling evidence of an evolutionary link between the distinctive tails used by these phages. The inability to trace the evolutionary link through sequence homology (gpVN and XkdM display no significant sequence similarity) might suggest that this structural similarity arises through convergent evolution; however, the lack of sequence similarity is not surprising, given that tube proteins within the same phage families often display no detectable sequence similarity. The apparent common structure and evolution of the contractile and noncontractile TTPs raises intriguing questions as to whether the sheath protein evolved to interact with the tail tube of a common progenitor phage, or whether a contractile-tailed progenitor lost its sheath at some point to give rise to the noncontractile tail family.
Phage λ Tails Are Structurally Related to the Hcp1 Type VI Secretion System Protein.
Although the structural similarity between gpVN and XkdM is striking, an even stronger structural similarity was found between gpVN and Hcp1 (PDB ID: 1Y12), which is an essential component of a type VI secretion system from Pseudomonas aeruginosa (24). Hcp1 and gpVN can be overlaid at 74 equivalent Cα positions with an RMSD of 2.1 Å. The topology and tertiary structure of these proteins are the same with only minor deviations (Fig. 5 A and B). The most prominent difference is the presence of 3 extra strands at the C terminus of Hcp1 where gpVN has a disordered region. Remarkably, within its crystal lattice, Hcp1 is assembled into hexameric rings that stack on each other to form tubes (Fig. 5C). The dimensions of the Hcp1 hexameric rings (90 Å, with a 40-Å central hole) are strikingly similar to the dimensions of the hexameric rings of gpV in λ tails (90 Å, with a 30-Å central hole; see ref. 13).
Fig. 5.
Structural similarities between gpVN and Hcp1. (A) Tertiary and (B) secondary structure comparison between gpVN and Hcp1. Each secondary structure element in Hcp1 is colored-coded based on its alignment with gpVN (β1, salmon; β2, orange; β3, yellow; β4, blue; α1, green; β5, sky blue; β6, purple; and β7, deep pink). Loops and secondary structure differences are colored in gray, with the exception of the β2-β3 loop that is colored red. In B, disordered regions in gpVN are denoted with a red star. (C) Superposition of gpV (blue) onto monomer A of the Hcp1 hexamer (yellow). Crystallographic symmetry was applied to build the stacked Hcp1 hexamers. The β2-β3 loop of Hcp1 is represented in red spheres, which are clearly located in the hexamer-hexamer interface. (D) Structural superpositions suggest that the disordered C-terminal residues of gpVN could form a strand. For ease of visualization, in the superposition of Hcp1 and gpVN, residues 32–56 and 138–162 from Hcp1 and 49–82 from gpVN have been removed, and in the superposition of XkdM with gpVN, residues 33–59 from XkdM and 49–82 from gpVN have been removed. The folding of the gpVN C terminus (deep pink) into the Hcp1 β8′ (deep pink) or the XkdM β7 position (deep pink) are likely conformational changes. (E) A side view of 3 monomers from the Hcp1 hexamer where monomer A has been replaced with gpVN (blue). Highly conserved hydrophobic residues are colored red, whereas relatively conserved residues are colored in cyan. Conserved residues in the β2-β3 loop are orange. Residues contributing to the hexameric interface are circled in black (Asp-44, Trp-86, Gln-93, Trp-123, Phe-112, Phe-120, Val-136, and Ile-137).
The structural similarity between gpVN and Hcp1 supports a growing body of data pointing to an evolutionary relationship between components of the type VI secretion system and phage tails. Leiman et al. report close structural similarities for VgrG, another key component of the type VI system, and the tail spike proteins, gp27 and gp5, from bacteriophage T4 (Myoviridae) (35). In Vibrio cholerae, the VgrG secreted proteins have been shown to interact with one another, and it has been speculated that they may form a trimeric complex analogous to the (gp27)3(gp5)3 membrane-puncturing tail spike complex from bacteriophage T4 (25). Hcp1 is absolutely required for both the assembly of the type VI secretion apparatus and for the localization of ClpV1, a AAA+ family protein that provides the energy for Hcp1 secretion (24). The proclivity for Hcp1 to form tail-like tubes in solution has been demonstrated by engineering a disulfide bond at the hexamer-hexamer interface observed in the crystal structure (26). Also, the accompanying article reports that a homologue of Hcp1 is able to form tubes spontaneously. Because of its tube-forming ability and its high level of expression, it is believed that Hcp1 forms the injection needle for secretion. Because gpV does form a long tube through which DNA and proteins are transported, the hypothesis that Hcp1 may assemble into a comparable structure for the intercellular transport of proteins and/or oligonucleotide-type substrates is strongly supported by the structural similarity between these 2 proteins.
