Abstract
Mono-ADP-ribosylation is a reversible posttranslational modification that modulates the function of target proteins. The enzymes that catalyze this reaction in mammalian cells are either bacterial pathogenic toxins or endogenous cellular ADP-ribosyltransferases. For the latter, both the enzymes and their targets have largely remained elusive, mainly due to the lack of specific techniques to study this reaction. The recent discovery of the macro domain, a protein module that interacts selectively with ADP-ribose, prompted us to investigate whether this interaction can be extended to the identification of ADP-ribosylated proteins. Here, we report that macro domains can indeed be used as selective baits for high-affinity purification of mono-ADP-ribosylated proteins, which can then be identified by mass spectrometry. Using this approach, we have identified a series of cellular targets of ADP-ribosylation reactions catalyzed by cellular ADP-ribosyltransferases and toxins. These proteins include most of the known targets of ADP-ribosylation, indicating the validity of this method, and a large number of other proteins, which now need to be individually validated. This represents an important step toward the discovery of new ADP-ribosyltransferase targets and an understanding of the physiological role and the pharmacological potential of this protein modification.
Keywords: ADP-ribosylation, ADP-ribosyltransferase, NAD, toxin, posttranslational modification
Mono-ADP-ribosylation is a posttranslational modification that is catalyzed by bacterial toxins and eukaryotic enzymes that transfer the ADP-ribose moiety from β-NAD+ to various cellular acceptor proteins (1–3). Several bacterial ADP-ribosyltransferases (ARTs) are potent virulence factors. After translocation into the host cell, they modify eukaryotic proteins with high specificity and affect their biological functions (4). Diphtheria toxin (DT), produced by Corynebacterium diphtheriae, was the first bacterial ART family member to be identified (5–7). Along with the related exotoxin A, which is produced by Pseudomonas aeruginosa, DT inhibits protein synthesis in intoxicated cells by catalyzing the ADP-ribosylation of diphthamide of elongation factor 2 (eEF2) (5–7). Cholera toxin and pertussis toxin (PT) are two other well-characterized bacterial ARTs that are produced by Vibrio cholerae and Bordetella pertussis, respectively, and that ADP-ribosylate the α subunits of the heterotrimeric G proteins (4, 8, 9). Other toxins disrupt the cytoskeleton of mammalian cells by ADP-ribosylating either monomeric GTP-binding proteins of the Rho family or actin (4). In addition to these, it is possible that many more ADP-ribosylating toxins and targets remain to be discovered (10–12).
Importantly, endogenous, toxin-related ARTs have also been found in mammals, where they appear to mediate both intracellular and extracellular reactions (1–3). The latter are due to glycosylphosphatidylinositol-anchored or secreted enzymes that can modify either soluble or plasma-membrane-associated protein targets, including the P2X7 purinergic receptor, human neutrophil protein 1, and α7 integrin (13–15). Intracellular, enzymatic mono-ADP-ribosylation modifies proteins with roles in cell signaling and metabolism, such as the chaperone GRP78/BiP, the β subunit of the heterotrimeric G proteins, and mitochondrial glutamate dehydrogenase (GDH) (16–19).
Despite its potential to have crucial roles in the regulation of complex biological processes, including cell signaling and metabolism, membrane trafficking, apoptosis, and gene regulation (2, 3, 20), protein mono-ADP-ribosylation in intact cells remains little explored. This is mainly because of the difficulties in the isolation of the mono-ADP-ribosylated target proteins, due to a lack of specific tools. To date, the antibodies produced in different laboratories can detect only specific proteins that are ADP-ribosylated in vitro by either cellular enzymes or toxins (21–24). Unfortunately, these antibodies do not have a wide application, because they do not recognize some of the well-characterized mono-ADP-ribosylated proteins. This may arise from recognition of the ADP-ribose environment by their specific recognition epitopes, thus limiting their use to their specific targets and their use for the detection of previously unknown ADP-ribosylated target proteins.
Recently, a protein module known as the “macro domain” was described that recognizes monomeric, polymeric, or both forms of ADP-ribose (25, 26). After determination of the crystal structure of the ADP-ribose–macro-domain complex (25, 26), we have explored the possibility that selected macro domains can interact not only with free ADP-ribose-like molecules but also with ADP-ribosylated proteins. We now demonstrate that the macro domain is a powerful tool that can specifically recognize this posttranslational modification on cellular proteins. By combining the use of macro domains with mass spectrometry, we show that this method can be applied to the identification of the protein targets within a host cell that are ADP-ribosylated by bacterial toxins and by endogenous ARTs. This new methodology has the potential for defining both the endogenous and the infection-induced ADP-ribosyl proteomes in mammalian and other cell systems.
Results
Macro-Domain Interactions with ADP-Ribosylated Proteins.
