Abstract
Drug-induced liver injury is a frequent side effect of many drugs, constitutes a significant threat to patient health and has an enormous economic impact on health care expenditures. Numerous efforts have been made to identify reliable and predictive markers to detect the early signs of drug-induced injury to the liver, one of the most vulnerable organs in the body. These studies have, however, not delivered any more informative candidates than the serum aminotransferase markers that have been available for ≈30 years. Using acetaminophen overdose-induced liver injury in the mouse as a model system, we have observed highly significant differences in the spectrum and levels of microRNAs in both liver tissues and in plasma between control and overdosed animals. Based on our survey of microRNA expression among normal tissues, some of the microRNAs, like messenger RNAs, display restricted tissue distributions. A number of elevated circulating microRNAs in plasma collected from acetaminophen-overdosed animals are highly expressed in the liver. We have demonstrated that specific microRNA species, such as mir-122 and mir-192, both are enriched in the liver tissue and exhibit dose- and exposure duration-dependent changes in the plasma that parallel serum aminotransferase levels and the histopathology of liver degeneration, but their changes can be detected significantly earlier. These findings suggest the potential of using specific circulating microRNAs as sensitive and informative biomarkers for drug-induced liver injury.
Keywords: acetaminophen overdose, plasma, miRNA, toxicity, ALT
Drug-induced liver injury is a serious clinical problem and is the leading cause of drugs being removed from the market (1, 2). Many studies have been conducted to identify more reliable and sensitive early blood markers for liver injury by using various high throughput technologies such as protein mass spectrometry and gene expression arrays (3, 4). These efforts have so far yielded candidates that perform no better than the existing aminotransferase-based markers.
Acetaminophen is one of the most commonly used medicines with a very high safety profile when used properly. If misused, either intentionally or accidentally, acetaminophen can cause significant liver injury (5, 6). In acute overdose, the normal sulfation and glucuronidation pathways involved in metabolizing acetaminophen become saturated, and the excess drug in the body is then processed via the monooxygenase cytochrome P450 enzymes, specifically by the CYP2E1, CYP1A2, and CYP3A4 subfamily members, to a toxic metabolite, N-acetyl-p-benzoquinone-imine (NAPQI) (7, 8). NAPQI is a highly reactive electrophile but can be quickly neutralized by binding with intracellular glutathione (GSH). In conditions such as acute acetaminophen overdose, where the level of GSH in the cell is insufficient to eliminate all of the toxic metabolite, the excess intracellular NAPQI forms cysteine adducts on various macromolecules, disrupts numerous critical cellular functions, and elevates innate immune responses (9), which collectively result in massive centrilobular hepatocyte death (5).
N-acetylcysteine (NAC) is an amino acid derivative that can be quickly converted into intracellular GSH in the body (10). NAC can effectively prevent and reduce acetaminophen-induced liver injury if it is promptly diagnosed and NAC is administered within 8–10 h after the initial ingestion (11). However, acetaminophen overdose usually produces either no immediate symptoms or nonspecific intestinal irritation during the first 24 h after ingestion, followed by the onset of liver failure. Thus, the need for early and accurate blood-based diagnosis for acetaminophen overdose is acute.
The most commonly used diagnostic test for acetaminophen overdose is to determine the activity of certain hepatocellular enzymes, aspartate aminotransferase (AST or SGOT) and alanine aminotransferase (ALT or SGPT), in the blood (12). Ideally, the patient may disclose the amount ingested and the time of ingestion; however, this information is often unreliable (13). The levels of acetaminophen in the blood combined with the Rumack–Matthew nomogram, a diagram to assess the risk of hepatotoxicity based on blood acetaminophen concentration (14), can also be used as guidance to predict tissue injury (15). Because the time of ingestion often is not reliable, and the blood acetaminophen level usually peaks during the first few hours after ingestion, a low serum acetaminophen level does not preclude a high level of acetaminophen exposure. The level of plasma NAPQI-protein adducts correlates well with the serum aminotransferase levels and has been demonstrated as a good indicator for acetaminophen toxicity (16). However, the protein adducts are measured by high-performance liquid chromatography or mass spectrometry (17) and are not used routinely in most clinics.
MicroRNAs (miRNA) are small regulatory, noncoding RNAs. It is believed that miRNAs primarily affect the stability of mRNA and/or the initiation and progression of protein translation, but broader regulatory roles have been suggested (18, 19). Even though the biological function of miRNA is yet to be fully understood, it has been shown that the tissue levels of specific miRNAs correlate well with the pathological development of several different cancers (20, 21). Recently, finding miRNA in the blood has suggested the potential for miRNA-based blood biomarkers in cancer detection (22, 23). We hypothesized that the levels of specific circulating miRNA species may also be used to detect and monitor the pathological development associated with drug-induced tissue injuries. Using a mouse model, we report here a set of plasma miRNAs whose levels are indeed associated with hepatocellular injuries induced by acetaminophen overdose.
