Abstract
Lymphocyte function-associated antigen 1 (LFA-1) plays important roles in immune cell adhesion, trafficking, and activation and is a therapeutic target for the treatment of multiple autoimmune diseases. Efalizumab is one of the most efficacious antibody drugs for treating psoriasis, a very common skin disease, through inhibition of the binding of LFA-1 to the ligand intercellular adhesion molecule 1 (ICAM-1). We report here the crystal structures of the Efalizumab Fab alone and in complex with the LFA-1 αL I domain, which reveal the molecular mechanism of inhibition of LFA-1 by Efalizumab. The Fab binds with an epitope on the inserted (I) domain that is distinct from the ligand-binding site. Efalizumab binding blocks the binding of LFA-1 to ICAM-1 via steric hindrance between its light chain and ICAM-1 domain 2 and thus inhibits the activities of LFA-1. These results have important implications for the development of improved antibodies and new therapeutic strategies for the treatment of autoimmune diseases.
Integrins are a family of large cell surface adhesion molecules composed of noncovalently linked α and β subunits that mediate cell-to-cell, cell-to-extracellular-matrix, and cell-to-pathogen interactions (1–5). They respond dynamically to a wide variety of signals and act as key regulators of many cellular processes. In the classical “outside-in” signaling process, ligand binding induces conformational changes of integrins and then transduces signals from the extracellular domain to the cytoplasm. In the “inside-out” signaling process (also called priming), intracellular signals impinge on the cytoplasmic domains of integrins and then alter their adhesiveness for extracellular ligands. These dynamic properties of integrins are critical to their proper functions.
Lymphocyte function-associated antigen 1 (LFA-1, αLβ2 or CD11a/CD18) consisting of an αL subunit of 180 kDa and a β2 subunit of 95 kDa, belongs to the β2 integrin subfamily, which contains 4 members characterized by a common β2 subunit (6). These β2 integrins are widely expressed in the immune system and play important roles in immune cell adhesion, trafficking, and activation (7, 8). LFA-1 is present on all leukocytes and recognizes intercellular adhesion molecules (ICAMs), which are members of the Ig superfamily (9). ICAM-1 is highly inducible on antigen-presenting cells and endothelium by cytokines in inflammation and is the most important ligand for LFA-1-dependent adhesion of B, T, and myeloid cells (10). The ligand-binding site for ICAM-1 in LFA-1 has been mapped to an ≈180-residue region of the αL subunit entitled inserted (I) domain (3, 11), which has also been shown to be a key ligand-binding domain in many other integrins (12, 13). The αL I domain assumes a typical Rossmann fold and contains a conserved metal ion-dependent adhesion site (MIDAS) consisting of residues Asp-137–Ser-141, Thr-206, and Asp-239 at the C-terminal end of the central β-sheet, which is the binding site for ligands (14, 15). The MIDAS binds an Mg2+ ion under physiological conditions, which mediates interactions with the ligands. The I domain can exist in 3 conformational states: a closed state, an open state, and an intermediate state, and the integrins can have at least 3 overall conformational states: A bent state, an extended-open state, and an extended-closed state (5, 11, 15–20).
Because LFA-1 plays important roles in many cellular processes, disorder of its functions can cause serious autoimmune and inflammatory diseases and is implicated in multiple cancers including myeloma, malignant lymphoma, and acute and chronic leukemias (21–25). Thus, it has become a therapeutic target for the treatment of multiple autoimmune and inflammatory diseases and cancers (8, 26, 27). Psoriasis is a very common skin disease that is characterized by red or salmon pink color, white or silver scaly and raised plaques (22). Although the cause of psoriasis remains an enigma, it has become increasingly clear that the activity of the lymphocytic infiltrate consisting primarily of T cells is the driving force for induction of the changes in psoriasis and is also required for maintenance of the plaques (28).