Until recently, no specific protein domains that recognize ADP-ribose had been described. The discovery of the macro domain as an ADP-ribose-binding module (25) prompted us to determine whether these domains can also recognize mono-ADP-ribosylated proteins. The macro domains from both the human poly(ADP-ribose)-polymerase (PARP9) enzyme BAL (mBAL) and the archaeobacterial protein Af1521 from Archaeoglobus fulgidus (mAf1521) recognize free ADP-ribose with high affinities and specificities (e.g., they bind NAD+ and adenosine with a >1,000-fold lower affinity) (25). Despite their specificities, the macro domains of BAL9 and Af1521 also recognize the poly(ADP-ribose) polymer in vitro, consistent with a binding pocket that can accommodate additional chemical groups beyond the core ADP-ribose (25). We therefore surmised that the macro-domain module can also selectively bind mono-ADP-ribosylated proteins.
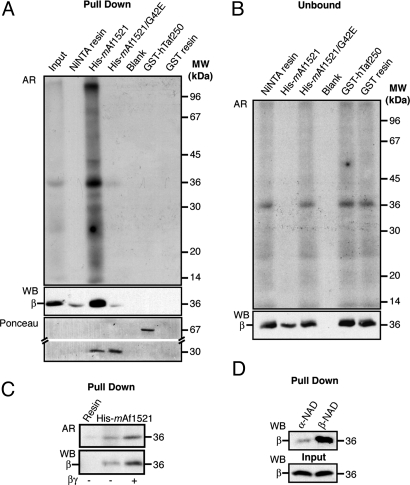
To examine whether we could reveal mono-ADP-ribosylated proteins by using the macro-domain module, we first used a pull-down assay based on the binding between the Ni2+-nitrilotriacetic acid (Ni-NTA)-resin-bound His-tagged mAf1521 (His6-mAf1521) and the mono-ADP-ribosylated β subunit of G proteins (2, 17), a well-characterized model target protein for ADP-ribosylation. Plasma membranes were isolated from Chinese hamster ovary (CHO) cells and used as the source of the endogenously ADP-ribosylated βγ dimer (19). His6-mAf1521 was incubated with radioimmunoprecipitation-assay-buffer-solubilized plasma membranes from CHO cells in which the endogenous β subunit had been [32P]-ADP-ribosylated, as described in ref. 19, and its binding with the β subunit was assessed (Fig. 1). His6-mAf1521 quantitatively retained the [32P]-ADP-ribosylated β subunit, whereas the Ni-NTA resin did not. In a control experiment, an unrelated GST-tagged hTaf250 double bromodomain module that does not bind ADP-ribose (25, 27) did not bind detectable levels of the [32P]-ADP-ribosylated β subunit (Fig. 1A; unbound protein in Fig. 1B). When the amount of the [32P]-ADP-ribosylated β subunit to be loaded onto the resin was increased by adding purified βγ dimer to the ADP-ribosylation assay [a condition in which βγ is known to associate with the plasma membrane and to be ADP-ribosylated (17, 28)], it was again quantitatively retained by His6-mAf1521 (Fig. 1C). Finally, to ascertain that these interactions represented specific binding between mAf1521 and the mono-ADP-ribosylated form of the β subunit, the pull-down assay was carried out with plasma membranes that had been incubated with α-NAD+ [which, unlike β-NAD+, cannot sustain enzymatic mono-ADP-ribosylation (28)]. Under these conditions, His6-mAf1521 did not retain the β subunit, as expected (Fig. 1D).
Fig. 1.
Macro module pull-down assay of mono-ADP-ribosylated G protein β subunit from CHO cells. (A–C) Plasma membrane proteins (10 μg) were ADP-ribosylated with [32P]-NAD+ (1 h at 37°C) in the absence (A–C) or presence (C) of the purified βγ dimer (250 ng) (see Materials and Methods), and solubilized and pulled down with 200 pmol of the indicated macro modules. The pulled-down samples (A, C) and the unbound fractions (B; one-eighth of the total volume) were analyzed by autoradiography (AR) and Western blot analysis (WB) with an anti-β subunit antibody. The Ponceau red staining (A, lower) of the pulled-down samples shows that equivalent amounts of the different macro modules were used. (D) Plasma membrane proteins (10 μg) were incubated with unlabeled β-NAD+ or α-NAD+ (12 h, 30°C). His6-mAf1521-pulled-down proteins and the corresponding input material were analyzed by Western blot with an antibody to the β subunit. When β-NAD+ was replaced with α-NAD+, the residual labeling of substrates can be ascribed to the contaminating β-NAD+ present in the commercial α-NAD+ preparations. The data shown are representative of two to six experiments.
To ascertain a specific role of the ADP-ribose binding pocket in the binding reaction, we also used a site-specific mutant (His6-mAf1521/G42E) as a further control. This mutation, from Gly to Glu, is sufficient to abrogate the ADP-ribose binding of Af1521 (25). As expected, this mAf1521 mutant failed to pull down the [32P]-ADP-ribosylated β subunit, confirming that the ADP-ribose binding pocket is essential for the interaction of mAf1521 with mono-ADP-ribosylated proteins (Fig. 1A).
