Abstract
The cyclin D1–cyclin-dependent kinase 4 (CDK4) complex is a key regulator of the transition through the G1 phase of the cell cycle. Among the cyclin/CDKs, CDK4 and cyclin D1 are the most frequently activated by somatic genetic alterations in multiple tumor types. Thus, aberrant regulation of the CDK4/cyclin D1 pathway plays an essential role in oncogenesis; hence, CDK4 is a genetically validated therapeutic target. Although X-ray crystallographic structures have been determined for various CDK/cyclin complexes, CDK4/cyclin D1 has remained highly refractory to structure determination. Here, we report the crystal structure of CDK4 in complex with cyclin D1 at a resolution of 2.3 Å. Although CDK4 is bound to cyclin D1 and has a phosphorylated T-loop, CDK4 is in an inactive conformation and the conformation of the heterodimer diverges from the previously known CDK/cyclin binary complexes, which suggests a unique mechanism for the process of CDK4 regulation and activation.
Keywords: cell cycle, kinase, X-ray, CDK4, cyclinD
Cyclin-dependent kinases (CDKs) are a conserved family of proline-directed serine/threonine kinases that perform critical roles in regulating the stepwise progression through the eukaryotic cell cycle. The activity of CDKs is regulated through phosphorylation by other upstream kinases such as CDK-activating kinases (CAKs) and most significantly by interaction with cyclins (1, 2). In turn, CDK/cyclin complexes are inhibited through the reversible binding of CDK inhibitors from the Cip/Kip and INK protein families and through the cyclical degradation of cyclins during the cell cycle (reviewed in ref. 3).
CDK4 and CDK6 associate with the D-type cyclins (D1, D2, D3) and phosphorylate and inactivate the retinoblastoma (Rb) protein family members (p107, p130, pRb). Phosphorylation of pRb by CDK4/6 then leads to the derepression and activation of E2F target genes, including the E-type cyclins, which facilitate progression through the G1 phase of the cell cycle.
Deregulation of the CDK4/cyclin D pathway has been identified in many cancers (refs. 4 and 5 and references therein and ref. 6). Notably, most genetic alterations target specifically CDK4 or cyclin D1, whereas alterations in other CDKs and cyclins are far less common. The CDK4 gene is amplified in a high percentage of liposarcomas (7), and breast cancers frequently exhibit high cyclin D1 levels, either through genetic amplification of the gene or overexpression (8). Translocation of cyclin D1 to the IgH promoter is a hallmark aberration in mantle cell lymphoma (9). Cyclin D1 translocations can also be detected in many cases of multiple myelomas (10). A mutation of CDK4 (Arg-24–Cys) that renders it refractory to inhibition by the tumor suppressor protein p16INK4a has also been identified, and, similarly, deletion or mutation of the p16INK4a gene results in defective CDK4 inhibition and dysregulated CDK4 activity (11). Finally, genetic inactivation of p16INK4 is among the most frequent tumor suppressor mutations found in human cancers. Taken together, these data indicate that an unchecked or hyperactivated CDK4/cyclin D1 pathway may be responsible for enhanced cellular proliferation in cancers and imply that CDK4 is a promising target for the development of anticancer therapies (reviewed in ref. 12).
The molecular basis of CDK activation has been the focus of many studies using cellular, biochemical, and structural approaches (reviewed in ref. 3). Maximal CDK activation requires both binding of a cognate cyclin and phosphorylation of residues within the CDK T-loop, and X-ray crystallographic studies of various CDKs and CDK/cyclin complexes have identified the conformational movements associated with CDK activation (13–16). Activation is characterized by movement of the αC-helix such that a highly-conserved glutamate residue moves into the kinase active site and an arginine residue is positioned to interact with the phosphothreonine of the T-loop. The T-loop moves away from the active site and is held in position by interactions with the cyclin and interaction of the phosphothreonine with a cluster of arginine residues.
Despite the significant interest in CDK4 its structure determination has eluded numerous efforts from many groups. Here, we describe the crystal structure of human CDK4 in complex with a cognate D-type cyclin.
