Abstract
Trace amine-associated receptors (TAARs) in mammals recently have been shown to function as olfactory receptors. We have delineated the taar gene family in jawless, cartilaginous, and bony fish (zero, 2, and >100 genes, respectively). We conclude that taar genes are evolutionary much younger than the related OR and ORA/V1R olfactory receptor families, which are present already in lamprey, a jawless vertebrate. The 2 cartilaginous fish genes appear to be ancestral for 2 taar classes, each with mammalian and bony fish (teleost) representatives. Unexpectedly, a whole new clade, class III, of taar genes originated even later, within the teleost lineage. Taar genes from all 3 classes are expressed in subsets of zebrafish olfactory receptor neurons, supporting their function as olfactory receptors. The highly conserved TAAR1 (shark, mammalian, and teleost orthologs) is not expressed in the olfactory epithelium and may constitute the sole remnant of a primordial, nonolfactory function of this family. Class III comprises three-fourths of all teleost taar genes and is characterized by the complete loss of the aminergic ligand-binding motif, stringently conserved in the other 2 classes. Two independent intron gains in class III taar genes represent extraordinary evolutionary dynamics, considering the virtual absence of intron gains during vertebrate evolution. The dN/dS analysis suggests both minimal global negative selection and an unparalleled degree of local positive selection as another hallmark of class III genes. The accelerated evolution of class III teleost taar genes conceivably might mark the birth of another olfactory receptor gene family.
Keywords: Danio rerio, evolution, in situ hybridization, shark, trace amine
Trace amine-associated receptors (TAARs) are close relatives of G protein-coupled aminergic neurotransmitter receptors (1). Initially, TAARs have been considered neurotransmitter receptors as well, based on the expression and effects of some family members in the central nervous system (1). However, recently, Liberles and Buck (2) reported for several mammalian taar genes, some of whom they could deorphanize, the expression in olfactory sensory neurons. Thus, the taar genes joined a growing number of GPCR families that serve as olfactory receptors (see ref. 3). Surprisingly, the fish taar gene repertoire appeared to be much larger than the mammalian repertoire (4, 5), whereas the opposite holds true for the other olfactory receptor families. After the cloning of the first TAAR receptors in mammals (6), TAAR genes have been found in genomes from lower vertebrate species (4), and recently, it has been suggested that the family occurs already in lamprey (5). However, in this study, the delineation from classical aminergic neurotransmitter receptors has not been investigated comprehensively, and consequently both the scope of the TAAR family and its evolutionary origin are still unknown. Because all further phylogenetic analysis critically depends on the correct delineation of the family, we have performed exhaustive data mining for taar genes in 14 vertebrate genomes: 5 teleosts, 2 basal fish, and 7 higher vertebrates.
We report a late evolutionary origin of the taar gene family after the divergence of jawed and jawless vertebrates. Taar genes segregate into 3 classes, with the third and youngest class emerging in teleost fish. Members of all 3 classes were found to be expressed in subsets of olfactory receptor neurons of zebrafish, with exception of TAAR1. Class III of the taar gene family is characterized by the loss of the aminergic ligand motif, extensive recent gene duplications, and 4 independent intron gain/loss events and is likely to be under unusually strong positive Darwinian selection. Thus, class III taar genes seem to have acquired a so far unknown set of ligands and appear poised to eventually become an olfactory receptor gene family.
Results
A Monophyletic Origin and Distinct Consensus Motifs Distinguish TAAR Genes from the Monophyletic Group of Aminergic GPCRs.
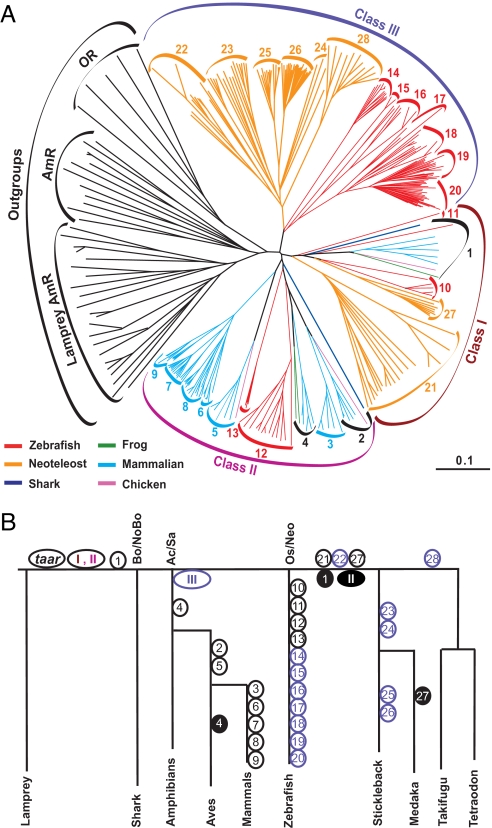
Using a recursive search strategy, we retrieved the complete taar gene repertoire of 5 teleost fish species, a shark, frog, chicken, 4 placental, and 1 marsupial mammalian species [supporting information (SI) Table S1 and Table S2]. All taar genes analyzed subdivide into 28 different subfamilies. Subfamilies 1 to 9 correspond to previously identified TAARs, with mostly mammalian members, whereas subfamilies 10 to 28 are fish-specific. The subfamilies segregate into 3 major clades (Fig. 1 and Fig. S1), which we designated class I to III, in analogy to corresponding subdivisions in the odorant receptor (OR) gene family (7). Class I (TAAR1, 10–11, 21, 27) and class II (TAAR2–9, 12–13) contain both tetrapod and teleost genes, but class III is restricted to teleosts (TAAR14–20, 22–26, 28).