Taken together, these data demonstrate that the ADP-ribose-binding mAf1521 interacts with the mono-ADP-ribosylated β subunit in a specific manner that depends on the β subunit ADP-ribosylation state. Thus, macro modules can, in principle, be exploited to bind mono-ADP-ribosylated proteins, indicating that they represent valuable tools for the study of endogenous mono-ADP-ribosylation of proteins.
Efficiency and Reversibility of the Binding of ADP-Ribosylated Proteins to mAf1521.
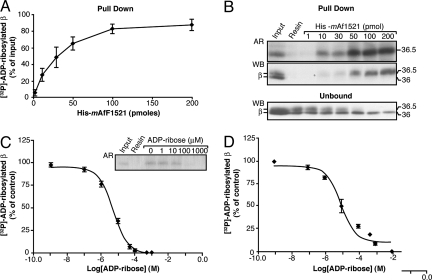
Affinity purification procedures ideally require that the binding between the proteins to be isolated and the affinity resin be both efficient and reversible. To investigate these characteristics in our system, CHO cell plasma membranes enriched with purified βγ dimer were first ADP-ribosylated in the presence of [32P]-NAD+ (see Materials and Methods and refs. 19 and 31). The amount of [32P]-ADP-ribosylated β subunit that was pulled down using increasing concentrations of His6-mAf1521 was then evaluated by autoradiography and Western blot analysis (Fig. 2 A and B; unbound fractions shown as control in Fig. 2B, lower panel). Approximately a ≈25-fold molar excess of resin-bound His6-mAf1521 was required to nearly completely (≈85%) adsorb the ADP-ribosylated βγ (Fig. 2A).
Fig. 2.
Efficiency and reversibility of binding to His6-mAf1521 of the ADP-ribosylated βγ dimer. CHO plasma membrane proteins (10 μg) and 250 ng of purified βγ dimer were ADP-ribosylated in the presence of [32P]-NAD+ for 12 h at 30°C. After solubilization, they were pulled down with His6-mAf1521. (A) Increasing concentrations of His6-mAf1521 in the [32P]-ADP-ribosylated β subunit pull-down assay, shown as percentage of the input of [32P]-ADP-ribosylated β subunit (≈4 pmol). Data are means ± SD of five independent experiments. (B) As for (A), showing pulled-down samples and unbound fractions (one-eighth of the total volume) from a representative experiment, as analyzed by autoradiography (AR) and by Western blotting (WB) with an antibody against the β subunit that recognizes both the unmodified (36 kDa) and the ADP-ribosylated (36.5 kDa) β subunit. The disappearance of the [32P]-ADP-ribosylated β subunit from the unbound fraction was also followed in parallel (B, lower). (C) Pull-down of the [32P]-ADP-ribosylated β subunit after its incubation with 30 pmol of His6-mAf1521 in the presence of increasing concentrations of ADP-ribose (1 nM to 1 mM) (“competition” conditions). Data are expressed as percentages of [32P]-ADP-ribosylated β subunit pulled down in the absence of ADP-ribose and are means ± SD from three independent experiments. Inset: Autoradiography of the pulled-down samples from a representative experiment. (D) As for (C), but with the addition of ADP-ribose after the pull-down assay of the [32P]-ADP-ribosylated β subunit by 30 pmol of His6-mAf1521 (“displacement” conditions). At 100 μM ADP-ribose (≈400 pmol under these incubation conditions), all of the His6-mAf1521-bound [32P]-ADP-ribosylated β subunit (≈4 pmol) was displaced. A ≈100-fold excess of ADP-ribose can displace the immobilized ADP-ribosylated β subunit from the His6-mAf1521-resin complex. Data are means ± SD from three independent experiments. Curve fitting analyses in (C) and (D) were performed with GraphPad PRISM, giving ADP-ribose EC50 values of 5 and 10 μM, respectively.
We also examined the reversibility of the binding of the [32P]-ADP-ribosylated β subunit to His6-mAf1521. Here, a range of ADP-ribose concentrations (1 nM to 10 mM) and 30 pmol of His6-mAf1521 were used. In the “competition” experiments (Fig. 2C), the His6-mAf1521 and ADP-ribose were added simultaneously to the labeled, solubilized, CHO plasma membranes. Here, the ADP-ribose competes with the [32P]-ADP-ribosylated β subunit for binding to the resin-bound His6-mAf1521. In the “displacement” experiments (Fig. 2D), the [32P]-ADP-ribosylated β subunit was initially pulled down with the resin-bound His6-mAf1521, and then ADP-ribose was added to displace the [32P]-ADP-ribosylated β subunit from the His6-mAf1521 (see Materials and Methods for further details). These assays demonstrated that free ADP-ribose can displace the [32P]-ADP-ribosylated β subunit from the resin with EC50 values of 5 and 10 μM, respectively. This indicates that our macro-domain-based procedure can yield highly pure and intact ADP-ribosylated proteins in solution.
Use of the Macro Module To Identify Substrates of Bacterial ART Toxins from Intact Cells.