Results
The blood ALT level, a widely used biomarker for liver function in both animal models and human patients, was determined in all plasma samples collected in the experiments. The samples from mice 24 h after i.p. injection with a single LD50 dosage at 300 mg/kg of acetaminophen showed a significant elevation of ALT levels (13,269 ± 1,253; n = 4) compared with the control plasma samples (37 ± 4; n = 4), and the ALT levels relate well with the degree of tissue injuries induced by acetaminophen as ascertained by histopathological examination.
Dynamic Range of miRNA Expression Levels Is Similar but the Spectra Are Different Between Liver and Plasma in Both Control and Acetaminophen-Overdosed Animals.
We adapted a microarray platform from Agilent to profile the miRNA spectra. The array contains probes representing 576 mouse and 10 viral miRNA sequences. The correlation coefficient of hybridization results among control samples either from plasma or liver tissues were high (ranged from 0.95 to 1.00, n = 4), as expected, and for treated samples, the correlation coefficients were slightly lower (r = 0.73–0.99, n = 4), which may have been caused by different degrees of response to acetaminophen administration among different animals. Like an mRNA gene expression array, the low-expressing miRNAs showed a much higher level of variation among different samples.
miRNA spectra in liver tissues.
The miRNA expression levels in liver tissues were distributed over 4 orders of magnitude, and a significant number of miRNAs were either not expressed or expressed at very low levels as described (24, 25). Excluding any miRNA with hybridization intensity <1.5 times the global mean intensity resulted in 38 oligonucleotide probes representing 23 miRNAs from the control liver samples and 96 oligonucleotide probes representing 40 miRNAs in the treated group [summary in Table 1 and list in supporting information (SI)]. There were 8 miRNA species that were observed in both control and treated liver samples. Mouse mir-122 showed the highest expression level in the control liver samples, whereas mir-1224 was highest in the treated group.
Table 1.
Summary of numbers of miRNA observed under different conditions
| Category | No. of observed miRNAs |
No. of changed miRNAs after acetaminophen treatment compared with control | ||
|---|---|---|---|---|
| Control | Acetominophen treated | Common between control and acetaminophen treated | ||
| Liver | 23 | 40 | 8 | 51 |
| Plasma | 43 | 53 | 29 | 44 |
| Common between liver and plasma | 10 | 12 | 5 | 20 |
miRNA spectra in plasma samples.
The hybridization intensity of miRNA circulating in the plasma was also distributed over 4 orders of magnitude, a similar range observed with various tissues and cell types. By using the same selection criterion as above, there were 84 oligonucleotide probes representing 43 miRNAs from the control plasma samples and 88 oligonucleotide probes representing 53 miRNAs in the treated group (summary in Table 1 and list in SI). Among these, there were 29 miRNA species that were represented in both groups, a much higher number than in liver tissue (Table 1).
Combining the detectable miRNA from liver and plasma samples revealed a total of 93 different miRNA species, where 67 of them are seen in plasma samples from either the control or treated animals and 55 of them in liver samples. With all 93 observed miRNAs, 8 of them were in all of the liver samples (control and treated-livers), 29 of them were in all of the plasmas, and only 5 miRNAs were seen in both liver and plasma samples under all conditions (Table 1). Because many different cell types and tissues in the body might contribute to the composition and the level of miRNAs in the plasma, comparison of the composition of detectable miRNAs between plasma and liver tissue revealed very little overlap, 10 between liver and plasma samples from control animals and 12 from treated animals (Table 1). Besides the different spectra, the number of observable miRNAs is also higher in plasma compared with the liver tissue.
Circulating miRNA in Plasma May Have a Complex Tissue Origin.
To further investigate the origin of circulating miRNA in both control and acetaminophen-overdosed plasma samples, we performed microarray-based miRNA profiling on 6 major organs, brain, heart, kidney, liver, lung, and spleen from normal mice (see SI). Among these 6 organs, 167 probes representing 100 different miRNAs showed an expression level >1.5 times the global mean in at least 1 of the tissues. A significant number of these probes, 71 of 167, were observed only in 1 tissue, suggesting that some miRNAs have relative tissue-specific distributions—at least with in the limits of the tissues examined.
Tissue Origin of miRNA in Plasma.
Comparing the list of the 43 observed miRNA species in control plasma (see SI) with the expression profiling of 6 different normal tissues, 6 of the 43 miRNAs were predominantly expressed in brain, 7 in heart, 2 in liver, 5 in lung, and 5 in spleen, and 18 of them were not expressed in any of the 6 tissues surveyed (see SI). We did not observe any circulating miRNA predominantly expressed in the kidney, even though there were quite a few miRNAs expressed in the kidney along with other tissues.