Fig. 1.
Phylogenetic tree of TAAR family members and estimated minimal evolutionary age. (A) Radial tree of teleost and tetrapod TAARs, species groups are color-coded. We analyzed 5 teleost genomes (Danio rerio, zebrafish; Gasterosteus aculeatus, 3-spined stickleback; Oryzias latipes, medaka; Takifugu rubripes, fugu; Tetraodon nigroviridis, tetraodon), 5 mammalian genomes (Monodelphis domestica, opossum; Bos taurus, cow; Mus musculus, mouse; Rattus norvegicus; rat, Homo sapiens, human), avian (Gallus gallus, chicken), amphibian (Xenopus tropicalis, clawed frog), lamprey (Petromyzon marinus), and elephant shark (Callorhinchus milii) genome. Zebrafish and mouse aminergic neurotransmitter receptors were used as outgroup together with a selection of ORs. A group of lamprey receptors considered TAARs by ref. 5 has aminergic receptors as closest neighbors, not TAARs. (Scale bar, 10% divergence.) For accession numbers and/or gene Ids, see Table S2. (B) The estimated minimal evolutionary age of TAAR subfamilies and genes is shown. Open circles represent the gene gain events in each lineage, and filled circles represent the gene loss events. Inside each circle is the name of the respective gene or subfamilies. Emergence of the taar gene family and of the 3 classes of taar genes is indicated by ovals. The major phylogenetic transitions are indicated: bo/nobo, bony fish/cartilaginous fish; ac/sa, actinopterygian/sarcopterygian split, i.e., between the ray-finned bony fish (teleosts) and the lobe-finned fish giving rise to tetrapods; os/neo, ostariophysii/neoteleostei segregation between less derived (zebrafish) and more modern fish (medaka, stickleback, pufferfish). The maximum-parsimony principle was followed, thus gene gains are depicted at the last possible stage before additional gains would become necessary for explanation but may in fact have occurred earlier. A gene gain implies a preceding gene duplication on the same branch of the species tree that gave rise to the new subfamily. Subsequent gene duplications generate the extant members of the subfamily.
All taar genes identified form a monophyletic group, clearly distinct from their close relatives, the aminergic neurotransmitter receptors (Fig. 1). The TAAR gene family also segregates with maximal bootstrap values from the ORs, which are less closely related, but belong to the same major family of GPCRs, the rhodopsin type GPCRs (8). We emphasize that the appropriate choice of outgroups is especially relevant for the proper delineation of the TAAR gene family. The classical aminergic neurotransmitter receptors are relatively close neighbors in the phylogenetic tree, but constitute a rather diverse group by themselves. Inclusion of just a few aminergic receptors (see ref. 5) can lead to erroneous conclusions. Thus, we found it necessary to include representatives from all major aminergic receptor subtypes (cholinergic, dopaminergic, histaminergic, noradrenergic, and serotinergic receptors) in the phylogenetic analysis to avoid spurious results.
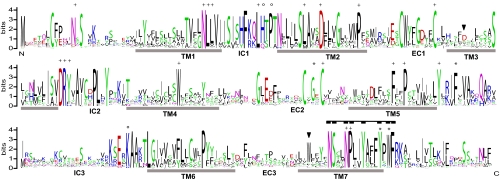
Taar genes frequently show low identity values <30% in pairwise comparisons. We have therefore analyzed the retention of consensus motifs to obtain a second line of evidence for proper delineation of the taar gene family. Of 48 aa positions absolutely conserved between human and rodent TAARs (1), the vast majority (41 aa) are highly conserved in fish TAARs. Besides general GPCR motifs many TAAR-specific motifs are in this group that are not present even in the closely related aminergic receptors (Fig. 2). The characteristic TAAR fingerprint motif, described to be 100% sensitive and specific for mammalian TAARs (1), is strikingly conserved in all fish taar genes analyzed (Fig. 2). In contrast, 2 of the TAAR-specific amino acids from this motif are absent in the lamprey receptors, and 2 others are only weakly conserved, further delineating the TAAR receptors from the group of aminergic receptors in general and from the lamprey aminergic receptor family in particular. As expected, amphibian and avian TAAR sequences share the great majority of conserved motifs as outlined above, supporting their assignment as taar genes. Some motifs distinguish the 3 classes of TAARs from 1 another, including the aminergic ligand motif (9), which is highly conserved in class I and II, but absent from all but one class III taar genes (Fig. S2).