On the basis of the pathogenesis of psoriasis, therapies targeted at T cells have been designed and applied. Among many anti-CD11a monoclonal antibodies (29–34), murine monoclonal antibody (mAb) MHM24 was developed to a recombinant, humanized monoclonal IgG1 antibody Efalizumab (Raptiva, Genentech) (35), which has become one of the most efficacious drugs for treating psoriasis. Efalizumab was shown to be efficacious in treating patients with psoriasis (36) and can inhibit the extravasation and (re-)activation of T lymphocytes, and their interactions with keratinocytes (37). MHM24 binds specifically to LFA-1 and blocks the binding of ICAM-1 (38, 39). The epitope of MHM24 was mapped to the region containing residues Lys-197 to His-201 of the αL I domain of LFA-1 (29).
To investigate the structural basis of the recognition and binding of Efalizumab with LFA-1 and the molecular mechanism of inhibition of LFA-1 by Efalizumab, we determined the crystal structures of the Efalizumab Fab fragment alone and in complex with the αL I domain of LFA-1. The Efalizumab Fab binds to the I domain mainly via the 3 heavy-chain complementarity determining regions (CDRs). The epitope on the I domain for Efalizumab is located nearby but does not overlap with the MIDAS. The binding of Efalizumab does not occlude the binding site for ICAM-1, but the light chain of the Fab occupies the spatial position of ICAM-1 domain 2 in the ICAM-1/I domain complex, thus preventing ICAM-1 domain 1 from accessing the ligand-binding site of the I domain. Our structural data suggest that Efalizumab binding blocks the binding of LFA-1 to ICAM-1 via steric hindrance and thus inhibits the activities of LFA-1. These findings provide a structural basis for the development of improved antibodies and new strategies for treatment of psoriasis and other autoimmune diseases.
Results and Discussion
Structures of the Efalizumab Fab Alone and in Complex with the LFA-1 αL I Domain.
The crystal structure of the Efalizumab Fab alone was determined by the molecular replacement (MR) method and refined to 1.8 Å resolution (Table 1). There is 1 Fab molecule in the asymmetric unit, consisting of residues 1–213 of the light chain and residues 1–136 and 142–219 of the heavy chain. The Efalizumab Fab has a canonical immunoglobin fold consisting of 4 β-barrel domains (Fig. 1A). The elbow angle of the Fab, defined as the subtended angle by the 2 pseudo 2-fold axes relating VH to VL and CH to CL, was calculated as 185.5°, consistent with the observation that Efalizumab has a κ light chain (35, 40). The complementarity determining regions (CDRs) are clearly defined in this antigen-free structure. The H3 loop lies between the light-chain CDRs L1, L2, and L3 and the heavy-chain CDRs H1 and H2, and has both hydrophobic and hydrophilic interactions with the other CDRs.
Table 1.
Summary of diffraction data and structure refinement statistics
| Fab | Complex | |
|---|---|---|
| Diffraction data | ||
| Wavelength (Å) | 1.5418 | 1.0000 |
| Space group | P3121 | P212121 |
| Resolution (Å) | 20.00–1.80 (1.86–1.80)* | 50.00–2.80 (2.90–2.80) |
| Cell parameters | ||
| a (Å) | 87.2 | 64.7 |
| b (Å) | 87.2 | 81.7 |
| c (Å) | 117.2 | 281.9 |
| Observed reflections | 340,383 (30,994) | 463,213 (15,067) |
| Unique reflections (I/σ(I) > 0) | 47,282 (4,699) | 37,165 (3,139) |
| Average redundancy | 7.2 (6.6) | 12.5 (4.8) |
| Average I/σ(I) | 6.3 (1.9) | 21.1 (2.7) |
| Completeness (%) | 98.1 (99.8) | 98.1 (84.7) |
| Rmerge (%)† | 8.3 (37.3) | 11.8 (34.3) |
| Refinement and structure model | ||
| Reflections (Fo ≥ 0σ(Fo)) | ||
| Working set | 44,966 | 35,307 |
| Test set | 2,347 | 1,850 |
| R factor‡ | 0.189 | 0.225 |
| Free R factor | 0.217 | 0.264 |
| No. of protein atoms | 3,275 | 9,442 |
| No. of water atoms | 625 | 356 |
| Average B factor (Å2) | ||
| All atoms | 22.0 | 31.4 |
| Fab main chain/side chain | 19.7/20.6 | 32.2/32.1 |
| I domain main chain/side chain | 29.4/30.3 | |
| Water | 33.2 | 31.5 |
| RMS deviations | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles (°) | 1.3 | 1.1 |
| Ramachandran plot (%) | ||
| Most favoured regions | 91.2 | 88.2 |
| Additional allowed regions | 8.3 | 10.5 |
| Generously allowed regions | 0.5 | 1.3 |
| Luzzati atomic positional error (Å) | 0.21 | 0.37 |
*Numbers in parentheses represent the highest resolution shell.