In acetaminophen-overdosed plasma samples, 40 of 53 (see SI) observed circulating miRNAs were also observed in the tissues that were surveyed. Among them, 9 were predominantly expressed in brain, 8 in heart, 1 in kidney, 7 in liver, 9 in lung, and 6 in spleen, a significant increase of miRNA species from brain, liver, and lung compared with the control plasma (see SI). This suggests, as expected, that acetaminophen overdose may be toxic for organs other than just the liver. In both control and acetaminophen-overdosed plasma samples, a significant number of the circulating miRNA species were not observed among the 6 organs surveyed. This suggests that the composition of circulating miRNA may originate from diverse tissues and cell types in the body.
Acetaminophen Exposure Induced Changes in the Expression of miRNAs in Liver and Plasma.
Acetaminophen overdose affected not only the total number of detectable miRNAs in the mouse liver and plasma but also the relative expression levels of a significant number of them. Like many transcription microarray experiments, a >2-fold change on microarray hybridization intensity was adopted as a selection criterion for differentially expressed miRNA.
Effects of acetaminophen overdose on miRNA spectra in liver tissues.
In the liver samples, a large fraction, 33 of 51 (65%), of differentially expressed miRNA species showed higher expression levels when compared with the control tissues. The top increased miRNA in the liver 24 h after acetaminophen overdose was mir-711, with a 124-fold increase (6.95 in log2 scale; Table 2), and the strongest decreased miRNA species was mir-29b with a8-fold decrease based on array hybridization results (3.06 in log2 scale; Table 2). The changes of miRNA levels in the liver could be due to cellular responses toward acetaminophen overdose. Using miRPortal (www.mirportal.net), a web tool summarizing the relationship between miRNA and metabolic pathways, pathways including antigen processing and presentation, apoptosis, B cell receptor signaling pathways, cytokine–cytokine receptor interaction, and cell cycles are all potentially affected by miRNA species with decreased levels in the liver after acetaminophen exposure. Pathways involved in various signaling transduction and cell–cell interactions such as gap-junctions, focal adhesion, and MAPK signaling pathways are potentially affected by miRNAs species with increased levels in the liver (Table 2). This may suggest a general breakdown of tissue integrity and an increase in immune responses and tissue repairing processes after acetaminophen overdose.
Table 2.
List of affected microRNA species by acetaminophen overdose
| Liver tissues (24 h after exposure) |
Plasma samples (24 h after exposure) |
||
|---|---|---|---|
| miRNA name | Fold change (log2) treated/control array hyb intensity | miRNA name | Fold change (log2) treated/control array hyb intensity |
| mmu-miR-29b | −3.06 | mmu-miR-710 | −4.69 |
| mmu-miR-122 | −2.71 | mmu-miR-720 | −4.32 |
| mmu-miR-15a | −2.65 | mmu-miR-205 | −3.77 |
| mmu-miR-29a | −2.65 | mmu-miR-711 | −3.70 |
| mmu-miR-130a | −2.28 | mmu-miR-486 | −3.63 |
| mmu-miR-29c | −2.14 | mmu-miR-133a | −2.92 |
| mmu-miR-192 | −2.12 | mmu-miR-124 | −2.87 |
| mmu-miR-194 | −2.11 | mmu-miR-133b | −2.76 |
| mmu-miR-212 | −2.06 | mmu-miR-135a* | −2.42 |
| mmu-let-7c | −2.04 | mmu-miR-202-3p | −2.13 |
| mmu-let-7b | −1.86 | mmu-miR-125a-3p | −2.12 |
| mmu-miR-487b | −1.82 | mmu-miR-125b-5p | −2.07 |
| mmu-miR-26a | −1.75 | mmu-miR-483 | −2.00 |
| mmu-miR-21 | −1.59 | mmu-miR-1224 | −1.91 |
| mmu-miR-101b | −1.48 | mmu-miR-23a | −1.67 |
| mmu-miR-19b | −1.38 | mmu-miR-712 | −1.57 |
| mmu-miR-193 | −1.28 | mmu-miR-451 | −1.41 |
| mmu-miR-30a | −1.17 | mmu-miR-26a | −1.20 |
| mmu-miR-297a | 2.58 | mmu-miR-721 | −1.15 |
| mmu-miR-483 | 2.89 | mmu-miR-680 | 1.02 |
| mmu-let-7d* | 3.09 | mmu-miR-365 | 1.14 |
| mghv-miR-M1-2 | 3.12 | mmu-miR-30c | 1.16 |