Fig. 2.
aa sequence conservation in the fish taar gene repertoire. Sequence logo representation of the alignment of all 223 fish full-length TAAR sequences, the height of the 1-letter amino acid code in the logo reflects the degree of conservation. Sequence logos were generated as described (32). TM, transmembrane region; IC, intracellular loop; EC, extracellular loop; plus signs, broadly conserved in rhodopsin type GPCRs; circles, conserved in some rhodopsin type GPCRs but not in aminergic receptors; asterisks, conserved in TAARs but not in other rhodopsin type GPCRs. Two triangles in TM 3 and TM 7 depict the aminergic ligand motif (9); filled rectangle motif in TM 7, the characteristic fingerprint for TAARs (1).
Taar Genes Are an Evolutionary Young Family: Class I and II Emerge After the Segregation of Jawed from Jawless Fish, and Class III Even Later, After the Divergence of Tetrapods from Bony Fish.
TAAR1 orthologs occur in both tetrapods and teleosts (Fig. 1), i.e., TAAR1 ought to have been present already in the most recent common ancestor (MRCA) of both lineages. To determine the evolutionary origin of the taar gene family, we have searched all currently available sequence information for cartilaginous fish and jawless fish. Two taar genes, both with a perfectly conserved TAAR-specific fingerprint motif (1), were uncovered in the elephant shark, one of them an ortholog of TAAR1 (Fig. 1). Cartilaginous fish are considered basal to all jawed vertebrates (10), so TAAR1 was present already in the MRCA of bony fish and cartilaginous fish and may be the ancestral member of class I. All tetrapod species analyzed contain a TAAR1 ortholog, as does the avian genome examined here. Interestingly, no orthologs for TAAR1 could be found in any of the neoteleost species analyzed, i.e., this ancestral gene appears to have been lost in neoteleosts.
The other shark gene exhibits a basal location in class II (Fig. 1) and may thus correspond most to the ancestral class II taar gene. Despite an extensive search, no taar genes were uncovered in the genome of a jawless vertebrate (sea lamprey). Thus, the taar gene family appears to have originated in the MRCA of cartilaginous and bony fish as a pair of genes that later expanded into class I and II genes. No shark representative of class III was found, consistent with a later evolutionary origin of this class, after the segregation of the tetrapod from the ray-finned bony fish lineage.
Genomic Arrangement of Teleost taar Genes Pinpoints the Evolutionary Origin of Class III.
Mammalian taar genes are found without exception in a single cluster in the genome (11). All newly identified mammalian, avian, and amphibian taar genes conform to this previously described pattern (Table S2). In contrast, teleost taar genes are found in 2 large clusters and a few solitary genes (chromosomal allocation for zebrafish and medaka, large scaffolds for stickleback). Within the clusters, genes are organized mostly in accordance to phylogenetic relationship (Fig. S3), consistent with a genesis of the clusters by recurrent local gene duplication. A few exceptions to the colinearity of phylogenetic relationship and genomic location do occur (see Table S2), possibly caused by recent genomic rearrangements involving these genes. Interestingly, taar1 gene is always located at one end of the cluster in tetrapod and avian species, consistent with an asymmetric process being responsible for at least some of the repeated gene duplications.
Average intergenic distance is 7.9 ± 0.5 kb (mean ± SEM, n = 97) in the zebrafish gene clusters, with exception of a large intervening region at approximately the same relative position in both clusters (see Table S2). This similarity in cluster structure is consistent with the 2 clusters resulting from the whole genome duplication known to have occurred in early teleosts (12). Indeed, the cluster positions for zebrafish and medaka are syntenic not only within and between species, but also to the human cluster (see Table S2) (12, 13). Class III taar genes are found in both genomic clusters and consequently, class III appears to be older than the whole genome duplication observed in early teleost evolution (12). Because, on the other hand, class III is restricted to teleosts, it appears to have originated shortly after the segregation of the teleost and tetrapod lineages.
Gene Duplication Rate and Gene Divergence Are Much Higher in Teleost Compared with Mammalian Species, Suggesting a Teleost-Restricted Rapid Evolution of taar Genes.