†Rmerge = ΣhklΣi|Ii(hkl)i−<I(hkl)>|/ΣhklΣiIi(hkl).
‡R = Σhkl||Fo|−|Fc||/Σhkl|Fo|.
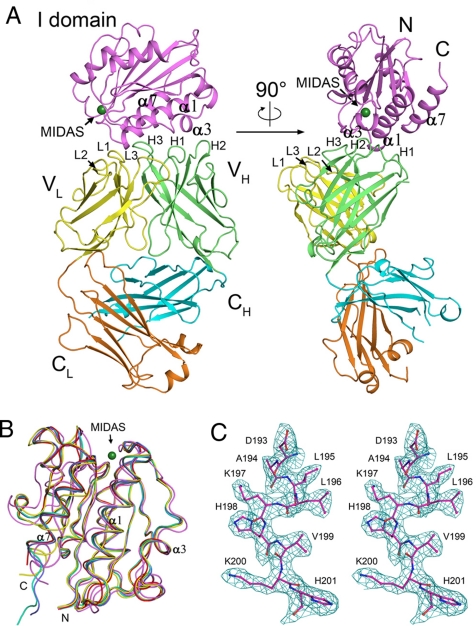
Fig. 1.
Structure of the Efalizumab Fab/LFA-1 αL I domain complex. (A) Overall structure of the Fab/I domain complex. The I domain is colored in magenta. The Fab is colored as the V and C domains of the light chain in yellow and orange, and the V and C domains of the heavy chain in green and cyan, respectively. The MIDAS is shown with the position of Zn2+ by a green ball. (B) Structural comparison of the αL I domain in different conformational states. The I domain adopts a closed conformation in the unliganded form (PDB code 1ZON, yellow) and an open conformation in the complex with ICAM-1 (PDB code 1MQ8, magenta). The major conformational differences occur at the MIDAS, which is the binding site for ICAM-1 and the C-terminal α-helix α7, which relays the conformational signals from the MIDAS to other domains of the integrin in the activation of LFA-1. The αL I domain in complex with the Efalizumab Fab (red) adopts the closed conformation as seen in the unliganded form. The αL I domain bound with Mg2+ (PDB code 1ZOO, green), or Mn2+ (PDB code 1ZOP, cyan), or the small molecule drug lovastin (PDB code 1CQP, blue) also assumes the closed conformation with slight conformational change in the region of the metal-binding site or the inhibitor-binding site. (C) A representative composite-omit-map (1.0 σ contour level) in the epitope region of the I domain. All figures were prepared by PyMOL (58).
The Fab/I domain complex can be crystallized in 2 crystal forms. Crystals of form I belong to space group P212121 and can diffract to 2.8 Å resolution (Table 1), and crystals of form II belong to space group P6122 and can diffract to 3.6 Å resolution (Table S1). Both structures of the protein complex were solved using the MR method and contain 2 Fab/I domain complexes in the asymmetric unit, consisting of the Fab light-chain residues 1–214 and heavy-chain residues 1–136 and 142–220, and the I domain residues 128–306 (Fig. 1A). There is a Zn2+ atom bound at the MIDAS of each I domain in the form II complex structure, but no metal ion was identified in the form I complex structure. Other than that, the overall structures of the 4 Fab/I domain complexes in the 2 crystal forms are almost identical [root mean square deviation (RMSD) of 0.27–0.35 Å for the I domain, RMSD of 0.32–0.72 Å for the Fab, and RMSD of 0.35–0.84 Å for the complex]. Therefore, we will use 1 of the 2 complexes in the high-resolution structure in the following structural analysis and discussion.