| mmu-miR-709 | 3.19 | mmu-let-7d* | 1.45 |
| mmu-miR-574-3p | 3.23 | mmu-miR-30e | 1.49 |
| mmu-miR-466 g | 3.27 | mmu-miR-574-5p | 1.49 |
| mmu-miR-466 h | 3.27 | mmu-miR-30a | 1.62 |
| mmu-miR-466f-3p | 3.34 | mmu-miR-15a | 1.73 |
| mmu-miR-1224 | 3.36 | mmu-miR-107 | 1.79 |
| mmu-miR-574-5p | 3.40 | mmu-miR-19b | 1.88 |
| mmu-miR-467a* | 3.42 | mmu-miR-294* | 2.10 |
| mmu-miR-689 | 3.44 | mmu-miR-27b | 2.23 |
| mmu-miR-467b* | 3.56 | mmu-miR-29a | 2.45 |
| mmu-miR-207 | 3.56 | mmu-let-7 g | 2.48 |
| mmu-miR-669c | 3.58 | mmu-miR-29b | 2.64 |
| mmu-miR-483* | 3.61 | mmu-miR-148a | 2.78 |
| mmu-miR-877* | 3.65 | mmu-miR-22 | 2.92 |
| mmu-miR-467e* | 3.68 | mmu-miR-21 | 3.40 |
| mmu-miR-468 | 3.74 | mmu-miR-130a | 3.41 |
| mmu-miR-466c-5p | 3.75 | mmu-miR-101b | 3.48 |
| mmu-miR-297b-3p | 3.84 | mmu-miR-29c | 3.71 |
| mmu-miR-672 | 4.02 | mmu-miR-193 | 4.02 |
| mmu-miR-671–5p | 4.07 | mmu-miR-685 | 4.82 |
| mmu-miR-197 | 4.27 | mmu-miR-192 | 6.32 |
| mmu-miR-485* | 4.33 | mmu-miR-122 | 8.89 |
| mmu-miR-188–5p | 4.34 | ||
| mmu-miR-328 | 4.37 | ||
| mmu-miR-669a | 4.52 | ||
| mmu-miR-721 | 4.63 | ||
| mmu-miR-466d-3p | 4.70 | ||
| mmu-miR-710 | 5.85 | ||
| mmu-miR-711 | 6.95 | ||
The common miRNA species that affected by acetaminophen overdose between tissue and plasma are listed in bold characters. hyb, hybridization.
Effects of acetaminophen overdose on miRNA spectra in plasma samples.
The effect of acetaminophen overdose on the spectra and levels of miRNA in plasma was dramatic. Hierarchical clustering analysis was performed on 8 plasma samples, 4 from control and 4 from 24 h after acetaminophen overdose, with 44 miRNAs that showed a >2-fold change after acetaminophen administration. The clustering clearly separated the samples into the control and treated group (see SI). Of the 44 altered miRNAs in plasma, 25 (57%) of them showed higher levels in treated plasma samples. The highest observed increase in acetaminophen-overdosed plasma samples was mir-122, with an average of a 470-fold increase (8.89 in log2 scale; Table 2), and the mir-710 showed an average of a 26-fold decrease based on microarray hybridization results (4.69 in log2 scale; Table 2). To validate the microarray hybridization results, we used SYBR green-based quantitative PCR (QPCR) on a few selected miRNA species with RNA isolated from plasma samples. The results from QPCR and array results are in agreement (see SI).
Plasma miRNA spectra and levels inversely reflect the changes of miRNA induced in liver by acetaminophen overdose.
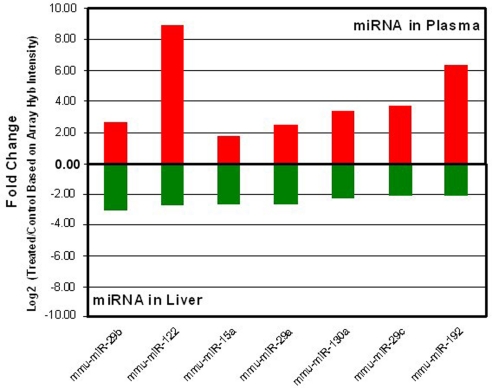
Comparing the list of miRNA species in plasma and liver samples that were significantly affected by acetaminophen overdose revealed 20 miRNAs in common (Tables 1 and 2). Most of these commonly changed miRNAs, 17 of 20, showed reciprocal changes between plasma and liver tissues. To further demonstrate the changes of specific miRNA species in opposite directions between liver and plasma, the top 7 down-regulated miRNA species in the liver (Table 2) were plotted (green bars in Fig. 1) with the changes of corresponding miRNA in the plasma (red bars in Fig. 1) induced by acetaminophen overdose. For example, mir-122 and mir-192 showed higher levels in plasma after acetaminophen treatment, but they were decreased in tissue. mir-710 and mir-711 were elevated in liver tissues but exhibited lower levels in plasma samples, after acetaminophen overdose (Table 2 and Fig. 1, note the log scale).
Fig. 1.
Selected miRNA shows opposite changes between liver and plasma samples based on microarray results. The miRNA species that are predominantly expressed in the liver are down-regulated (green bars) in liver upon acetaminophen exposure; however, the level of these miRNA species showed a higher level in the corresponding plasma (red bars). The fold changes (on a log 2 scale) are based on microarray hybridization intensity.
Specific Circulating miRNA Can Be Used to Detect Liver Injury.