The teleost TAAR repertoires range from 112, 48, 25, to 18 genes (zebrafish, stickleback, medaka, and pufferfish, respectively), whereas mammalian families just reach minimal fish family size, and avian and amphibian families are minuscule, with only 3 genes each (see Table S1 and Table S2). Most of these differences are caused by massive recent gene expansions in teleosts that led to 30 members within a single zebrafish-specific subfamily, TAAR20, and 28 genes in the stickleback-specific subfamily TAAR26. Only TAAR11 and TAAR24 have not undergone recent gene duplications. In contrast, mammalian gene expansions are less frequent, and also much smaller, maximally to 6 genes in opossum TAAR9. No recent gene expansions were found for TAAR1, 2, 3, and 5. No recent gene duplications have been observed in amphibian and avian species (Fig. 1).
Individual teleost TAAR genes rarely possess any orthologs. Thirteen of 19 subfamilies are restricted to a single species each, i.e., all gene duplications giving rise to these genes appear to have occurred after the respective species diverged from the other 4 (Fig. 1B). Only 2 subfamilies contain genes from all 4 neoteleost species examined, and none contain genes from zebrafish and neoteleosts (see Table S1). Even in the case of subfamilies containing orthologs, a gene expansion may occur in one species but not another, e.g., TAAR27 has expanded to 7 genes in tetraodon but remains a single gene in both stickleback and fugu (see Table S2). Thus, most gene duplications have occurred rather recently, after the divergence of the teleost and neoteleost species analyzed here (Fig. 1B) and many even after the 2 pufferfish species diverged ≈20–30 million years ago (14).
In contrast, orthologs are readily identifiable between all mammalian species analyzed. We uncovered bovine orthologs for all 9 previously identified mammalian taar subfamilies (Table S1 and Table S2). In humans, all 9 subfamilies are represented by 1 member each, albeit 3 of them by pseudogenes (Table S2). Seven of the 9 subfamilies are detected also in opossum, a marsupial mammal (Table S1), i.e., should be present already in the MRCA of marsupials (15) and modern mammals. Although very small, with 3 genes each, the amphibian and avian taar gene repertoires are not located at the base of the tetrapod tree and clearly belong to different mammalian subfamilies. Thus, gene losses appear to have shaped the avian and amphibian gene families.
We selected a mammalian and a fish species pair with approximately equal evolutionary distance for an initial comparison of evolutionary rates. Rat and mouse diverged ≈23 million years ago (16), very similar to the 18–30 million years given for tetraodon and fugu (14). For both pairs of species, many orthologs or ortholog subfamilies are observed. Differences between orthologs accumulate only after the separation of the respective species, thus larger divergence in 1 pair of species indicates a faster evolutionary rate. We find that maximal ortholog divergence is, without exception, higher for pufferfish than for rodent pairwise comparisons, maximally 68% for pufferfish, but only 16% for the rodents (Fig. S4). These data suggest a faster evolutionary rate in bony fish compared with tetrapods.
Strong Local Positive Selection in Teleost taar Genes Is Masked by Global Negative Selection.
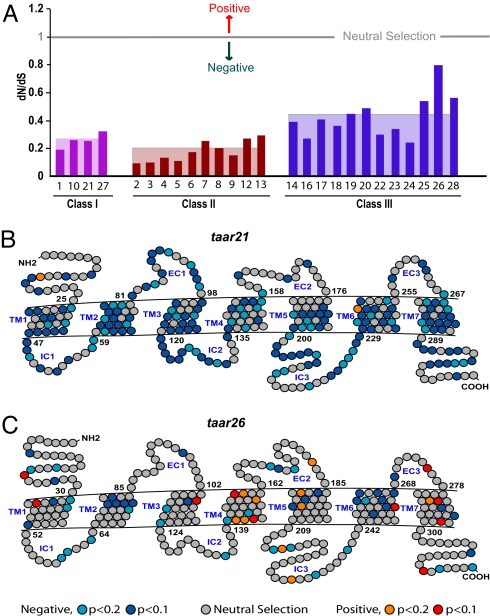
To better understand the evolutionary dynamics of the taar genes, we analyzed the selective pressure on these genes using both global and local analysis of substitution rates in synonymous vs. nonsynonymous base positions. The global dN/dS values calculated for each of the ortholog groups show that all of the gene groups are under negative selection (Fig. 3 and Table S3), but the extent varies considerably, from 0.09 (pronounced negative selection) up to 0.8 (close to neutral selection). The average dN/dS value for the teleost-restricted class III is by far the highest, more than double the value for class II taar genes and significantly different from both class I and class II values (Fig. 3).
Fig. 3.
Evolutionary distances and selective pressure on taar genes. (A) dN/dS ratios of the TAAR ortholog groups in which this analysis was possible (>2 genes per group). Genes are arranged by class, the class average is indicated by background shading. (B and C) A representation of site-by-site selective pressure is shown for 2 TAAR sequences (negative selection in light blue, P < 0.2 or blue, P < 0.1, neutral selection in gray, positive selection in orange, P < 0.2, and red, P < 0.1). (B) Results for TAAR21, a class I subfamily, which includes ortholog genes of all 4 neoteleost species. (C) Results for stickleback-specific TAAR26, a class III subfamily.