Structure comparison of the Efalizumab Fab alone and in complex with the αL I domain indicates that the elbow angle of the Fab undergoes a dramatic change (185.5° in the free form and 132.5°–137.5° with a slight variation in the complex structures). Crystal packing analyses indicate that both the V and C domains are involved in interactions with symmetry-related molecules in both the free form and the complex. In addition, superposition of the V domains (residues 1–107 of the light chain and 1–120 of the heavy chain) of the Fab in the free form and the complex reveals an RMSD of 0.51 Å and superposition of the C domains (residues 108–213 of the light chain and 121–219 of the heavy chain) an RMSD of 0.65 Å, respectively. Furthermore, the 6 CDRs of the Fab in the free form and the complex adopt almost identical conformations. These results indicate that the binding of the I domain does not cause evident conformational changes of the CDRs and the V and C domains. Thus, it is very likely that the change of the elbow angle is caused by crystal packing due to the inherent flexibility of the elbow (40).
The LFA-1 αL I domain in the complex assumes the classic Rossmann fold, consisting of a central parallel β-sheet of 5 β-strands and an additional antiparallel β-strand, surrounded on both sides by amphipathic α-helices (Fig. 1A). Comparison with the previously reported structures of the αL I domain shows that the I domain in this complex assumes a conformation similar to the closed, low-affinity conformation of the unliganded I domain (an RMSD of 0.40–0.60 Å) (18) rather than the open conformation of the I domain in complex with ICAM-1 (an RMSD of 1.65 Å) (11) (Fig. 1B). Specifically, both the MIDAS (residues Asp-137–Ser-141, Thr-206, and Asp-239) which is the binding site for ICAM-1 and the C-terminal α-helix α7, which undergoes major conformational change to activate LFA-1 when ICAM-1 binds, adopt the conformations as seen in the unliganded I domain.
Interactions Between the Efalizumab Fab and the LFA-1 αL I Domain.
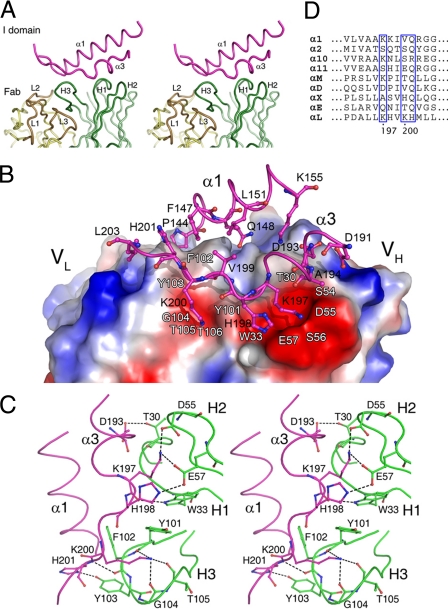
The interactions between the Efalizumab Fab and the LFA-1 αL I domain involve primarily the 3 heavy-chain CDRs of the Fab and 2 α-helices α1 (residues 144–155) and α3 (residues 191–203) of the I domain (Fig. 1A and Fig. 2A). In total, there are 111 van der Waals contacts and 10 hydrophilic interactions between 14 Fab residues and 14 I domain residues. The interaction interface buries 4.7% (1319.7 Å2) of the total solvent accessible surface area (9135.9 Å2 for the I domain and 19109.0 Å2 for the Fab), which is within the range for other antibody–antigen interaction (41). The shape complementarity statistics score (Sc) is 0.78, which is also higher than the average value (0.64–0.68) for other antibody–antigen complexes (42). These results are in agreement with the high binding affinity of the Efalizumab Fab with the αL I domain (KD = 2.2 ± 0.5 nM) (Fig. S1), which is comparable to that of the MHM24 Fab with the αL I domain (KD = 1.9 ± 0.4 nM) (43). It is noteworthy that the antibody–antigen interaction in the Efalizumab Fab/I domain complex relies almost exclusively on the 3 VH CDRs. In most of the antibody–antigen complexes, usually both VH and VL CDRs are involved in the antibody–antigen interaction with the VH CDRs playing a dominant role. A similar recognition mode of the antibody–antigen interaction with exclusive reliance on the VH CDRs has been observed in some other antibody–antigen complexes (44, 45).
Fig. 2.