Results
Coexpression of CDK4 and cyclin D1 in insect cells yielded an active, but heterogeneously-phosphorylated, complex that did not crystallize. To facilitate crystallization of the CDK4/cyclin D1 complex a number of modifications were required (see SI Text). The C-terminal 24 residues of cyclin D1, containing a polyglutamate region and phosphorylation site, were removed to reduce predicted conformational flexibilty and phospho-heterogeneity. Residues 42–48 of CDK4, consisting of 7 glycine residues, were replaced with the equivalent GEEG sequence from CDK6. Combining these changes gave a small, but significant, increase in the thermal stability of the complex and proved crucial for obtaining crystals that diffracted to high resolution. In different complexes, the site of CDK4 T-loop phosphorylation (Thr-172) was either phosphorylated (T172Ph), changed to alanine (T172A, phospho knockout) or changed to aspartate (T172D, phosphomimetic) (see Fig. S1). The constructs and their relative enzymatic activities are described in Table 1. All constructs that yielded crystals diffracting to high resolution gave structures that were highly homologous to each other. In each case the N-terminal 20 residues of cyclin D1 were disordered, thus a new construct was generated in which residues 1–14 of cyclin D1 were removed. This process resulted in a complex that crystallized readily from a variety of conditions, but did not result in higher-resolution data. Because all of the structures were equivalent to each other the key features of the structure will be described for the complex that gave the highest-resolution data (CDK4 T172D, Gly-42–Glu-43′–Glu-44′–Gly-48/cyclin D11–271).
Table 1.
Relative activities of constructs expressed as a percentage relative to wild-type CDK4/cyclin D1
| Construct | Activity relative to wild type, % |
|---|---|
| Wild type (CDK4/CycD1) | 100 |
| CDK4EET172Ph/CycD11–271 | 290 |
| CDK4EET172D/CycD11–271 | 2 |
| CDK4EET172A/CycD11–271 | 12 |
| CDK4EET172Ph/CycD115–271 | 26 |
| CDK4EET172A/CycD115–271 | 2 |
| CDK2(T160Ph)/CycA | 175 |
The relative activities of the constructs were determined by measuring the initial rates of reaction for each construct in the presence of 2 mΜ ATP. Rates were then normalized relative to the wild-type construct. CDK4EE represents CDK4 with residues 42–48 replaced by Gly42–Glu43′–Glu44′–Gly48, the sequence found in CDK6. Ph represents phosphorylation of the indicated residue.
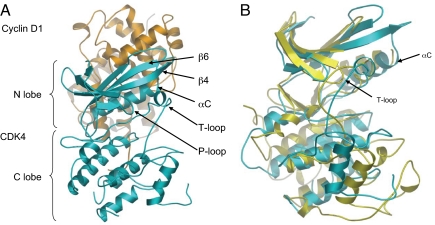
CDK4 has the typical bilobal structure seen in other kinases, comprising a 5-stranded β-sheet, N-terminal domain (residues 1–96), in which helix-αC (sequence PISTVRE in CDK4 and PSTAIRE in CDK2) is packed against the β-sheet, and a predominantly helical C-terminal domain (residues 97–303) (Fig. 1A). The ATP binding site is located in the cleft between the domains. Although this structure is unliganded, it suggests that only minimal conformational rearrangement of residues Asp-99, Asp-140, Lys-142, and Tyr-17 would be necessary to accommodate ATP binding in the active site.
Fig. 1.
Ribbon diagram of the CDK4 (cyan)/cyclin D1 (orange) heterodimer. (A) The N- and C-terminal lobes of the kinase are labeled as are key secondary structural elements. (B) CDK7 (yellow) (Protein Data Bank ID code 1UA2) and CDK4 (cyan) (rmsd 1.053 Å). Both the αC-helix and T-loop of CDK7 adopt inactive conformations that are similar to the conformations of the equivalent secondary structural elements observed in CDK4.