The relaxed negative selection observed especially for class III TAAR subfamilies may result from an overall pronounced negative selection masking positive selection at some sites. To clarify this point, we analyzed the dN/dS values for each individual codon position for all genes of a particular taar subfamily. As predicted by the analysis of the previously calculated global dN/dS values, negatively selected sites were found without exception throughout all of the taar gene families, with some preponderance in the transmembrane regions (Fig. 3). Consistent with the results of the global analysis, class III taar genes contain only approximately half as many negatively selected sites as the other 2 classes (Table S3).
Excitingly, the site-by-site analysis suggested a significant number of sites under positive Darwinian selection that were masked by the predominance of negative selection in the global analysis. Although there are few such sites in class I and II taar genes (0–2 sites per gene), several genes in class III show much higher values of up to 20 sites per gene (Fig. 3 and Table S4). The values for class I and II taar genes are comparable with those reported for other olfactory receptor gene families (1–2 sites, see refs. 17 and 18). We repeated the analysis for zebrafish OR genes (7) using the identical algorithm and obtained a range of 0–5 sites, on average 1 site per gene (see Table S5). To the best of our knowledge, the much larger number of such sites in class III taar genes is without precedent in olfactory receptor gene families. We conclude that the teleost-restricted class III, which is evolutionary much younger than class I and class II, is likely to have undergone extensive positive selection. The more rapid evolution of class III has resulted in massive expansion of gene families beyond that observed in the older classes I and II.
Dynamic Loss and Gain of Introns Restricted to the Class III of Neoteleost taar Genes.
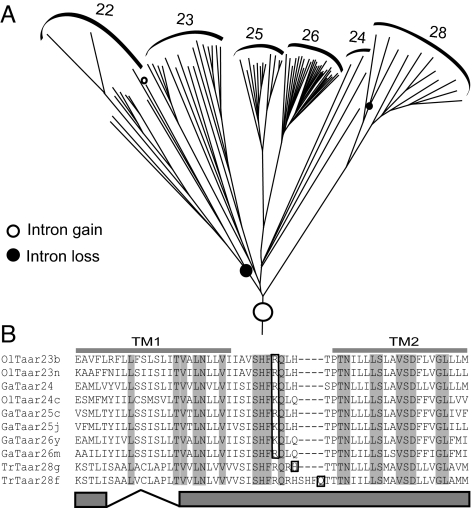
Generally taar genes are monoexonic, like the related ORs (but see ref. 5). We report that, without exception, all class I, class II, and class III zebrafish taar genes are monoexonic. However, from class III, all taar genes of neoteleost subfamilies 23–26 and some genes from subfamily 28 contain an intron between TM1 and TM2 (Fig. 4). The intron is rather short, in the range of 76 to 373 nt, with an average value of 155 nt. Homologies between introns parallel those of the corresponding coding regions. The intron/exon border is strictly conserved (Ol_taar23d and Tr_taar28f show a slightly extended first exon), consistent with a single phylogenetic event early in the neoteleost lineage subsequent to the segregation from the more basal ostariophysan fish (Fig. 4). Consequently, the most parsimonious explanation for the absence of this intron in subfamily 22 and some genes of subfamily 28 is a secondary loss, which must have happened at least 2 times independently. The intron loss in subfamily 28 occurred very late, after the segregation of the 2 pufferfish species (Fig. 4), indicative of the unusually high intron dynamics in the taar gene family compared with the tiny average frequency of intron losses after the divergence of fugu and tetraodon (19).
Fig. 4.
Intron dynamics in class III neoteleost taar genes. (A) By using maximum parsimony, predictions for all independent events of intron gain or loss are depicted in the phylogenetic tree detail. (B) A representative subset of taar genes sharing an early intron gain exhibits a strictly conserved intron/exon border (boxed). The intron interrupts a loop between TM 1 and TM 2.
Another intron gain is predicted in an individual stickleback gene (Ga_taar22a, class III), but not in its pufferfish or medaka orthologs, i.e., late in the neoteleost evolution (Fig. 4). It is caused by insertion of a short repeat that leads to the expansion of a short, conserved poly CV stretch (see Fig. 2) into much of TM4. In total, at least 4 independent intron gain/loss events have occurred after the neoteleosts emerged. Because genome-wide searches so far have failed to identify a single intron gain in vertebrates (19), the 2 gain events documented here appear to be an extremely rare case and may be related to the selection for divergence of class III taar genes.
Most taar Genes Are Expressed in Sparse Olfactory Sensory Neurons.