Interactions between the Efalizumab Fab and the LFA-1 αL I domain. (A) A stereoview showing the interaction interface of the Fab/I domain complex and the relative role of each CDR loop in the interaction with the I domain. (B) An electrostatic potential surface of the Fab at the interaction interface. There are 2 negatively charged surface patches in the paratope to accommodate several important residues of the I domain. Residues of the I domain are shown side chains. The locations of some Fab residues are indicated with white labels for references. (C) A stereoview showing the hydrogen-bonding interactions between the Fab heavy chain and the epitope of the I domain. The Fab residues are colored in green and the I domain residues in magenta. Hydrogen bonds are indicated by dashed lines. (D) Sequence alignment of the I domains of all 9 known α subunit-containing integrins in the region containing the specificity determining residues. Residues corresponding to Lys-197, Lys-200, and His-201 of the LFA-1 αL I domain are indicated by blue boxes.
The 3 heavy-chain CDRs of the Fab form a large pocket to accommodate the epitope (Fig. 2A) and make extensive hydrophobic interactions with the I domain (Table S2). In particular, residues of the H3 loop contribute more than half of the total van der Waals contacts (58 out of 111). In contrast, the light chain (Tyr-49L of the L2 loop) has only 2 van der Waals contacts with Pro-144I of the I domain. (Residues of the I domain are designated by a superscripted suffix I, those of the Fab heavy chain by H, and those of the Fab light chain by L, respectively. The Efalizumab Fab uses the sequential numbering scheme and the Kabat numbering scheme of the Fab is provided in Table S3 for comparison.) Moreover, several aromatic residues including Trp-33H, Tyr-101H, Phe-102H, and Tyr-103H make 60 van der Waals contacts with residues Tyr-101H, Phe-102H, and Tyr-103H each having more than 15 contacts. These observations are consistent with the results that the heavy-chain CDRs usually make more contributions than the light-chain CDRs in antigen binding (46, 47) and aromatic residues usually make the majority of the hydrophobic interactions (48).
In addition to these hydrophobic interactions, 2 negatively charged surface patches in the pocket play important roles in the recognition and binding of the I domain (Fig. 2 B and C). One patch is formed by residues Tyr-101H–Thr-106H of the H3 loop. The side-chain Nζ of Lys-200I forms 3 hydrogen bonds with the main-chain carbonyls of Tyr-101H (2.7 Å), Gly-104H (2.8 Å), and Thr-105H (3.4 Å); the main-chain amide of His-201I forms a hydrogen bond with the main-chain carbonyl of Phe-102H (3.0 Å); and the side-chain Nδ1 of His-201I forms a hydrogen bond with the side-chain hydroxyl of Tyr-103H (2.7 Å). The other patch consists of residues Thr-30H and Trp-33H of the H1 loop and Asp-55H and Glu-57H of the H2 loop. The side chain of Lys-197I forms 2 salt bridges with the side chains of Asp-55H (3.2 Å) and Glu-57H (2.7 Å); the main-chain carbonyl of Lys-197I makes a hydrogen bond with the side-chain Nε1 of Trp-33H (2.9 Å); the side-chain Nε2 of His-198I forms a hydrogen bond with the side-chain Oε2 of Glu-57H (3.1 Å); and the side-chain Oδ2 of Asp-193I has a hydrogen bond with the side-chain Oγ1 of Thr-30H (2.6 Å).
Previous alanine scanning mutagenesis studies of Efalizumab have shown that the 3 heavy-chain CDRs and the light-chain CDR L3 are involved in the binding of Efalizumab to the LFA-1 I domain (35). Our structural results are in agreement with and can explain very well the biochemical data. The mutagenesis studies showed that mutations of Trp-33H of the H1 loop, Asp-55H and Glu-57H of the H2 loop, and Tyr-101H of the H3 loop have severe effects on the I domain binding. In the complex structure, these residues are involved in direct interactions with the αL I domain and therefore their mutations would severely affect the binding of the I domain. Mutations of Tyr-107H of the H3 loop and Lys-74H of the FWR-3 have moderate effects on the I domain binding. These 2 residues are located near the Efalizumab Fab/I domain interface, but have no direct interaction with the I domain. They appear to contribute to the conformational stabilization of the CDR loops. Furthermore, Gln-62H and Lys-65H of the H2 loop are suggested to be involved in the interactions with the I domain because their mutations have notable effects on the I domain binding (35). These 2 residues are located distantly from the interaction interface and may contribute indirectly to the stabilization of the conformations of the CDR loops. The mutagenesis data also showed that in addition to the heavy-chain CDRs, 3 light-chain L3 residues His-91L, Asn-92L, and Tyr-94L are involved in the antibody–antigen interaction as well because their mutations can substantially impair the binding of Efalizumab to the I domain. In the complex structure, these residues have no direct interaction with the I domain, but have interactions with the H3 loop. Mutations of these residues would alter the conformation of the H3 loop and thus affect the binding of the I domain.