To further investigate the effects of exposure duration and dose of acetaminophen on the level of circulating miRNA, 32 mice were randomly grouped into 8 groups with 4 mice in each group. The mice were injected with either PBS (control) or a single dose of acetaminophen at concentrations of 75 mg/kg, 150 mg/kg, and 300 mg/kg in PBS. After either 1 or 3 h of acetaminophen exposure, the animals were killed, and plasma samples were collected for both ALT enzyme and miRNA QPCR assays. As expected, the ALT levels showed a dose- and time-dependent change, especially in samples from 3 h after treatment; however, the variation of ALT measurements is quite large (Fig. 2 A and B, blue line). The amount of RNA isolated from plasma also showed an upward trend along with the increased acetaminophen dosage and time after exposure, especially at samples from 3 h after various doses of acetaminophen injection (see SI).
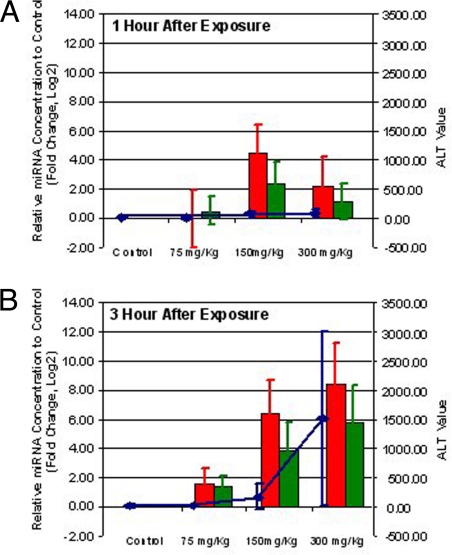
Fig. 2.
MicroRNAs are more sensitive markers than ALT for liver injury. The comparison among the levels of mir-122 (red bars), mir-192 (green bars), and ALT (blue line) in plasma samples collected from mice at 1 (A) and 3 (B) h after exposure to different doses of acetaminophen (indicated on x axis). The relative change of miRNA expression levels (ratio in log 2 compared with control) is indicated on the left side of the figure and the scale of ALT level is on the right. The relative change of miRNA levels is expressed in log 2 ratio of each treatment condition compared with the corresponding control. The values of miRNA fold change and ALT levels are the average of 4 independent samples from each time point, and the standard derivations are shown as error bars.
To better understand the time and dose dependency of specific miRNA levels, ≈50 ng of total RNA isolated from each plasma sample were used to generate cDNA for QPCR assay on selected miRNA species, mir-22, mir-101b, mir-122, mir-133a, mir-135*, mir-192, mir-193, and mir-486. Like the ALT levels, the selected miRNA species all showed dose- and time-dependent changes in plasma (see SI). The acetaminophen overdose-induced miRNA change was readily apparent, especially in plasma samples obtained from animals 3 h after 300 mg/kg of drug. To better compare the changes with ALT levels, data from mir-122 (red bar in Fig. 2) and mir-192 (green bar in Fig. 2) were plotted together with ALT levels (blue line in Fig. 2) to illustrate the similarity in changes between these 2 different types of biomolecules during the progression of drug-induced liver injury.
The data further suggested that specific circulating miRNAs, such as mir-122 and mir-192, might be more sensitive biomarkers than the current ALT test to detect liver injuries induced by acetaminophen because an unambiguous elevation of these miRNA levels was observed in the plasma as early as 1 h after exposing the animal to a dose of only 150 mg/kg of acetaminophen (Fig. 2A). The variations of miRNA levels in the same treatment group were also smaller than the variations associated with measured ALT enzyme activities (Fig. 2). This may suggest that the plasma miRNA levels individually may be more reliable and reproducible plasma biomarkers relative to the ALT levels. Taken together, these miRNA-based markers represent a potentially far more sensitive and reliable test for drug-induced tissue injury.
Discussion
Besides being recognized as key molecules in intracellular regulatory networks for gene expression, the spectra and levels of some miRNAs are emerging as biomarkers for various pathological conditions (26). Recent findings suggest that circulating miRNAs may be plasma biomarkers for the diagnosis of lung (23), colorectal (23), and prostate cancers (22). This report demonstrates the dynamic changes of circulating miRNAs in the plasma resulting from external toxin exposure. We used drug-induced liver injury in the mouse as a model system and showed changes in the spectra and levels of circulating miRNA as a result of acetaminophen overdose (Tables 1 and 2). The level of specific plasma miRNA species is likely to be even more sensitive than the current markers for detecting and monitoring drug-induced tissue injury (Fig. 2). In addition, this study has also demonstrated the diverse tissue origin of circulating miRNA (see SI).
Changes in the Spectrum and Level of miRNA in Liver and Plasma in Response to Toxin Exposure Are Different.
Acetaminophen overdose induces a significant change in the expression levels of a number of miRNAs in liver tissue. Comparing the injured tissues with controls revealed 51 miRNA species, 18 with decreased levels and 33 with increased levels (a >2-fold change.) However, only 20 of those changed miRNA species in the tissue also changed in the plasma (Tables 1 and 2).