The rapid evolution and positive selection observed in the taar gene family in teleosts are consistent with expectations for olfactory receptor genes (see ref. 18), because efficient adaptation to changing environmental stimuli may require high evolutionary rates. Another requirement for olfactory receptor genes is an expression in the olfactory epithelium. This was analyzed by in situ hybridization using a representative subset of 8 taar genes from all 3 classes (class I, TAAR1, 10; class II, 12f, 13c; class III, 14d, 15a, 19l, 20t). Probes were chosen to minimize cross-reactivity with related taar genes as far as possible.
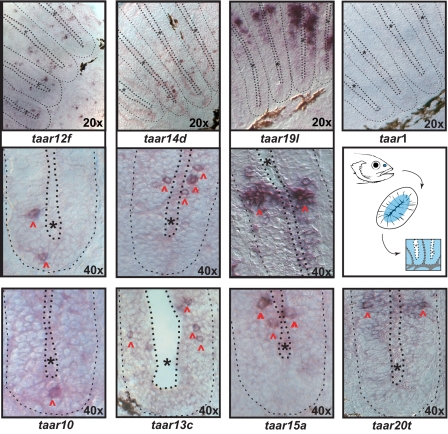
All genes tested were expressed in the adult zebrafish olfactory epithelium (Fig. 5), except TAAR1. Labeled cells were sparsely distributed within the sensory area of the olfactory epithelium. A higher density of labeled cells for genes 19l and 20t (Fig. 5) presumably is caused by unavoidable cross-reactivity in these large and highly homologous subfamilies. No expression was observed in the outer, nonsensory ring of the nasal epithelium. Within the sensory surface individual taar genes are expressed in overlapping, but clearly distinct, concentric expression domains (Fig. S5). Taar genes 19l and 20t occupy the most distal positions, with peak expression frequencies rather close to the border between sensory and nonsensory epithelium, and show a correspondingly skewed distribution, whereas taar genes 10 and 12f show more medial and more symmetrical radial distributions (Fig. 5, Fig. S5). These spatial patterns are reminiscent of the ring-like expression domains observed for zebrafish ORs (20, 21) (see also ref. 22). Thus, the spatial expression patterns observed for TAARs support an expression in olfactory sensory neurons, consistent with an expression of most or all taar genes in these neurons. Furthermore, the frequency of labeled cells [10–50 per section, without taar (19, 20)] is within the range observed for ORs and the V1R-related ORAs (20, 21).
Fig. 5.
Expression of taar genes in the zebrafish olfactory epithelium (OE). A schematic representation shows the approximate position of the olfactory epithelium in the zebrafish, the morphology of a horizontal section (lamellae are cut perpendicular to their flat face) and finally an enlargement of 2 lamellae. The central blue-colored area in the lamellae indicates the location of the sensory neuroepithelium (see ref. 20); gray areas and thin dotted line, basal lamina; black dots and asterisk, lumen. In situ hybridization was performed in horizontal sections with antisense RNA probes. The top row depicts the sensory region of several lamellae, whereas the other 2 rows show enlargements of 1 lamella, corresponding approximately to one-half of the schematical representation (Center Right). Red arrowheads point to labeled neurons, other symbols as above. Taar genes 10, 12f, 13c, 14d, and 15a are expressed in sparse cells, whereas taar 19l and 20t label a somewhat larger subset of cells within the sensory surface, probably because of cross-hybridization in the large and closely related subfamilies taar 19 and taar 20.
Discussion
TAARs, unlike the other 3 families of olfactory receptor genes (OR, V1R, V2R), have not undergone major radiation in mammals. We were thus interested in defining the characteristic properties of the family responsible for the extensive ramification observed in teleosts. Currently, rather completely sequenced genomes are available for several teleost species, and we have taken advantage of this large improvement in data bank quality to establish the complete taar gene repertoire in 5 teleost fish species.
Previous estimates of family size have been either too low (see ref. 4), presumably because of incomplete databases or too high because of inadequate delineation of the taar gene family from the related aminergic neurotransmitter receptors (see ref. 5). In our experience, it is necessary to include representatives from all major aminergic receptor families to obtain a proper delineation of the taar gene family, which is supported by the presence of the characteristic TAAR fingerprint motif (1). In this analysis, all lamprey receptors previously considered TAARs (5) clearly segregate with teleost and tetrapod aminergic receptors and not with teleost or tetrapod taar genes. Despite an extensive search, no further lamprey taar genes were found. Consequently, the origin of the TAAR family appears to be more recent than previously thought. The discovery of shark taar genes allows us to place the origin within the MRCA of cartilaginous and bony fish.
Unexpectedly, the major clade of taar genes, class III, emerged even later, within the teleost lineage of bony fishes, i.e., after the segregation from the tetrapod lineage. This clade shows several exceptional properties that stand out from class I and II taar genes (and, incidentally, from all other known olfactory receptor gene families). Class III contains three-fourths of all teleost taar genes and exhibits no evidence of gene loss, in contrast to the loss of class II and TAAR1 in neoteleosts.