The main-chain density of the T-loop for the Thr-172–Ala and Thr-172–Asp substitutions is well defined, but the side-chain density is poor in this region (residues170–192). In the structure obtained with a phosphorylated T-loop there is no density for residues 170–175. The presence of CDK4 phosphorylated on Thr-172 was confirmed by mass spectrometry of dissolved crystals (see SI Text). However, in each case the T-loop has a short section of well-defined α-helical structure (residues 161–171) and is in the “loop in” conformation more characteristic of an “inactive” CDK (Fig. 1B). This ensemble of structures suggests that for CDK4 neither phosphorylation of the T-loop nor binding to its cognate cyclin is sufficient to trigger the conformational change typically associated with a fully-activated CDK, although enzymatic data demonstrate that the phosphorylated complex used to obtain the structure has robust activity against Rb protein substrate (see below and Table 1). Our data and lambda-phosphatase incubation studies (see Fig. S2 and Nick Brown, Oxford University, Oxford, United Kingdom, personal communication) suggest that the phosphate on the T-loop is solvent accessible and that T-loop phosphorylation is required for enzymatic activity.
The cyclin D1 molecule has a classical double cyclin box domain fold, comprising 11 α-helices (Fig. 2A). Intercomparison of cyclin D1 with the structures of other cyclins (A, E, K, M, vCyc) (Fig. S3 and Table S1) reveals the expected general equivalence between secondary structural elements. Cyclin D1 is known to contain 2 Rb binding sites (17) comprised of an N-terminal LxCxE consensus motif and a peptide recruitment site. The cyclin D1 structure exhibits N-terminal disorder, so the LxCxE motif cannot be visualized. Comparison of the cyclin D1 structure with cyclin A suggests that the peptide recruitment site, which recognizes a consensus RxL motif, is conserved in cyclin D1 and should be capable of comparable peptide binding (18) (Fig. S4D). This observation has been confirmed by peptide binding studies.
Fig. 2.
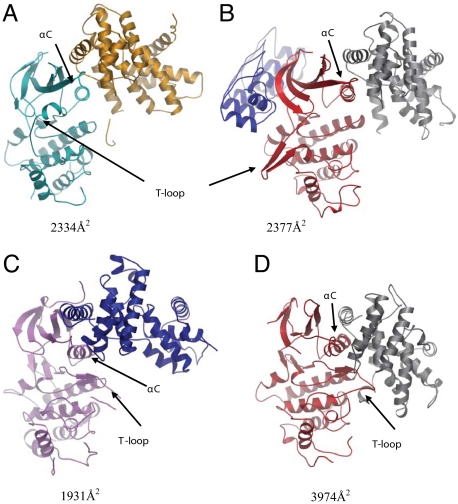
Superpositions of various CDK/cyclin complexes or CDKs with CDK4/cyclin D1. Only the kinases were used for the superpositions and rmsd values are given (see Table S2 and Fig. S5). For clarity each panel shows an individual CDK/cyclin complex. (A) Ribbon diagram of the CDK4 (cyan)/cyclin D1(orange) complex. The αC-helix is viewed end on. It can be seen that cyclin D1 primarily contacts the N-lobe and αC-helix of CDK4. The buried surface area of the complex is 2,334 Å2. (B) CDK6 (red)/cyclin K (gray)/INK4c (blue) ternary complex (Protein Data Bank ID code 1G3N) (rmsd 0.888 Å). INK binding maintains the αC-helix in an inactive conformation, triggers a reconfiguration of the T-loop, and induces a rotation of the CDK6 N-lobe. Cyclin K mainly contacts the CDK6 N-lobe and αC-helix, resulting in a relatively small contact interface (2,377-Å2 buried surface area) compared with CDK/cyclin binary complexes (see D). Although the CDK6 N-lobe is rotated with respect to CDK4 the cyclin K and cyclin D1 are similarly engaged and disposed in each complex. (C) CDK9 (pink)/cyclin T1(blue) (Protein Data Bank ID code 3BLH) (rmsd 1.387 Å). Even though cyclin T1 only forms sparse contacts (buried surface area 1,931 Å2) with the kinase N-lobe the CDK αC-helix and T-loop adopt active conformations. The relative disposition of the CDK and cyclin deviate from the conformations typically seen in active CDK/cyclin complexes. (D) CDK6 (red)/ vCyclin (gray) (Protein Data Bank ID code 1JOW) (rmsd 1.114 Å). Although the kinases broadly overlay the relative disposition of their cognate cyclins is substantially different. The vCyclin engages with both the N- and C-terminal lobes of CDK6 and forms extensive contacts with the T-loop of CDK6. The CDK6 T-loop has a loop out conformation that is characteristic of a fully-activated CDK, as is the αC-helix conformation that is rotated to an in position. The buried surface area of the CDK6/vCyclin interface is 3,974 Å2. This is a high value for CDK/cyclin binary complexes and helps to explain how the viral cyclin is able to activate the CDK6 even in the absence of phosphorylation of the CDK6 T-loop.