We made the unexpected observation that most of the miRNAs with decreased levels in the liver (12 of 13) exhibit increases in the plasma in overdosed mice (Table 2 and Fig. 1). This could imply that the cellular damage in the liver tissue resulted in transport or release of cellular miRNAs into the plasma, which may be a similar process by which cellular enzymes are released after cellular damage. We cannot, however, rule out the possibility of a specific transport mechanism, as part of an inherent cellular process that releases specific miRNAs as a response to the acetaminophen overdose.
Like the miRNAs with decreased levels in overdosed liver samples, most of the miRNAs with increased levels in overdosed liver samples (5 of 7) also exhibited changes in the opposite direction in the plasma samples (Table 2). The increase of specific miRNA levels in tissue may reflect the initiation and progression of drug-induced liver injury. However, the reason for their lower levels in the plasma remains unknown at the moment.
Changes in the Composition of Circulating miRNA After Acetaminophen Overdose May Reflect a Complex Tissue-Injury Pattern.
The liver appears to be the primary targeted organ for acetaminophen-induced tissue injury, as indicated by the increase of liver-specific enzyme levels—ALT and AST in the plasma. Therefore, we might expect that the levels of circulating miRNAs originated from the liver should have higher levels in treated plasma samples. Indeed, based on our limited tissue miRNA profiling results, several miRNAs including, mir-15a, mir-21, mir-101b, mir-122, mir-148a, mir-192, and mir-193 that are predominantly expressed in the liver are elevated in plasma samples from toxin-treated mice (Table 1 and Fig. 1). Tissue injuries other than in the liver have also been reported in acetaminophen overdose (27). Several miRNA species that are predominantly expressed in the brain, heart, lung, and spleen, based on our 6-tissue survey, were also observed at higher levels in acetaminophen-overdosed plasma samples. Through the analysis of organ-specific blood proteins, we also have preliminary indications that organs other than the liver are also perturbed during acetaminophen overdose in mice (B. Sun, personal communication). The finding of elevated levels of relatively organ-specific proteins and miRNA species from other tissues may further indicate that the effect of acetaminophen overdose may affect multiple organs and cell types despite the majority of pathological damage being reported to be confined to the liver. The diverse origin of plasma miRNA also strongly suggests that using circulating miRNA as a specific window into the physiopathological condition of the body, perhaps in concert with certain plasma protein profiles, will be a very powerful approach to diagnostics.
Dose- and Duration-Dependent Changes on the Circulating miRNA.
We have also demonstrated in this well-established model that tissue injuries can be precisely detected and monitored with a small number of circulating miRNA species. In our study, this miRNA-based method is more sensitive and probably more reliable than the current method-ALT level for detecting liver injuries as illustrated in Fig. 2 by using mir-122 and mir-192 as examples.
Even though ALT is fairly abundant in the liver, and elevated levels in the plasma usually imply hepatic injuries or diseases, elevated ALT levels from extrahepatic sources have been reported from patients with burns (28) or individuals with muscle inflammation (29), hypothyrodism (30), or myopathies (31). This may make the proper differential diagnosis based on ALT levels alone for liver related diseases more difficult. In contrast, some of the affected circulating miRNAs by acetaminophen overdosing, as demonstrated in our study, are highly restricted to the liver, such as mir-122 (32). We intend to obtain samples from cardiomyopathies and muscle injuries to determine the specificity of these circulating miRNA for liver injury.
Acetaminophen overdose induces injuries primarily to centrilobular hepatocytes because of the enrichment of cytochrome P450 isozymes for bioactivation and less intercellular GSH for detoxification (33). Like the ALT level, most of the circulating miRNAs identified in this report probably are not centrilobular-specific but, rather, reflects a general liver injury. We intend to test this with samples from other kinds of liver injury. In addition, we are planning to obtain human samples to investigate the feasibility of using the orthologous miRNA species identified for the same diagnostic objectives (note that some of the “miRNA orthologs” are actually identical in sequence between mouse and human.) Because of the similarity of pathology between human and mouse in acetaminophen-induced liver injury, we believe the miRNA species identified in our study would show similar results in human samples.
MicroRNA May Be Excellent Blood-Based Biomarkers.
Challenges for developing protein-based biomarkers from body fluids, such as plasma, serum, and urine, include the complexity of protein composition, the assorted posttranslational modifications, the low abundance of proteins of interest, the difficulty of developing suitable high-affinity capture agents, the complexities of proteolysis and protein denaturation, and potentially complex assay methods (34, 35). All of these make the discovery and development of protein-based biomarkers with proper specificity, sensitivity, and predictive value an expensive, time consuming, and difficult task. The biological function of circulating miRNA is largely unknown; however, unlike proteins, there are far fewer known miRNA species, so obtaining a complete profile is relatively easy. Currently, there are 866 known miRNAs for human and 627 for mouse (based on the latest miRBase release 12.0 http://microrna.sanger.ac.uk/sequences/index.shtml) compared with perhaps a million or more serum proteins, including various processing variants and posttranslationally modified proteins. In addition, miRNAs do not have known postprocessing modifications, and with their size, their chemical composition is much less complex than most other biological molecules. Detecting specific miRNA species, although somewhat challenging, is inherently a much easier task than detecting proteins. A synthetic complementary oligonucleotide should deliver sufficient specificity in most cases, and a standard PCR assay can be used to increase the detection sensitivity. It has also been demonstrated by us and others that the circulating miRNAs are stable and can be reliably extracted and assayed in either serum or plasma (22). However, the effects of different sample preparation protocols need to be investigated. It is likely that miRNAs will be far more stable than their protein counterparts in the long-term storage of biological samples.