A hallmark of class III taar genes is the strong positive selection suggested by the unusually high dN/dS ratios observed in this clade. Three species-specific subfamilies of class III show dN/dS ratios >1 at many individual sites, 10-fold above the maximal number determined for class I and II genes, which are comparable with ORs and V2R-like OlfC genes in this respect (17, 18). Not a single positively selected site was found in another group of olfactory receptor genes, the V1R-like ORAs (21). Positive selection is a rare event genome-wide (23); thus, its large frequency in class III taar genes high above that found in other olfactory receptor genes is very significant.
A high dN/dS ratio is usually taken as evidence for a selective pressure on sequence divergence. However, because of several confounding influences, among them saturation of mutations and nucleotide bias, calculated dN/dS ratios may not accurately reflect the factual selective pressure. Nevertheless, with the possible exception of very closely (>90% amino acid homology; see ref. 24) or very distantly related genes, high dN/dS ratios appear to be a reliable indicator of positive selection (see refs. 23 and 25). The average homology for groups of taar genes analyzed here was nearly always in the range between 90% and 60%, predominantly <80%. Thus, the dN/dS ratios >1 obtained for several class III taar genes appear likely to reflect positive Darwinian selection. Once ligands become available for class III TAARs, it will be informative to directly examine the adaptive value of the divergence observed in class III taar genes. For ORs, positive selection has been argued as a mechanism to maximize the odor space recognizable by the receptor repertoire. The likely presence of extensive positive selection in the teleost taar gene family supports a role as olfactory receptor genes.
Two independent intron gains and 2 independent intron losses, all exclusively in the neoteleost taar genes of class III, underscore an evolutionary dynamics unprecedented for olfactory receptors (see ref. 7) and beyond. Although there has been some controversy surrounding intron gains in higher eukaryotes (see ref. 26), it is now commonly thought that very few, if any, intron gains occurred during vertebrate evolution (19, 27). Thus, the independent gain of 2 introns in a single subclade of a single gene family constitutes an extraordinary finding. Intron retainment may be favored by the selective pressure toward divergence as evidenced by dN/dS ratios >1. Taken together, the accelerated evolution of class III teleost taar genes conceivably might mark the birth of another olfactory receptor gene family.
Teleost taar genes from all 3 classes are expressed in generally sparse olfactory receptor neurons. The frequency of expression appears to lie in the range of that described for ORs (20) and would be consistent with monogenic expression, which already has been demonstrated for mammalian TAARs (2). The mostly intermediate position of labeled neurons in the apical–basal dimension of each lamella is consistent with an expression in ciliated receptor neurons (see ref. 22), which again would be analogous to the mammalian situation. TAARs are expressed in ring-like domains similar to those described for teleost ORs (20), possibly suggesting some similarity in regulation of expression of ORs and TAARs. The ligands of teleost TAARs from class I and class II may include amines (see refs. 1 and 2) for mammalian TAARs, consistent with the presence of the aminergic ligand motif (9) and the detection of amines by the fish olfactory system (28). A comprehensive analysis of ligand spectra for a representative subset of taar genes will be required to obtain a robust understanding of olfactory representation of the amine group of odors at the peripheral level.
The absence of the aminergic ligand motif in class III genes suggests an evolutionary shift in ligands, away from amines, for this largest class of teleost TAARs. An understanding to what extent the rapid evolution of class III taar genes may enable rapid adaptation to changing ecologies both within and between species will have to await the identification of ligands for these receptors. The genesis of class III appears to be already the second shift in function in the evolution of the TAAR family. The earlier shift occurred during the genesis of the class I and class II genes, because the most ancient of all extant taar genes found in teleosts and tetrapods, TAAR1, is not an olfactory receptor and not detected in either zebrafish or mouse olfactory epithelium (2). Thus, the TAAR family appears to have begun its existence with a function different from the one currently emphasized.
Materials and Methods
Data Mining.
All annotated TAAR sequences were compiled and used as query in TblastN searches in the National Center for Biotechnology Information (NCBI) and Ensembl data banks. Additionally blastP searches were performed in the NCBI databanks and automated ortholog prediction was used in the Ensembl data bank (29). For shark, lamprey, and zebrafish, also EST databanks were searched, in addition, for elephant shark WGS sequences with 1.4-fold genomic coverage were analyzed. Search was recursive until no new candidates were found. Validation of candidates as proper taar genes required: (i) position within the TAAR clade in the phylogenetic analysis; (ii) application of the BLASTP algorithm in the NCBI nonredundant database should result in confirmed TAARs as first hits; (iii) presence of typical TAAR family motifs; (iv) CDS length between 800 and 1300 aa; (v) presence of 7 transmembrane domains (regions assignment according to conserved position as described in ref. 1. For accession numbers see Table S2.