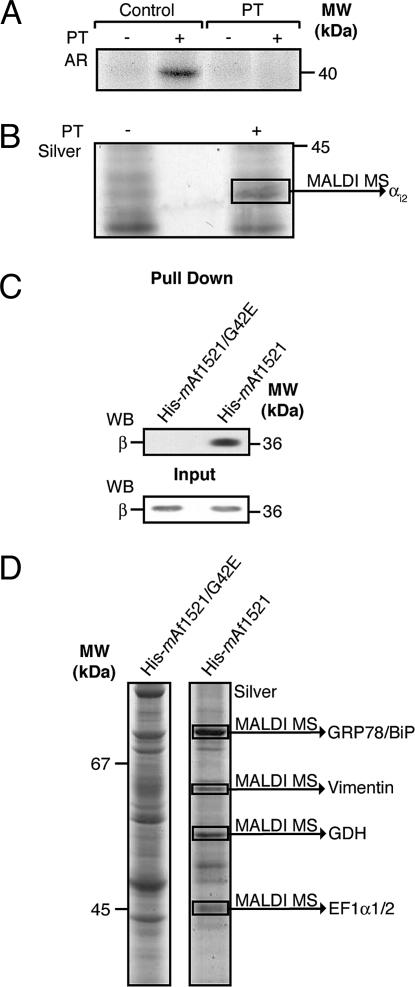
We next investigated whether this method can be used to identify substrates of bacterial toxins that display ART activity. We focused our attention on the cellular substrates of PT, a cysteine-specific ART. For this, we used CHO cells that were either left untreated (control) or were intoxicated with PT. First, to verify that the αi subunit of the heterotrimeric G proteins was indeed fully modified by the treatment of intact cells with PT, membranes from control and intoxicated cells were analyzed in an in vitro ADP-ribosylation assay using PT and radiolabeled NAD, according to established procedures (see Materials and Methods and ref. 34). Although in control membranes αi was radiolabeled by PT, in the intoxicated membranes this Gi subunit was not, confirming that it had already been fully modified in situ by the PT pretreatment and was therefore not available for further in vitro labeling (Fig. 3A).
Fig. 3.
(A, B) GST-mAf1521 pull-down assay of proteins from CHO cell membranes that are either ADP-ribosylated by PT or endogenously ADP-ribosylated. (A) Cells were either untreated (control) or intoxicated with 5 nM PT for 15 h (PT), and then their solubilized membrane proteins (25 μg) were ADP-ribosylated with [32P]-NAD+ and in the absence (−) or presence of PT (+) (see Materials and Methods). The samples were analyzed by SDS/PAGE and visualized by autoradiography (AR). (B) Cells were untreated (−) or intoxicated with PT (+), and their solubilized membrane proteins (2 mg) were pulled down with 2 nmol of GST-mAf1521. The samples were analyzed by SDS/PAGE and visualized by silver staining. The indicated band was identified by MALDI-TOF-MS (see Materials and Methods and Table 1). (C, D) His6-mAf1521 pull-down assay of endogenous ADP-ribosyltransferase substrates. (C) Solubilized HL60 cell membrane proteins (2 mg) were pulled down with 2 nmol of His6-mAf1521. The samples were analyzed by SDS/PAGE followed by Western blot analysis (WB) with an anti-β subunit antibody. The β subunit relative content in the input was revealed by Western blot analysis (C, lower, WB). (D) Solubilized CHO postnuclear membrane proteins (5 mg) were pulled down with 2 nmol of His6-mAf1521, resolved by SDS/PAGE, and visualized by silver staining. The indicated bands were identified by MALDI-TOF-MS (see Materials and Methods and Table 1). The data shown are representative of two to six experiments.
These PT-treated membranes were next subjected to the macro-domain pull-down protocol with GST-mAf1521. A band in the PT-treated samples with a molecular mass of 40 kDa, as revealed by silver staining (Fig. 3B), was indeed identified by MALDI-TOF-MS analysis as the G protein αi2 subunit (Table 1).
Table 1.
Protein identification
| NCBI accession no. | Name | Score* | Matched peptide, no. | Sequence coverage, % | Molecular mass, kDa | |
|---|---|---|---|---|---|---|
| Maldi-TOF-MS | 77386129 | Gαi2 | 112 | 10 | 27 | 41 |
| 121570 | GRP78 | 101 | 12 | 22 | 70 | |
| 1220484 | Elongation factor-1α | 109 | 13 | 37 | 50 | |
| 6680027 | GDH1 | 134 | 18 | 35 | 62 | |
| 62414289 | Vimentin | 138 | 19 | 40 | 54 | |
| 48145673 | HNRPH1† | 106 | 11 | 30 | 49 | |
| 130781 | PARP1 | 1135 | 23 | 31 | 113 | |
| LC-MS/MS | 14916999 | GRP78/BiP | 436 | 72 | 18 | 72 |
| 56757569 | Tubulin | 265 | 5 | 22 | 50 | |
| 120649 | GAPDH | 835 | 8 | 65 | 41 | |
| 2507460 | PDI | 238 | 4 | 15 | 57 | |
| 730922 | CCT-zeta | 219 | 4 | 12 | 58 |
*Protein scores ≈65 are significant (P < 0.05). See Methods for details.