The most striking feature of the CDK4/cyclin D1 complex is the interaction between the 2 molecules. Although cyclin D1 has engaged with the αC-helix (PISTVRE) of CDK4, the helix has not undergone the conformational switch associated with CDK activation, as observed in all other CDK/cyclin binary complexes solved to date (Fig. 2) (14–16, 19). Instead the CDK4 structure more closely resembles the structures of inactive and non-cyclin-bound CDK2 and CDK7 (Fig. 1B and Table S2) (13, 20), or an INK-inhibited CDK6, particularly the CDK6/p19INK4d structure (21). Furthermore, the CDK4 T-loop is stabilized in an inactive conformation caused by interactions with both the N- and C-terminal lobes of the kinase (Fig. 1B). The residues immediately following the Asp158–Phe–Gly160 (DFG) motif have condensed into a helix (Leu161–Leu171) that is stabilized by interactions with αC-helix, β4-strand, β6-strand, and the apex of the P-loop (Fig. 1). Again, broadly similar T-loop architectures are observed with CDK7, where the T-loop is phosphorylated (Fig. 1B), and with CDK6 in the CDK6/p19INK4d complex. The interconnectivity of these secondary structural elements in CDK4 indicates how activation signals could be transmitted throughout the kinase upon movement of the αC-helix.
Although the CDK4 αC-helix appears to have been engaged and bound by cyclin D1 in a comparable fashion to other heterodimeric complexes, the C-lobe of CDK4 has not rotated to maximally associate with the cyclin N terminus. This lack of rotation reduces the buried surface area of the CDK4/cyclin D1 interface (2,334 Å2) relative to the majority of other CDK/cyclin binary complexes (typically in the 3,000–4,000 Å2 range; Fig. 2, Table S3, and Fig. S5). This conformation of the CDK4/cyclin D1 complex is observed in all of our structures, even though they were crystallized under a diverse set of conditions. In terms of relative subunit disposition and CDK/cyclin interface, the most analogous structures are the ternary complex comprising CDK6/cyclin K bound to the protein inhibitor p18INK4c (22) (Fig. 2B) and the binary CDK9/cyclinT1 structure (19) (Fig. 2C). INK4c binding is distal to the CDK6/cyclin K interface and induces a rotation of the CDK6 N-terminal lobe relative to the C-terminal domain. This rotation distorts the CDK6 active site, induces the T-loop to adopt a distinct, inhibited conformation, and drives the αC-helix into an inactive, or “out,” configuration. These CDK6 conformational changes also remodel the CDK6/cyclin K interface such that cyclin K only engages with the CDK6 αC-helix and the N-terminal lobe (Fig. 2B), which is highly reminiscent of what is observed in the CDK4/cyclin D1 structure (Fig. 2A). Inspection of the CDK4/cyclin D1 crystal packing reveals an absence of lattice contacts capable of mimicking INK binding, thereby reducing the possibility of the conformation of the complex being an artifact of crystallization (Fig. S6). In the CDK9/cyclin T1 structure cyclin binding drives the αC-helix into an active, in, conformation; however, the relative disposition of the cyclin and CDK and the buried surface area of the complex deviate significantly from other activated CDK/cyclin heterodimers (Fig. 2C, Fig. S5, and Table S3). Cyclin T1 is pivoted away from the CDK by ≈26°, reducing the buried surface area to ≈60% of that observed in the CDK2/cyclin A complex (19).