Lower complexity, no postprocessing modification, synthetic high-affinity “capture” reagent, tissue-restricted expression profile, and “amplifiable” signals make circulating miRNA ideal candidates as biomarkers to reflect various physiopathological conditions in the body. Even though we have demonstrated the use of specific circulating miRNAs as biomarkers to detect acetaminophen-induced liver injury, this finding is but a preliminary indication of the abundant possibilities of using miRNA to detect specific pathological conditions. A more comprehensive program in mice and humans is needed to determine the specificity and sensitivity of selected miRNA species. Nevertheless, our study demonstrates the complexity of circulating miRNA and the use of specific circulating miRNA species as biomarkers to detect drug-induced liver injury. Our results, although preliminary, strongly suggest that a more comprehensive study of circulating miRNAs and their association with various physiopathological conditions may lead to another dimension in the discovery of biomarkers in the blood for many physiological and pathological conditions, including toxicity.
Materials and Methods
Animals and the Animal Model.
All animal experimental protocols were in agreement with the guidelines established by the Institute and preapproved by the institutional review board. Six-month-old male Balb/C mice were fasted for 24 h before treatments (see description in SI).
RNA Isolation.
Total RNA, including miRNA, was extracted from ≈100 mg of tissue by using the mirVana total RNA extraction kit and following the instructions from the manufacturer (Applied Biosystems/Ambion). The RNA from plasma was isolated by using the miRNeasy kit (Qiagen) with minor modifications (see description in SI).
Array Hybridization and Data Analyses.
To assess the level and composition of miRNA, microarrays from Agilent Technologies were used and were described in detailed in SI.
Real-Time Quantitative RT-PCR Analysis.
The expression levels of miRNA were confirmed with a SYBR-based quantitative PCR (QPCR) using individual miRNA-specific primers (Qiagen). The detailed protocol is described in the SI.
Supplementary Material
Acknowledgments.
We acknowledge stimulating discussions with Clay Marsh concerning circulating miRNAs and their potential roles and uses. We thank Richard Gelinas for a careful reading and comments on the manuscript. This work was supported by a Systems Biology Center grant from the National Institutes of Health and research contracts from the Battelle Biology and Health Science Initiative (Battelle OP46250) and the Department of Defense (W911SR-07-C-0101).
Footnotes
The authors declare no conflict of interest.
Data deposition: Array data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSM352577).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813371106/DCSupplemental.
References
- 1.Chang CY, Schiano TD. Review article: Drug hepatotoxicity. Alimentary Pharmacol Therapeutics. 2007;25:1135–1151. doi: 10.1111/j.1365-2036.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- 2.Halegoua-De Marzio D, Navarro VJ. Drug-induced hepatotoxicity in humans. Curr Opin Drug Discovery Dev. 2008;11:53–59. [PubMed] [Google Scholar]
- 3.Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Amacher DE, Adler R, Herath A, Townsend RR. Use of proteomic methods to identify serum biomarkers associated with rat liver toxicity or hypertrophy. Clin Chem. 2005;51:1796–1803. doi: 10.1373/clinchem.2005.049908. [DOI] [PubMed] [Google Scholar]
- 5.Larson AM. Acetaminophen hepatotoxicity. Clinics Liver Dis. 2007;11:525–548. vi. doi: 10.1016/j.cld.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Amar PJ, Schiff ER. Acetaminophen safety and hepatotoxicity—Where do we go from here? Exp Opin Drug Safety. 2007;6:341–355. doi: 10.1517/14740338.6.4.341. [DOI] [PubMed] [Google Scholar]
- 7.Thummel KE, Lee CA, Kunze KL, Nelson SD, Slattery JT. Oxidation of acetaminophen to N-acetyl-p-aminobenzoquinone imine by human CYP3A4. Biochem Pharmacol. 1993;45:1563–1569. doi: 10.1016/0006-2952(93)90295-8. [DOI] [PubMed] [Google Scholar]
- 8.Manyike PT, Kharasch ED, Kalhorn TF, Slattery JT. Contribution of CYP2E1 and CYP3A to acetaminophen reactive metabolite formation. Clin Pharmacol Therapeutics. 2000;67:275–282. doi: 10.1067/mcp.2000.104736. [DOI] [PubMed] [Google Scholar]