†Identified from A375 cells (melanoma cell line).
Thus, the cellular targets of bacterial toxins can be identified using this macro domain, further supporting the potential of this method to identify mono-ADP-ribosylated proteins in intact cells.
Purification and Identification of Substrates of Intracellular ART Activities by Macro Domains.
Intracellular mono-ADP-ribosylation in mammals modifies proteins that have important roles in cell signaling and metabolism, including, among those best characterized, the chaperone GRP78/BiP, the β subunit of G proteins, and GDH (16–19, 29).
We evaluated whether macro-domain-based pull-down assays coupled to MALDI-TOF-MS analysis can be used to identify such proteins that carry an endogenous ADP-ribosyl modification under resting conditions in an intact-cell model. Postnuclear and membrane fractions isolated from CHO and HL60 cells were used in parallel, where they were first subjected to a nonspecific pull-down step using the His6-tagged mAf1521/G42E mutant; this thus initially eliminated proteins that bound to the resin or to the macro domain in an ADP-ribose-independent manner (Fig. 3 C and D). Next, the unbound material underwent a second pull-down assay with wild-type His6-mAf1521 to specifically retain the ADP-ribosylated proteins (Fig. 3 C and D).
This two-step procedure resulted in the separation of >20 visible bands in well-resolved gels, all representing potential ADP-ribosylated substrates (Fig. 3D). Among the 10 major bands from CHO cells, four were identified by MALDI-TOF-MS analysis as GRP78/BiP, vimentin, GDH, and the elongation factor α1/2 (Fig. 3D and Table 1). Moreover, the endogenously ADP-ribosylated β subunit from HL60 membranes, which was at too low a level to be revealed by colloidal Coomassie blue staining, was readily identified by a specific antibody against the β subunit (Fig. 3C). These proteins are well-known substrates of enzymatic ADP-ribosylation, and so they provide the “proof of principle” that our two-step procedure can successfully isolate different ADP-ribosylated proteins. Importantly, previously unknown putative substrates for endogenous ART activity were also identified. One of these is the intermediate filament protein vimentin (Fig. 3D), which has not been demonstrated to be a substrate of endogenous mono-ADP-ribosylation, although it has been described as a cellular target of the bacterial toxin exoenzyme S (ExoS) from P. aeruginosa (30).
An additional interesting feature of the macro-domain recognition of ADP-ribosylated substrates is its independence from the nature of the modified amino acid. The mAf1521 not only binds the G protein β subunit that we have shown to be modified on arginine (17) but also GDH that is modified on cysteine (18), eEF2 on diphthamide (7, 31), and GRP78/BiP on an unidentified amino acid, clearly indicating that the recognition of the ADP-ribose is independent of the modified amino acid. This highlights the potential of our methodology and its relevance for the definition of the role of the ADP-ribosylation reaction.
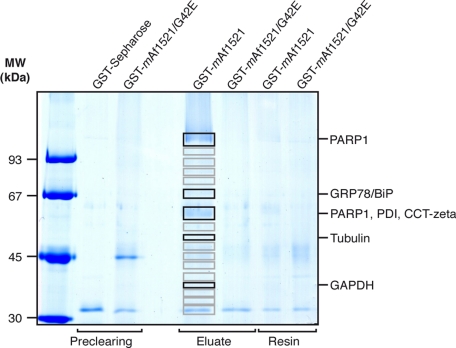
To analyze in more detail the total mammalian ADP-ribosylation proteome, we introduced some further modifications to this method. The pattern of endogenously ADP-ribosylated proteins was determined by examining HL60 cells (Fig. 4). To minimize nonspecific resin interaction with proteins, we used GST-mAf1521 and GST-mAf1521/G42E cross-linked to the Sepharose resin, instead of His6-mAf1521 and His6-mAf1521/G42E. The HL60 cell postnuclear fractions were thus first incubated with glutathione-Sepharose resin to remove endogenous GST. Then the unbound proteins were subjected to the two-step procedure, with the first pull-down assay with the GST-tagged mAf1521/G42E mutant as above, to initially eliminate proteins that bound to the macro domain in an ADP-ribose-independent manner. The unbound material was then divided into two aliquots for the second pull-down assay, one using GST-mAf1521/G42E (control) and the other wild-type GST-mAf1521 (sample), to specifically retain the ADP-ribosylated proteins. Finally, to add further specificity to this method, the proteins bound to both the control and the sample columns were eluted with ADP-ribose, and the eluates were separated by SDS/PAGE. The corresponding bands from both the control and the sample lanes were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS), and only the proteins specific to the sample were considered ADP-ribosylated [Fig. 4 and scheme in supporting information (SI) Fig. S1].
Fig. 4.