Discussion
The CDK4/cyclin D1 structure suggests that there may be 2 general mechanisms for CDK/cyclin regulation. The first is inhibition of CDK/cyclin complexes, through the binding of proteinaceous inhibitors. This would apply to CDK/cyclin complexes that spontaneously adopt an active conformation upon heterodimerization (22). The second mechanism would require activation of CDK/cyclin heterodimers, such as CDK4/cyclin D1, that do not spontaneously remodel into an active conformation upon association. This activation would be via cofactor (e.g., Cip/Kip proteins) and/or substrate binding.
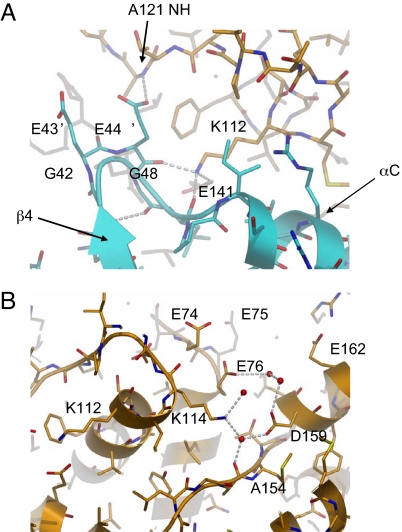
To facilitate crystallization, all 4 of our CDK4/cyclin D1 complexes contain CDK4 in which the loop [Gly-42–(Gly)5–Gly-48, Gly7] immediately preceding the αC-helix has been truncated and modified to the equivalent sequence found in CDK6 [Gly-42–Glu-43′–Glu-44′–Gly-48, GEEG) (Fig. 3A). An intercomparison of the structure of this modified loop in CDK4/cyclin D1 with the equivalent loops in CDK2/cyclin A and CDK6/vCyc reveals a high degree of structural parity in this region (Fig. S4). In all 3 systems the loop conformation is stabilized by main-chain hydrogen bonds from the loop to a highly-conserved lysine (LysD1112) and glutamate (GluD1141) on the cyclin. The glutamates from the engineered CDK4 GEEG replacement mimic the interactions formed by the identical loop in CDK6 and help to further stabilize this region of the CDK4/cyclin D1 structure. Although we cannot rule out the possibility that the introduced substitutions have stabilized an intermediate conformational state, Coleman et al. (23) have demonstrated that truncation of the CDK4 Gly7 loop does not perturb either cyclin D1 or p16 binding. The enzymatic activity of phosphorylated CDK4 GEEG in complex with truncated cylin D1 was comparable to that of CDK4/cyclin D1 in which the Gly7 loop had not been modified (Table 1), suggesting that the crystallized complex is structurally and functionally equivalent to the wild-type complex. This assertion is further supported by the structure of native CDK4 in complex cyclin D3 described by Takaki et al. (40) in this issue, which does not contain substitutions in this region but yields a homologous complex structure. Furthermore, the interactions observed in this region are consistent with previous studies (24–26) that have shown that mutation of LysD1112 or LysD1114 (Fig. 3) results in cyclins that are defective in their ability to activate and/or bind CDK4. The LysD1112–Glu mutation would disrupt the stabilizing H-bond interactions with the β4–αC helix loop and GluD1141. The LysD1114–Glu mutation would introduce an additional, potentially destabilizing, negative charge into the acidic environment formed by GluD174–Glu–GluD176 and AspD1159 and GluD1162 (Fig. 3B). This mutation, through its proximity to the CDK/cyclin interface and αC-helix, could again disrupt complex activation.
Fig. 3.