- 9.Liu ZX, Govindarajan S, Kaplowitz N. Innate immune system plays a critical role in determining the progression and severity of acetaminophen hepatotoxicity. Gastroenterology. 2004;127:1760–1774. doi: 10.1053/j.gastro.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 10.Atkuri KR, Mantovani JJ, Herzenberg LA, Herzenberg LA. N-acetylcysteine—A safe antidote for cysteine/glutathione deficiency. Curr Opin Pharmacol. 2007;7:355–359. doi: 10.1016/j.coph.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong VH. Acetaminophen overdose and N-acetylcysteine therapy. Ann Acad Med Singapore. 2007;36:704. [PubMed] [Google Scholar]
- 12.James O, et al. Liver damage after paracetamol overdose. Comparison of liver-function tests, fasting serum bile acids, and liver histology. Lancet. 1975;2:579–581. doi: 10.1016/s0140-6736(75)90170-1. [DOI] [PubMed] [Google Scholar]
- 13.Waring WS, Robinson OD, Stephen AF, Dow MA, Pettie JM. Does the patient history predict hepatotoxicity after acute paracetamol overdose? Q J Med. 2008;101:121–125. doi: 10.1093/qjmed/hcm139. [DOI] [PubMed] [Google Scholar]
- 14.Rumack BH, Matthew H. Acetaminophen poisoning and toxicity. Pediatrics. 1975;55:871–876. [PubMed] [Google Scholar]
- 15.Lewis RK, Paloucek FP. Assessment and treatment of acetaminophen overdose. Clin Pharm. 1991;10:765–774. [PubMed] [Google Scholar]
- 16.Bond GR. Acetaminophen protein adducts: A review. ClinToxicol. 2008;47:2–7. doi: 10.1080/15563650801941831. [DOI] [PubMed] [Google Scholar]
- 17.Damsten MC, et al. Liquid chromatography/tandem mass spectrometry detection of covalent binding of acetaminophen to human serum albumin. Drug Metab Dispos. 2007;35:1408–1417. doi: 10.1124/dmd.106.014233. [DOI] [PubMed] [Google Scholar]
- 18.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14:1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 19.Tang G, et al. The art of microRNA: Various strategies leading to gene silencing via an ancient pathway. Biochim Biophys Acta. 2008;1779:655–662. doi: 10.1016/j.bbagrm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2008;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dillhoff M, Wojcik SE, Bloomston M. MicroRNAs in solid tumors. J Surg Res. 2008 doi: 10.1016/j.jss.2008.02.046. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, et al. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waldman SA, Terzic A. MicroRNA signatures as diagnostic and therapeutic targets. Clin Chem. 2008;54:943–944. doi: 10.1373/clinchem.2008.105353. [DOI] [PubMed] [Google Scholar]
- 27.Mazer M, Perrone J. Acetaminophen-induced nephrotoxicity: Pathophysiology, clinical manifestations, and management. J Med Toxicol. 2008;4:2–6. doi: 10.1007/BF03160941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halkes S, van den Berg A, Hoekstra M, du Pont J, Kreis R. Transaminase and alkaline phosphatase activity in the serum of burn patients treated with highly purified tannic acid. Burns. 2002;28:449–453. doi: 10.1016/s0305-4179(02)00041-4. [DOI] [PubMed] [Google Scholar]
- 29.Korones DN, Brown MR, Palis J. “Liver function tests” are not always tests of liver function. Am J Hematol. 2001;66:46–48. doi: 10.1002/1096-8652(200101)66:1<46::AID-AJH1007>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Shimoda SI, Kasai K. [A clinical evaluation of the increased serum myoglobin: Creatine phosphokinase and lactic dehydrogenase in patients with thyroid disorders (author's transl)] Nippon Naibunpi Gakkai Zasshi. 1980;56:1096–1106. doi: 10.1507/endocrine1927.56.8_1096. [DOI] [PubMed] [Google Scholar]
- 31.Harr KE, Allison K, Bonde RK, Murphy D, Harvey JW. Comparison of blood aminotransferase methods for assessment of myopathy and hepatopathy in Florida manatees (Trichechus manatus latirostris) J Zool Wildl Med. 2008;39:180–187. doi: 10.1638/2007-0134R.1. [DOI] [PubMed] [Google Scholar]
- 32.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 33.Anundi I, Lahteenmaki T, Rundgren M, Moldeus P, Lindros KO. Zonation of acetaminophen metabolism and cytochrome P450 2E1-mediated toxicity studied in isolated periportal and perivenous hepatocytes. Biochem Pharmacol. 1993;45:1251–1259. doi: 10.1016/0006-2952(93)90277-4. [DOI] [PubMed] [Google Scholar]
- 34.Cowan ML, Vera J. Proteomics: Advances in biomarker discovery. Exp Rev Proteomics. 2008;5:21–23. doi: 10.1586/14789450.5.1.21. [DOI] [PubMed] [Google Scholar]
- 35.Ebert MP, Korc M, Malfertheiner P, Rocken C. Advances, challenges, and limitations in serum-proteome-based cancer diagnosis. J Proteome Res. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.