Model for gpV Polymerization Based on Hcp1.
In light of the structural and putative functional similarities between gpVN and Hcp1, we believe that the oligomerization mode of gpV within the phage tail is similar to that seen for Hcp1 within the crystal lattice. However, it is important to note that gpVN is monomeric in solution, under the conditions of the NMR experiments, whereas Hcp1 is hexameric in solution (24). Also, gpVN possesses significant regions of disorder. We predict that some or all of these disordered regions would gain structure on gpV polymerization and have a key role in facilitating this process. For example, the large disordered region from residues 50–78 in gpVN corresponds to a long loop in Hcp1 that interacts with the neighbouring subunit in the hexameric ring sits on the upper surface of the ring (Fig. 5C), where it can interact extensively with the next hexamer sitting on top. The unstructured loop in gpVN likely becomes ordered on gpV polymerization in a manner similar to Hcp1, a supposition which is supported by our mutagenesis data (Fig. 3 A and B), suggesting that residues within this loop may have a key role in the polymerization process. The importance of this loop in gpV function is also underscored by the occurrence of several highly conserved positions in this region (residues Tyr-55, Leu-60, and Gly-73) and the minimal degree of loop length variation observed here compared with other loops in the alignment (Fig. 2). The C-terminal residues of gpVN are disordered. Because this region forms a β-strand lying on the outside of the ring in Hcp1, a strand may also form here in gpV on assembly (Fig. 5D). This idea is bolstered by the computational prediction of a β-strand in this region of the gpVN alignment (Fig. 2), and by the occurrence of a strand at this position in the structure of XkdM (Fig. 5D). Last, the C-terminal domain of gpV has been seen as a protrusion on the exterior of the λ tail (13). The formation of the proposed strand at the end of gpVN would bring the terminus of this domain to the outside surface of the hexameric ring, where the C-terminal domain would be located.
Our model for the oligomerization of gpV based on the Hcp1 structure is supported by the gpVN sequence alignment. We detected 5 highly conserved hydrophobic positions in structured portions of gpVN that were >20% exposed in the monomeric structure (Fig. 2). Remarkably, the superposition of the gpVN monomer onto a subunit of Hcp1 reveals that each of these residues is positioned close to the hexamerization interface (Fig. 5E). Another 8 positions display relatively high conservation in that at least half of the residues at each position are identical, or they maintain charge or aromatic character. Five of these positions lie in the large central unstructured loop (Fig. 2), and 3 others are found at the putative hexamerization interface (Figs. 2 and 5E). The gpVN alignment has 2 regions exhibiting large insertions and/or deletions. Both of these regions comprise loops that would lie on the outside of the putative gpV ring in positions where Hcp1 also possesses large loops. In summary, the structural and bioinformatic data available supports the hypothesis that gpV forms a hexameric ring in a manner similar to Hcp1.
The presence of large unstructured regions within gpVN, which we hypothesize become structured on polymerization, provides a putative mechanism for the strict control of tail assembly during λ morphogenesis. Because gpV has the potential to polymerize into aberrantly long nonfunctional structures (18), premature association between gpV monomers must be prevented. The unstructured regions could serve this purpose such that gpVN remains monomeric even at the very high concentrations used in this NMR study (≈1 mM). On encountering the tail initiator complex, gpV commences polymerization in a process that is expected to include the ordering of unstructured regions. This ordering is likely facilitated by the initiator complex, and results in the formation of a surface that can participate in further polymerization until the end of the TMP is reached (Fig. S1B). Interestingly, when the λ tail is dissociated at low pH, individual hexameric rings of gpV are obtained, indicative of a major structural change occurring in gpV on oligomerization that prevents dissociation into monomers. The conformational changes in gpV may involve multiple steps of disordered to order transitions, because the 3 disordered regions may not all fold in the same morphogenic step.