Architecture of the engineered loop preceding the αC-helix in the CDK4 (cyan)/cyclin D1 (orange) structure. (A) Akin to CDK2 and CDK6 (see Fig. S4) the apex of the loop is stabilized by hydrogen bonds from the loop main chain to a highly-conserved lysine (LysD1112) and glutamate (GluD1141) on the cyclin. The second glutamate (GluK444′) from the GE′E′G insertion mimics the interactions formed by the glutamate in the CDK6 structure. The loop is further stabilized by intramolecular H-bonds, which are not observed in either the CDK2 or CDK6 structures. A cyclin D1 LysD1112–Glu mutation results in aberrant CDK4/cyclin D1 complex assembly and activation. (B) Residues in the vicinity of cyclin D1 (orange) LysD1114. The LysD1114–Glu mutation results in defective CDK4/cyclin D1 complex formation. LysD1114 sits within an acidic environment formed by GluD174, GluD175, GluD176, AspD1159, and GluD1162. It would be anticipated that introduction of an additional negative charge into this environment would be highly destabilizing and significantly perturb correct CDK/cyclin association.
The activation and kinetics of the CDK4/cyclin D1 complex differ from other CDK/cyclin complexes. p50cdc37 and Hsp90 have been shown to be important for CDK4 maturation (ref. 27 and references therein), and members of the (Cip/Kip/INK) protein families can modulate the assembly, stability, activity and correct nuclear localization of CDK4/cyclin D complexes (1, 28–33). Consistent with these data it has been shown that active CDK4/CycD1 complex cannot be readily reconstituted by mixing recombinant CDK4 and cyclin D (2). Unlike CDK2/cyclin A (34), CDK4 does not appear to spontaneously activate when bound to cyclin D and exhibits ordered addition kinetics (35, 36). Phosphorylation studies using peptidic or Rb-protein substrates reveal a low specific activity of CDK4/cyclin D1 versus peptides and a modulation of the enzymatic Km for ATP (36–39). On the basis of these data and our structure it can be speculated that protein substrate and/or cofactor binding may be necessary to promote remodeling of the CDK4/cyclin D1 interface and αC-helix into active conformations. The observation that deletion of the N-terminal region of cyclin D1 resulted in a large reduction in catalytic activity of the complex perhaps suggests that this region may play a role in this remodeling process (see SI Text). It can also be speculated that the CDK/cyclin conformation of the remodeled CDK4/cyclin D1 complex may more closely mimic the conformation of CDK9/cyclin T1 rather than CDK2/cyclin A.
We propose that the CDK4/cyclin D1 complex presented here represents a stable, and previously structurally uncharacterized, intermediate state in which the CDK is potentiated for final activation. It is distinct from most other CDK/cyclin complexes in which cyclin binding triggers spontaneous remodeling and activation of the CDK. Furthermore, complete activation of the CDK4/cyclin D1 complex may only occur with Cip/Kip ternary complex formation, nuclear translocation, substrate recruitment/binding, and T-loop phosphorylation. This mechanism would provide an elegant means for tightly regulating CDK4 activity until the CDK4/cyclin D1 complex was correctly localized within the cell and productively bound to substrate.
Materials and Methods
IMAGE Clones.
IMAGE consortium cloneID MGC2316 (cyclin D1) and IMAGE consortium cloneID MGC19704 (CDK4) (Geneservice Ltd.) were used for initial construct generation.
Protein Purification.
CDK4 and cyclin D1were expressed in SF21 insect cells and harvested 72 h after infection. After sonication and clarification, the complex was purified by using Ni-NTA agarose batch binding, HitrapQ FF ion exchange chromatography, and gel filtration on a Sephacryl S200HR(26/60) column. The purified complex was typically concentrated to 10–12 mg/mL in 20 mM Tris·HCl (pH 8), 10% glycerol, 150 mM NaCl, and 5 mM DTT for crystallization. All work was carried out at 4 °C.
Crystallization of the CDK4 T172D/Cyclin D11–271 Complex.
Protein was mixed 1:1 with reservoir solution composed of 0.1 M Hepes (pH 7.5), 0.5–0.8 M trisodium citrate in hanging drops and incubated at 20 °C. Large plates appeared in 1–3 days. Crystals were cryoprotected in 0.1 M Hepes (pH 7.5), 0.75 M trisodium citrate, and 20% (vol/vol) glycerol before freezing in liquid nitrogen and data collection.