The Efalizumab Epitope on the αL I Domain Is Distinct from the MIDAS.
Analyses of the free Fab structure and the Fab/I domain complex structure indicate that the Efalizumab epitope on the LFA-1 αL I domain is composed of the α-helices α1 and α3, and the conformation of the epitope is not altered upon the Fab binding. The MIDAS, which is the binding site for ICAM-1 comprises a DxSxS sequence (residues 137–141) from the β1-α1 loop, Thr-206 from the α3-α4 loop, and Asp-239 from the β4-α5 loop. The epitope is located nearby but does not overlap with the MIDAS (the distance between Lys-200I of the epitope and Zn2+ at the MIDAS is about 15 Å) (Fig. 1A). In particular, residues Lys-197I, Lys-200I, and His-201I of α-helix α3 contribute almost half of the hydrophobic interactions (50 out of 111) and more than half of the hydrophilic interactions (8 out of 10) and are the determinants for the specific recognition of the αL I domain by Efalizumab. These results are consistent with and provide a structural basis for the biochemical data in mapping the epitope of the αL I domain recognized by MHM24, the originating murine antibody of Efalizumab, showing that mutation K197D in the αL I domain completely abolished the binding with MHM24 and mutations K200D and H201A significantly diminished the binding, whereas mutations of other residues had very minor effects on the binding (29). Sequence alignment of the I domains of all 9 known α subunit-containing integrins (5) shows that Lys-197Ι has a counterpart only in the α1, α10, and αM I domains and Lys-200Ι and His-201Ι do not exist at the equivalent positions in the other I domains (Fig. 2D). This explains why Efalizumab can only bind specifically to the αL I domain but not other I domains.
Molecular Mechanism of Inhibition of LFA-1 by Efalizumab.
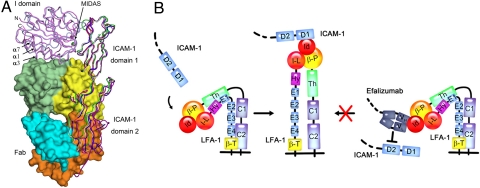
Previous structural data have shown that the MIDAS of the LFA-1 αL I domain is the binding site for ICAM-1 (11). Our structural data together with the previous biochemical data have revealed that the epitope of the LFA-1 αL I domain for Efalizumab consists of α-helices α1 and α3, which are located nearby but do not overlap with the MIDAS. The epitope has no spatial conflict with and/or structural constraint on the MIDAS. Thus, the inhibition of LFA-1 by Efalizumab cannot be through direct blockage of the ICAM-1 binding site. Then, how does the binding of Efalizumab to the I domain block the binding of ICAM-1 and then inhibit the activities of LFA-1? To understand this, we superimposed the structure of the Efalizumab Fab/I domain complex onto that of the ICAM-1/I domain complex (11) based on the I domain. The result clearly shows that although domain 1 of ICAM-1 has no serious spatial conflict with the Fab, domain 2 of ICAM-1 overlaps with the light chain of the Fab involving a large portion of the VL and CL domains and the linker (Fig. 3A). In other words, Efalizumab does not directly block domain 1 of ICAM-1 to access the ligand-binding site of the I domain, but the Fab light chain occupies the spatial position of domain 2 of ICAM-1 and thus prevents domain 1 from reaching the MIDAS. Thus, with Efalizumab bound to the I domain, ICAM-1 cannot bind to the I domain. Because the interactions between domain 1 and domain 2 of ICAM-1 are not very strong and the interdomain conformation can vary slightly in free form and in complex with the αL I domain (11, 49, 50), to examine the possibility of whether the steric clashes between the Efalizumab Fab and the ICAM-1 domain 2 could be avoided by interdomain movement of ICAM-1, we superimposed all structures of ICAM-1 with different interdomain conformations available in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) onto that of the aforementioned superimposed ICAM-1/I domain complex based on domain 1 of ICAM-1 (residues 1–82) (Fig. 3A). The results clearly show that for these interdomain conformations observed in the structures of ICAM-1 reported so far, the spatial clashes between the Fab light chain and the ICAM-1 domain 2 cannot be avoided.