Conclusions
The 3D structure of gpVN provides direct evidence that contractile and noncontractile phage tails evolved from a common ancestor. The structural similarities between the TTPs described here indicate that all long phage tails may be assembled from a common repository of components, and a common assembly mechanism may exist. The structural similarity observed between gpVN and the hexameric type VI secretion protein, Hcp1, strongly supports the hypothesis that phage tails and type VI secretion systems are structurally, functionally, and evolutionarily related, and that Hcp1 forms the tube of this bacterial secretion apparatus, just as gpVN forms the tubular structure of the phage tail. Because bacterial secretion systems are attractive drug targets, understanding the structural components and mechanisms of their assembly are imperative to the development of successful inhibitors. New possibilities for the application of knowledge from studies of tailed phages to the investigation of type VI secretion will dramatically increase the rate of knowledge acquisition in this emerging field. The inability of Hcp1 and its homologues to readily form tubes under normal solution conditions implies that, just as in the phage system, other factors, which remain to be discovered, are necessary for efficient polymerization of this protein.
Last, it is interesting to note that type VI secretion systems are not the first known example of bacteria evolving to use phages or parts of phages for their own advantage. Most strains of P. aeruginosa produce phage tail-like entities, known as Pyocins, that are able to selectively kill other P. aeruginosa strains (27). The operons encoding Pyocins are undoubtedly derived from phage genomes, because significant sequence similarity is observed between Pyocin proteins and various phage tail proteins. Also, phage-derived particles, called gene transfer agents, have an important role in mediating lateral gene transfer in many species of alpha-proteobacteria (28). Together, these observations emphasize the importance of phage-derived elements in the evolution and function of various complex bacterial systems.
Methods
Plasmid Construction and Protein Purification.
A region encoding gpV residues 1–153 was cloned into a modified pET32b expression vector (Novagen) to produce a 6-His tagged fusion with thioredoxin. Quikchange mutagenesis (Stratagene) was used to make all mutations. For in vivo complementation assays, full-length gpV and mutants were cloned into a pTrc99c vector (29). The gpVN construct was expressed in BL21 CodonPlus (Stratagene) with overnight incubation at 25 °C after induction. Purification was performed under native conditions by using Ni-NTA agarose following the standard protocol (Qiagen).
NMR Spectroscopy and Structure Calculation.
NMR studies on gpVN were carried out at protein concentrations of ≈1.0 mM in 50 mM Na2HPO4, pH 6.8/200 mM NaCl or in 50 mM Na2HPO4, pD 7.2/200 mM NaCl made by using 99.9% D2O. All NMR spectra were collected at 20 °C on a Varian INOVA 500 or 800 MHz spectrometer equipped with pulsed field gradients at the Québec/Eastern Canada High Field NMR Facility. Structure calculations were performed by using CYANA 2.1 (30). PROCHECK-NMR (31) was used for structure validation.
In Vivo Assay for gpV Activity.
BL21 (DE3-Δtail) (32) cells freshly transformed with either empty pTrc99c vector, or plasmids expressing WT or mutant gpV, were suspended in molten 0.7% top agar supplemented with IPTG to a final concentration of 0.5 mM, then poured onto LB/Agar plates containing 10 mM MgSO4. Serial dilutions of a λ Vam lysate were spotted in 4-μL aliquots on top of the cells, and the degree of complementation was scored by observing plaque formation after 24-h incubation at 37 °C. The activity of D61A/D62A was tested in the context of full-length gpV.
Electron Microscopy.