Data were collected to 2.3-Å resolution at European Synchrotron Radiation Facility (Grenoble, France) beamline ID29. The crystals belonged to space group P212121 (a = 55.9, b = 64.7, c = 168.7 Å) with 1 heterodimer per asymmetric unit. The structure was solved by molecular replacement (see SI Text) and refined to give R = 0.204 and Rf = 0.261 (see SI Text). Data relating to the crystallization, data collection, and refinement of the other constructs can be found in Table S4.
Measurement of CDK Enzyme Activity.
CDK enzyme activity was measured by using an ELISA format. Briefly, plates were coated with GST-pRb769–921 (purified as described in ref. 7), washed with TBST [100 mM Tris (pH 7.5), 150 mM NaCl, 0.5% Tween-20], and blocked with Superblock (Perbio Science). Assay buffer [15 mM MgCl2, 50 mM Hepes (pH 7.4), 1 mM DTT, 1 mM EGTA (pH 8.0), 0.02% Triton X-100, and 2.5% DMSO] and enzyme [CDK4/cyclin D1 or CDK2(T160Ph)/cyclin A] were added to each well, and the reaction was initiated with the addition of ATP. At various time points reactions were stopped by the addition of 0.5 M EDTA, pH 8.0. Plates were then washed with TBST and incubated for 1 h with the primary antibody (CDK4: anti-p-Rb Serine 780, New England Biolab; Cdk2: p-Rb Threonine 821, Biosource) diluted in Superblock. Excess antibody was washed off and plates were then incubated with secondary antibody [alkaline phosphatase-linked anti-rabbit (New England Biolab) for another hour. After removal of excess secondary antibody, plates were developed by using the Attophos system (Promega), and the fluorescence was read on a Spectramax Gemini plate reader (Molecular Devices) at excitation 450 nm and emission 580 nm.
Supplementary Material
Acknowledgments.
We thank European Synchrotron Radiation Facility beamline staff and Glyn Williams, John Lyons, Suzanne Brewerton, Paul Mortenson, Steve Howard, David Rees, Pamela Williams, John Lyons, Nicola Wallis, Bill Sellers, Ian Hunt, Christopher Brain, Sunkyu Kim, and Tim Ramsey for fruitful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in Protein Data Bank, www.pdb.org (PDB ID codes 2W96, 2W99, 2W9F, 2W9Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809645106/DCSupplemental.
References
- 1.Bockstaele L, et al. Regulated activating Thr172 phosphorylation of cyclin-dependent kinase 4(CDK4): Its relationship with cyclins and CDK “inhibitors.”. Mol Cell Biol. 2006;26:5070–5085. doi: 10.1128/MCB.02006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato JY, Matsuoka M, Strom DK, Sherr CJ. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol. 1994;14:2713–2721. doi: 10.1128/mcb.14.4.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockstaele L, Coulonval K, Kooken H, Paternot S, Roger PP. Regulation of CDK4. Cell Div. 2006;1:25. doi: 10.1186/1747-1028-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors, and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 6.Malumbres M, Barbacid M. To cycle or not to cycle: A critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 7.Binh MB, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: A comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol. 2005;29:1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 8.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 9.Amin HM, et al. Characterization of 4 mantle cell lymphoma cell lines. Arch Pathol Lab Med. 2003;127:424–431. doi: 10.5858/2003-127-0424-COMCLC. [DOI] [PubMed] [Google Scholar]
- 10.Specht K, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood. 2004;104:1120–1126. doi: 10.1182/blood-2003-11-3837. [DOI] [PubMed] [Google Scholar]
- 11.Wolfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 12.Sridhar J, Akula N, Pattabiraman N. Selectivity and potency of cyclin-dependent kinase inhibitors. AAPS J. 2006;8:204–221. doi: 10.1208/aapsj080125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bondt HL, et al. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 14.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 15.Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 16.Schulze-Gahmen U, Kim SH. Structural basis for CDK6 activation by a virus-encoded cyclin. Nat Struct Biol. 2002;9:177–181. doi: 10.1038/nsb756. [DOI] [PubMed] [Google Scholar]