GST-mAf1521 pull-down assay and protein identification from HL60 cells. Solubilized HL60 cell proteins (20 mg) were pulled down with resin-bound GST-tagged mAf1521/G42E or mAf1521, as indicated, eluted with 20 mM ADP-ribose, then resolved by SDS/PAGE, and visualized by Coomassie blue staining. The indicated bands (in the sample and in the control lanes) were identified by LC-MS/MS analyses (see Materials and Methods and Table 1). The data shown are representative of two experiments.
Some of the main bands identified with this two-step procedure revealed PARP1, GRP78/BiP, protein disulfide isomerase (PDI), T-complex protein 1 subunit zeta (CCT-zeta), tubulin (both α and β), and GAPDH (Fig. 4 and Table 1). In the case of tubulin, this was initially proposed to be regulated by mono-ADP-ribosylation by cellular enzymes, although only in an in vitro analysis (32). More recently, evidence of poly-ADP-ribosylation of this protein has also been reported in mouse brain homogenates (33). However, consistent with the single narrow band seen around the expected molecular mass (Fig. 4), this tubulin modification should involve a single ADP-ribose molecule, suggesting that tubulin is indeed a cellular target of mono-ADP-ribosylation in intact human cells.
Finally, of particular note, this procedure also identified proteins that have not been associated with either bacterial toxin or endogenous ADP-ribosylation, such as heterogeneous nuclear ribonucleoprotein H1 (HNRNPH1) (Table 1).
In summary, these data show that a two-step procedure with first the mutant and then the wild-type mAf1521 provides a very effective method for the isolation and identification of substrates of endogenous ART activities.
Discussion
In this study, we report on an affinity-based method for the analysis of endogenous and toxin-induced ADP-ribosylation of proteins in intact cells. The method combines the use of a specific macro-domain module, mAf1521 (which binds ADP-ribosylated proteins potently and selectively), with multistage mass spectrometry analysis (see scheme in Fig. S1). It should be noted that among the different macro domains that we have tested so far His6-mAf1521 is the most efficient for pulling down ADP-ribosylated protein σ from the mammalian cells analyzed; however, the procedure might require further adjustments according to the macro domain used (e.g., detergents or incubation times), when different cells (e.g., from plants) or tissue extracts are to be analyzed, or both. Nevertheless, the methods can be considered to be of wide applicability to a large number of biological samples, which can all be analyzed following the principle of the protocol schematically summarized in Fig. S1. This should lead to the identification of a number of unidentified proteins that can be regulated by ADP-ribosylation, which will help us to define the various functional roles of this posttranslational modification in cellular processes and organelles and in the whole organism.
Here, we have explored the use of this methodology in more than one way. The first was to purify known and previously unknown ADP-ribosylated proteins and to separate modified from unmodified substrates. This is clearly shown for the mono-ADP-ribosylated β subunit, which can be eluted from resin-bound His6-mAf1521 by some 100-fold excess of ADP-ribose, whereas the unmodified protein can easily be collected in the unbound fraction. The ability to separate the modified from the unmodified protein represents a significant advantage in studies of the function of mono-ADP-ribosylated substrates, and it solves a technical aspect that to date could not be addressed by classical chromatography or immunoprecipitation.
The other specific use of the macro-domain method that we have investigated is in the field of bacterial toxins. There is significant interest in the role of mono-ADP-ribosylation in human pathology, because many bacterial toxins are ARTs that disrupt cellular functions and affect human health (4, 7). The number of known toxins with ART activity is growing (10–12), and the new candidates require the characterization of their cellular targets. With the tools that have been available to date, this has been far from straightforward [e.g., the characterization of the mammalian proteins that are ADP-ribosylated by the P. aeruginosa toxin ExoT required the collection of silver-stained spots from 12 two-dimensional gels for MALDI-TOF-MS analysis (34)]. This technical complexity in part explains why it has not been possible so far to define a compendium of all of the bacterial ART targets in mammalian host cells. This macro-domain method will greatly accelerate the analysis of this important class of toxins.
Finally, this new strategy has been used to identify ART targets in mammalian cells. The main proteins to be ADP-ribosylated under basal conditions are cytoskeletal proteins, including actin and tubulin, and other proteins involved in cell metabolism, including GAPDH and GDH (e.g., see Figs. 3 and 4). Of further interest, although the same major bands can be identified with the His and GST mAf1521 systems (e.g., GRP78/BiP), the patterns of the major protein bands seen upon ADP-ribosylation are indeed different (e.g., compare Figs. 3D and 4). This will reflect functional differences across cellular systems and the specific patterns of protein expression in any given cell type.
These data therefore represent the beginning of the long list of proteins that can now be identified, with analyses not only including different organelles and tissues but also evaluating the changes occurring in the ADP-ribosyl proteome under different experimental conditions (e.g., stimuli versus basal or pathological versus normal). This also becomes more evident considering the various patterns of ADP-ribosylation that we have seen across different cell systems.