Samples were prepared by freshly transforming the Escherichia coli strain 594 carrying a λ prophage with wild-type morphogenic genes (λcI857Sam7), with either empty pTrc99c or pTrc99c plasmids expressing gpV-WT or gpV-D61A/D62A. Cells were grown to a final density of ≈0.8, and phage production and protein overexpression were induced simultaneously. Cells were then grown at 37 °C for 3 h, harvested by centrifugation, and lysed by vortexing in the presence of CHCl3. The resulting phage lysates were applied to the surface of a continuous carbon film coated EM grid and stained with 2% (wt/vol) uranyl acetate. Grids were examined with a Hitachi H-7000 microscope.
Construction of a Sequence Alignment of gpVN Homologues.
A PSI-BLAST (33) search was initiated by using residues 1–153 of gpV. Convergence was reached after 13 iterations. Sequences were aligned by using MUSCLE (34). The number of sequences in the alignment was iteratively reduced by taking single representatives of identified sequence subfamilies. Ultimately, a set of sequences with maximal diversity that could still be aligned easily by automated procedures was produced. Secondary structure prediction was carried out by using JPred (15).
Additional details regarding cloning and purification, NMR, electron microscopy, and sequence alignment construction can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Tara Sprules and Ranjith Muhandiram for NMR data acquisition, Andrew Chong for helpful discussions, and Paul Sadowski and Karen Maxwell for careful reading of the manuscript. This work was supported by the Canadian Institutes of Health Research Operating Grants Fund MOP-77680 (to A.R.D.) and MT13337 (to P.L.H.). P.L.H is the recipient of a Canada Research Chair. L.G.P. and V.K. were supported in part by a University of Toronto Open scholarship and a Canadian Cystic Fibrosis Foundation fellowship, respectively. NMR experiments were recorded at the Québec/Eastern Canada High Field NMR Facility, supported by grants from the Canada Foundation for Innovation, the Québec Ministère de la Recherche en Science et Technologie, and McGill University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates for the gpVN structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2K4Q). Chemical shift assignments have been deposited in the BioMagResBank database (accession no. 15807).
See Commentary on page 4067.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900044106/DCSupplemental.
References
- 1.Brussow H, Hendrix RW. Phage genomics: Small is beautiful. Cell. 2002;108:13–16. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann HW, Kropinski AM. Curated list of prokaryote viruses with fully sequenced genomes. Res Microbiol. 2007;158:555–566. doi: 10.1016/j.resmic.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Rohwer F, Edwards R. The Phage Proteomic Tree: A genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray JE, Todd AE, Pearl FM, Thornton JM, Orengo CA. The CATH Dictionary of Homologous Superfamilies (DHS): A consensus approach for identifying distant structural homologues. Protein Eng. 2000;13:153–165. doi: 10.1093/protein/13.3.153. [DOI] [PubMed] [Google Scholar]
- 6.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: The pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr Biol. 2006;16:R11–R13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 11.Katsura I. Lambda II. Plainview, NY: Cold Spring Harbor; 1983. Tail Assembly and Injection; pp. 331–346. [Google Scholar]
- 12.Katsura I, Tsugita A. Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail. Virology. 1977;76:129–145. doi: 10.1016/0042-6822(77)90290-2. [DOI] [PubMed] [Google Scholar]
- 13.Katsura I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J Mol Biol. 1981;146:493–512. doi: 10.1016/0022-2836(81)90044-9. [DOI] [PubMed] [Google Scholar]
- 14.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 15.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: A consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell KL, et al. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, et al. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 18.Bleviss M, Easterbrook KB. Self-assembly of bacteriophage lambda tails. Can J Microbiol. 1971;17:947–954. doi: 10.1139/m71-151. [DOI] [PubMed] [Google Scholar]
- 19.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 20.Ramelot TA, et al. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 21.Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- 22.Mauel C, Karamata D. Characterization of proteins induced by mitomycin C treatment of Bacillus subtilis. J Virol. 1984;49:806–812. doi: 10.1128/jvi.49.3.806-812.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero P, Lopez R, Garcia E. Genomic organization and molecular analysis of the inducible prophage EJ-1, a mosaic myovirus from an atypical pneumococcus. Virology. 2004;322:239–252. doi: 10.1016/j.virol.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 24.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 28.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 30.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs KA, et al. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol Microbiol. 2004;53:1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiman , et al. Type VI secretion apparatus and phage-tail associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.