- 17.Pan W, Cox S, Hoess RH, Grafstrom RH. A cyclin D1/cyclin-dependent kinase 4 binding site within the C domain of the retinoblastoma protein. Cancer Res. 2001;61:2885–2891. [PubMed] [Google Scholar]
- 18.Leng X, Noble M, Adams PD, Qin J, Harper JW. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol Cell Biol. 2002;22:2242–2254. doi: 10.1128/MCB.22.7.2242-2254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumli S, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure (London) 2004;12:2067–2079. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Brotherton DH, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 22.Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev. 2000;14:3115–3125. doi: 10.1101/gad.851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman KG, et al. Identification of CDK4 sequences involved in cyclin D1 and p16 binding. J Biol Chem. 1997;272:18869–18874. doi: 10.1074/jbc.272.30.18869. [DOI] [PubMed] [Google Scholar]
- 24.Benzeno S, et al. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006;25:6291–6303. doi: 10.1038/sj.onc.1209644. [DOI] [PubMed] [Google Scholar]
- 25.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Boschelli F, Caplan AJ, Arndt KT. Identification of a conserved sequence motif that promotes Cdc37 and cyclin D1 binding to Cdk4. J Biol Chem. 2004;279:12560–12564. doi: 10.1074/jbc.M308242200. [DOI] [PubMed] [Google Scholar]
- 28.Blain SW, Montalvo E, Massague J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 29.Chang F, McCubrey JA. P21(Cip1) induced by Raf is associated with increased Cdk4 activity in hematopoietic cells. Oncogene. 2001;20:4354–4364. doi: 10.1038/sj.onc.1204564. [DOI] [PubMed] [Google Scholar]
- 30.Cheng M, et al. The p21(Cip1) and p27(Kip1) CDK “inhibitors: are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaBaer J, et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 32.Parry D, Mahony D, Wills K, Lees E. Cyclin D-CDK subunit arrangement is dependent on the availability of competing INK4 and p21 class inhibitors. Mol Cell Biol. 1999;19:1775–1783. doi: 10.1128/mcb.19.3.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto M, et al. Activation of cyclin D1-kinase in murine fibroblasts lacking both p21(Cip1) and p27(Kip1) Oncogene. 2002;21:8067–8074. doi: 10.1038/sj.onc.1206019. [DOI] [PubMed] [Google Scholar]
- 34.Morris MC, Gondeau C, Tainer JA, Divita G. Kinetic mechanism of activation of the Cdk2/cyclin A complex. Key role of the C-lobe of the Cdk. J Biol Chem. 2002;277:23847–23853. doi: 10.1074/jbc.M107890200. [DOI] [PubMed] [Google Scholar]
- 35.Clare PM, et al. The cyclin-dependent kinases cdk2 and cdk5 act by a random, anticooperative kinetic mechanism. J Biol Chem. 2001;276:48292–48299. doi: 10.1074/jbc.M102034200. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinidis AK, Radhakrishnan R, Gu F, Rao RN, Yeh WK. Purification, characterization, and kinetic mechanism of cyclin D1. CDK4, a major target for cell cycle regulation. J Biol Chem. 1998;273:26506–26515. doi: 10.1074/jbc.273.41.26506. [DOI] [PubMed] [Google Scholar]
- 37.Grafstrom RH, Pan W, Hoess RH. Defining the substrate specificity of cdk4 kinase–cyclin D1 complex. Carcinogenesis. 1999;20:193–198. doi: 10.1093/carcin/20.2.193. [DOI] [PubMed] [Google Scholar]
- 38.Pan W, Sun T, Hoess R, Grafstrom R. Defining the minimal portion of the retinoblastoma protein that serves as an efficient substrate for cdk4 kinase/cyclin D1 complex. Carcinogenesis. 1998;19:765–769. doi: 10.1093/carcin/19.5.765. [DOI] [PubMed] [Google Scholar]
- 39.Wallace M, Ball KL. Docking-dependent regulation of the Rb tumor suppressor protein by Cdk4. Mol Cell Biol. 2004;24:5606–5619. doi: 10.1128/MCB.24.12.5606-5619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takaki T, et al. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci USA. 2009;106 doi: 10.1073/PNAS.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.