Among the targets for which the specific mono-ADP-ribosylation remains to be confirmed, further molecular chaperones have been identified, including PDI and CCT-zeta, and further cytoskeletal proteins, including vimentin (Table 1). Importantly, in addition to these targets that belong to protein families that include members that are already known to be mono-ADP-ribosylated, we have also identified completely new protein family members, for example, the ubiquitously expressed HNRNPH1. The HNRNPs appear to shuttle between the nucleus, where they are associated with pre-mRNAs, and the cytoplasm; they also appear to have a role in pre-mRNA processing and other aspects of mRNA metabolism and transport.
Clearly, an extensive characterization of targets of mono-ADP-ribosylation is still required for each of these proteins to determine whether they are indeed modified themselves or rather are parts of complexes that include at least one modified protein. The latter could indeed be the case for the different molecular chaperones, because an interaction between GRP78/BiP and PDI has been reported (35). In contrast, the confirmation of HNRNPH1 as a target of mono-ADP-ribosylation will open a new field of investigation, because it is possible that this posttranslational modification of HNRNPH1 affects its ability to bind to RNA and thus to regulate mRNA metabolism and transport.
In conclusion here, with this method now available, there are sure to be discoveries of numerous previously unknown targets of this posttranslational modification, which becomes even more important now that previously unknown ART enzymes have been identified.
Indeed, although endogenous mono-ADP-ribosylation is believed to have important roles in cellular signaling and regulation, the enzymatic activities responsible for this intracellular mono-ADP-ribosylation are only now beginning to be identified. The human genome, for example, encodes 17 so-called poly(ADP-ribose)-polymerase-like genes. Many of these are, however, unlikely to carry out genuine enzymatic ADP-ribose polymer formation, and although some have been proposed to act as cellular mono-ADP-ribosyltransferases (20, 36, 37), for PARP10 this has indeed been demonstrated (38). However, their cellular functions and targets remain unknown.
Another NAD+-using family of proteins, the sirtuins, are enzymes that can deacetylate protein substrates, such as acetylated histones (39, 40). The yeast member, silent information regulator 2 (Sir2), catalyzes mono-ADP-ribosylation of the removed acetyl group (39); moreover, SirT6 has been shown to have a mono-ADP-ribosyltransferase activity directed toward itself (41), and SirT4 mono-ADP-ribosylates mitochondrial GDH, thus repressing its activity (19) and resulting in the regulation of insulin secretion in pancreatic β cells. Moreover, to add to this complexity, several enzymes that catalyze the reverse reaction, the ADP-ribosyl hydrolases, have also been reported, although their specific roles and substrates also remain unknown. These examples give us a glimpse of the type of physiologically relevant information that can be gathered by fully exploring the endogenous mono-ADP-ribosylation machinery. Over the next few years, this macro-domain-based methodology should allow a systematic study of the ADP-ribosyl proteome in mammalian systems and should lead to the definition of the ADP-ribosylation-dependent pathways in physiology and pathology.
Materials and Methods
Detailed methods for CHO and HL60 cell culture and fractionation, ADP-ribosylation and immunoblot analysis, and expression and purification of macro modules are provided as SI Materials and Methods.
Pull-Down Assay.
Plasma membranes, total membranes, or postnuclear material were solubilized, incubated with the macro modules, and pulled down with Ni-NTA or glutathione-Sepharose resins. Detailed procedures, also for the competition, displacement, and affinity purification assays are in SI Materials and Methods.
Protein Identification by MALDI-TOF-MS and LC-MS/MS Analyses.
The protein bands of interest were isolated by SDS/PAGE and analyzed as detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Alberto Luini and M. Antonietta De Matteis for useful discussions and critical reading of this manuscript. We greatly appreciate and acknowledge Piero Pucci (CEINGE, Biotecnologie Avanzate, Napoli) for protein identification by LC-MS/MS analyses. We thank Oliviano Martella and Miriam Bortfeld for technical help, Christopher Paul Berrie for editorial assistance, and Elena Fontana for preparation of figures. This work was supported by Telethon (Italy), the Italian Association for Cancer Research (AIRC), Ministero dell'Università e Ricerca, the European Molecular Biology Laboratory, EU FP6 Marie Curie Research Training Network “Chromatin Plasticity,” and the Human Frontier Science Program. Fellowship support was provided by AIRC (N.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900066106/DCSupplemental.