Phylogenetic Analysis.
MAFFT, version 5.8 (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/), was used for multiple protein alignments using the E-INS-i strategy with the default parameters. Phylogenetic trees were constructed by using neighbor joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods (30, 31). Subclades within the taar gene family were determined from the tree as the largest clades that fulfilled 2 criteria: the clade had >70% bootstrap support in the NJ analysis (except the closely related families 18–20), was supported in the MP and ML, and all members within the clade had at least 40% protein identity to each other (except taar23 and 24, which cannot be resolved well and have to be considered provisional). Twenty-eight such subclades or subfamilies were identified, comprising both previously uncharacterized subfamilies and genes from previously known subfamilies.
dN/dS Analysis.
The global dN/dS ratios for the full-length ORF of the 223 fish TAAR receptor coding sequences were determined by using the HyPhy package (www.datamonkey.org), which implements a previously published method (25). The nucleotide alignment was manually edited, and gap positions present in >85% of the sequences were removed. To make inferences about selective pressure (positive and negative selection) on individual codons (sites) within the TAAR coding sequences, the Single Likelihood Ancestor Counting (SLAC) package was used (www.datamonkey.org), which implements the Suzuki–Gojobori method (25).
Cloning of Full-Length DNA for 8 taar Genes and in Situ Hybridization.
Zebrafish genomic DNA (strain Ab/Tü) was generated by using standard protocols. Full-length taar clones were generated by PCR using 16–25-base-long primers. All genes were cloned into pDrive (Qiagen) and confirmed by sequencing. The templates for the probes were amplified from the cloned DNA by using the same forward primers as above and reverse primers with a T3 promoter site (TATTAACCCTCACTAAAGGGAA) attached to their 5′ end. Reverse primers were chosen to obtain probes of ≈600-bp length and minimal cross-reactivity within the TAAR family. For primer sequences and PCR conditions, see Table S6. Digoxigenin (DIG) probes were synthesized according to the DIG RNA labeling kit supplier protocol (Roche Molecular Biochemicals). Sections were fixed in 4% paraformaldehyde for 10 min at room temperature. Hybridizations were performed overnight at 60 °C by using standard protocols. Anti-DIG primary antibody coupled to alkaline phosphatase (Roche Molecular Biochemicals) and NBT-BCIP (Roche Molecular Biochemicals) were used for signal detection.
Supplementary Material
Acknowledgments.
We thank Mehmet Saltürk for taking good care of the fish. Support from the Deutsche Forschungsgemeinschaft (S.I.K.) and the IGS-GFG (A.H. and L.R.S.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803229106/DCSupplemental.
References
- 1.Lindemann L, Hoener MC. A renaissance in trace amines inspired by a novel GPCR family. Trends Pharmacol Sci. 2005;26:274–281. doi: 10.1016/j.tips.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 3.Buck LB. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- 4.Gloriam DE, et al. The repertoire of trace amine G-protein-coupled receptors: Large expansion in zebrafish. Mol Phylogenet Evol. 2005;35:470–482. doi: 10.1016/j.ympev.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Hashiguchi Y, Nishida M. Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: Lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- 6.Borowsky B, et al. Trace amines: Identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci USA. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 9.Huang ES. Construction of a sequence motif characteristic of aminergic G protein-coupled receptors. Protein Sci. 2003;12:1360–1367. doi: 10.1110/ps.0305603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatesh B, Erdmann MV, Brenner S. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proc Natl Acad Sci USA. 2001;98:11382–7. doi: 10.1073/pnas.201415598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindemann L, et al. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85:372–385. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods IG, et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Peer Y. Tetraodon genome confirms Takifugu findings: Most fish are ancient polyploids. Genome Biol. 2004;5:250. doi: 10.1186/gb-2004-5-12-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: Candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh YH, Brenner S, Venkatesh B. Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon) Mol Biol Evol. 2008;25:526–535. doi: 10.1093/molbev/msm278. [DOI] [PubMed] [Google Scholar]
- 20.Weth F, Nadler W, Korsching S. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci USA. 1996;93:13321–6. doi: 10.1073/pnas.93.23.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc Natl Acad Sci USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–5. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki Y, Gojobori T. A method for detecting positive selection at single amino acid sites. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- 26.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;10:10. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in the goldfish Carassius auratus. J Exp Biol. 2003;206:1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–7. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. Seattle, WA: Department of Genome Sciences, Univ of Washington; 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. [Google Scholar]
- 32.Kraemer AM, Saraiva LR, Korsching SI. Structural and functional diversification in the teleost S100 family of calcium-binding proteins. BMC Evol Biol. 2008;8:48. doi: 10.1186/1471-2148-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.