Fig. 3.
Inhibition mechanism of LFA-1 by Efalizumab. (A) Structural comparison of the Fab/I domain complex, the ICAM-1/I domain complex, and ICAM-1. The structures of the Fab/I domain complex and the ICAM-1/I domain complex are superimposed on the basis of the I domain, and the structures of ICAM-1 are superimposed on the basis of domain 1 (residues 1–82). The Fab is shown with a surface representation in the same colors as in Fig. 1A. The I domain and ICAM-1 are shown with coiled ribbons. The MIDAS is shown with the position of Zn2+ by a green ball. The I domain in the Fab/I domain complex is colored in pink and the I domain in the ICAM-1/I domain complex in light blue. ICAM-1 in the ICAM-1/I domain complex (PDB code 1MQ8) is colored in blue (molecule A) and red (molecule B), ICAM-1 in the unliganded form (PDB code 1IAM) in silver, and ICAM-1 in the unliganded form (PDB code 1IC1) in green (molecule A) and magenta (molecule B), respectively. (B) A schematic diagram showing the inhibition mechanism of LFA-1 by Efalizumab. Upon ICAM-1 binding to the αL I domain, LFA-1 undergoes conformational changes and thus transforms the integrin from the inactive, bent conformation to the active, extended conformation. Binding of Efalizumab to the LFA-1 αL I domain blocks the binding of ICAM-1 via the steric hindrance between the Fab light chain and the ICAM-1 domain 2 and thus inhibits the activities of LFA-1.
On the basis of these results, we propose the inhibition mechanism of LFA-1 by Efalizumab (Fig. 3B). The binding of Efalizumab to the LFA-1 αL I domain does not occlude the binding site for ICAM-1; instead it blocks the binding of ICAM-1 via steric hindrance between the Fab light chain and the ICAM-1 domain 2 and thus inhibits the activities of LFA-1. This inhibition mechanism is reminiscent of that of epidermal growth factor receptor (EGFR) by Matuzumab (51). The binding of Matuzumab to domain III of EGFR does not occlude the binding site for EGF, but causes steric clashes between the light chain of the Matuzumab Fab and the N-terminal region of domain I of EGFR and thus prevents the local conformational changes and domain rearrangement that are required for high-affinity ligand binding and receptor dimerization. However, the inhibition mechanism of LFA-1 by Efalizumab is different from that of the α1β1 integrin by the AQC2 antibody (52). AQC2 binds to the MIDAS of the α1 I domain, and thus prevents directly the binding of the natural ligand at the MIDAS and inhibits the activities of the integrin.
In addition to Efalizumab, a number of antibodies have been reported that can bind specifically to the LFA-1 I domain and inhibit the LFA-1 activity. In the Efalizumab Fab/I domain complex the epitope for Efalizumab consists of residues of α1 (residues 144–155) and α3 (residues 191–203). Previous biochemical studies have shown that F8.8 and CBR-LFA-1/9 recognize Pro-144I, Lys-200I, and His-201I (30, 33, 34). BL5 recognizes Pro-144I, Lys-197I, and His-201I, and May.035 recognizes Lys-197I and His-201I. TS1/11 and TS1/12 also recognize Lys-197I, His-201I, and Leu-203I. These inhibitory antibodies share their epitope residues with those for Efalizumab and thus are likely to use a similar inhibition mechanism as that by Efalizumab. In addition, several other inhibitory mAbs also recognize the same epitope regions as those for Efalizumab, including CLB-LFA-1/2, 32E6, and 50G1 (29, 31). Furthermore, there are a number of other inhibitory mAbs that bind to the LFA-1 I domain but recognize epitopes different from that for Efalizumab. For example, antibodies TS1/22, TS2/14, and 25-3-1 recognize residues of the β5-α6 loop and helix α6 of the I domain (34). These mAbs appear to employ a different inhibition mechanism from that by Efalizumab.