References
- 1.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 3.Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): Emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- 4.Krueger KM, Barbieri JT. The family of bacterial ADP-ribosylating exotoxins. Clin Microbiol Rev. 1995;8:34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J Biol Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- 6.Collier RJ, Cole HA. Diphtheria toxin subunit active in vitro. Science. 1969;164:1179–1181. doi: 10.1126/science.164.3884.1179. [DOI] [PubMed] [Google Scholar]
- 7.Yates SP, Jorgensen R, Andersen GR, Merrill AR. Stealth and mimicry by deadly bacterial toxins. Trends Biochem Sci. 2006;31:123–133. doi: 10.1016/j.tibs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Cassel D, Pfeuffer T. Mechanism of cholera toxin action: Covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc Natl Acad Sci USA. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katada T, Ui M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc Natl Acad Sci USA. 1982;79:3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pallen MJ, Lam AC, Loman NJ, McBride A. An abundance of bacterial ADP-ribosyltransferases–implications for the origin of exotoxins and their human homologues. Trends Microbiol. 2001;9:302–307. doi: 10.1016/s0966-842x(01)02074-1. discussion 308. [DOI] [PubMed] [Google Scholar]
- 11.Wilde C, Chhatwal GS, Schmalzing G, Aktories K, Just I. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J Biol Chem. 2001;276:9537–9542. doi: 10.1074/jbc.M011035200. [DOI] [PubMed] [Google Scholar]
- 12.Masignani V, et al. NarE: A novel ADP-ribosyltransferase from Neisseria meningitidis. Mol Microbiol. 2003;50:1055–1067. doi: 10.1046/j.1365-2958.2003.03770.x. [DOI] [PubMed] [Google Scholar]
- 13.Seman M, et al. NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity. 2003;19:571–582. doi: 10.1016/s1074-7613(03)00266-8. [DOI] [PubMed] [Google Scholar]
- 14.Paone G, et al. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci USA. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zolkiewska A, Moss J. Integrin α7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J Biol Chem. 1993;268:25273–25276. [PubMed] [Google Scholar]
- 16.Leno GH, Ledford BE. ADP-ribosylation of the 78-kDa glucose-regulated protein during nutritional stress. Eur J Biochem. 1989;186:205–211. doi: 10.1111/j.1432-1033.1989.tb15196.x. [DOI] [PubMed] [Google Scholar]
- 17.Lupi R, Corda D, Di Girolamo M. Endogenous ADP-ribosylation of the G protein β subunit prevents the inhibition of type 1 adenylyl cyclase. J Biol Chem. 2000;275:9418–9424. doi: 10.1074/jbc.275.13.9418. [DOI] [PubMed] [Google Scholar]
- 18.Herrero-Yraola A, et al. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 2001;20:2404–2412. doi: 10.1093/emboj/20.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab CJ, Colville MJ, Fullerton AT, McMahon KK. Evidence of endogenous mono-ADP-ribosylation of cardiac proteins via anti-ADP-ribosylarginine immunoreactivity. Proc Soc Exp Biol Med. 2000;223:389–396. doi: 10.1046/j.1525-1373.2000.22355.x. [DOI] [PubMed] [Google Scholar]
- 22.Meyer T, Hilz H. Production of anti-(ADP-ribose) antibodies with the aid of a dinucleotide-pyrophosphatase-resistant hapten and their application for the detection of mono(ADP-ribosyl)ated polypeptides. Eur J Biochem. 1986;155:157–165. doi: 10.1111/j.1432-1033.1986.tb09471.x. [DOI] [PubMed] [Google Scholar]
- 23.Eide B, Gierschik P, Spiegel A. Immunochemical detection of guanine nucleotide binding proteins mono-ADP-ribosylated by bacterial toxins. Biochemistry. 1986;25:6711–6715. doi: 10.1021/bi00369a058. [DOI] [PubMed] [Google Scholar]
- 24.Osago H, Terashima M, Hara N, Yamada K, Tsuchiya M. A new detection method for arginine-specific ADP-ribosylation of protein—A combinational use of anti-ADP-ribosylarginine antibody and ADP-ribosylarginine hydrolase. J Biochem Biophys Methods. 2008;70:1014–1019. doi: 10.1016/j.jprot.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 28.Lupi R, et al. Endogenous mono-ADP-ribosylation of the free Gβγ prevents stimulation of phosphoinositide 3-kinase-γ and phospholipase C-β2 and is activated by G-protein-coupled receptors. Biochem J. 2002;367:825–832. doi: 10.1042/BJ20020660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laitusis AL, Brostrom MA, Brostrom CO. The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J Biol Chem. 1999;274:486–493. doi: 10.1074/jbc.274.1.486. [DOI] [PubMed] [Google Scholar]
- 30.Coburn J, Dillon ST, Iglewski BH, Gill DM. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fendrick JL, Iglewski WJ, Moehring JM, Moehring TJ. Characterization of the endogenous ADP-ribosylation of wild-type and mutant elongation factor 2 in eukaryotic cells. Eur J Biochem. 1992;205:25–31. doi: 10.1111/j.1432-1033.1992.tb16748.x. [DOI] [PubMed] [Google Scholar]
- 32.Raffaelli N, Scaife RM, Purich DL. ADPRibosylation of chicken red cell tubulin and inhibition of microtubule self-assembly in vitro by the NAD+-dependent avian ADPRibosyl transferase. Biochem Biophys Res Commun. 1992;184:414–418. doi: 10.1016/0006-291x(92)91209-9. [DOI] [PubMed] [Google Scholar]
- 33.Satchell MA, et al. A dual role for poly-ADP-ribosylation in spatial memory acquisition after traumatic brain injury in mice involving NAD+ depletion and ribosylation of 14–3-3γ. J Neurochem. 2003;85:697–708. doi: 10.1046/j.1471-4159.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J Biol Chem. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 35.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto H, et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 38.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 41.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.