Implications for Therapeutic Applications.
The structure of the Efalizumab Fab/I domain complex has revealed the detailed interactions between the antibody and the epitope. These structural data provide valuable information for the development of improved antibodies against psoriasis. On the basis of the complex structure, we could design changes of some residues on the CDRs of the Fab that could make favorable interactions and/or avoid unfavorable interactions with residues of the I domain to improve its binding affinity and specificity with LFA-1. For example, mutation of Ser-54H to Asp, Asn, Glu, or Gln could make more favorable hydrophilic interactions with Lys-197I. Replacement of Gly-31H with a polar residue might introduce hydrogen-bonding interactions with Asp-193I and/or Lys-155I. Substitution of Thr-30H with a basic residue (Arg or Lys) could generate hydrophilic interactions with Asp-191I and/or Asp-193I. Change of Ser-28H to a residue with a longer polar side chain could produce hydrogen-bonding interactions with Asp-152I and/or Asp-193I. Besides, our structural results could be used in the development of new therapeutic antibodies that target a similar region of the α I domain of other integrins against other autoimmune diseases and cancers. In addition, our findings could be exploited in designing new therapeutic strategies for the treatment of psoriasis and other autoimmune diseases and cancers. Since the epitope of the LFA-1 αL I domain by Efalizumab is distinct from the MIDAS and the binding sites for some other antibodies and antagonists, it might be advantageous to apply Efalizumab in combination with other drugs (antagonists or antibodies) that target different sites of the αL I domain to achieve synergistic or optimal therapeutic effect.
Methods
Protein Preparation.
The Fab fragment of Efalizumab was obtained by papain digestion of the Raptiva antibody (Genentech/Xoma). The digested protein sample was loaded onto a Protein A Sepharose 4 FF column (GE Healthcare), and the Fab fragment eluted in the flow through was separated from the Fc fragment and further purified with ion exchange chromatography using a Q-Sepharose FF column (GE Healthcare). The protein sample was concentrated to about 10 mg/mL and then exchanged to a stock buffer (10 mM Tris-HCl, pH 8.0, and 100 mM NaCl) for crystallization.
The cDNA sequence encoding residues Gly-128–Val-308 of human LFA-1 αL I domain with an enterokinase (EK) cleavage site (DDDDK) at the 5′-end was cloned into the pET32a vector (Novagen), which contains a thioredoxin (Trx) tag at the N terminus. The plasmid was transformed into Escherichia coli BL21(DE3) cells (Novagen). The transformed cells were grown in LB medium at 37 °C until OD600 reached 1.5 and expression of the protein was induced with 0.5 mM IPTG for 4 h at 20 °C. The bacterial cells were incubated in a lysis buffer (PBS, PBS) containing 1 mg/mL lysozyme, 1 mM PMSF, and 1% Triton X-100 for 20 min at 0 °C followed by sonication. The cell lysate was precipitated by centrifugation (10,000 g) and filtration (0.45 μm). The supernatant was purified with an Ni-NTA column (Novagen). The target protein was eluted with the lysis buffer supplemented with 0.4 M imidazole and the eluted sample was exchanged into 50 mM Tris-HCl, pH 8.0, and 50 mM NaCl with a Sephadex G-25 desalting column. For EK digestion, CaCl2 was added to the purified Trx-I domain fusion protein solution to a final concentration of 2 mM, and the mixture was incubated with recombinant EK (light chain) (Zhangjiang Biotech). The Trx-I domain fusion protein was digested by EK at 1 mg protein/1 U enzyme and incubated overnight at 21 °C. The digested protein sample was loaded onto an Ni-NTA column (Novagen) and the I domain eluted in the flow through was separated from the Trx tag and undigested fusion protein. The I domain was further purified with ion exchange chromatography using a Q-Sepharose FF column (GE Healthcare).
To prepare a homogenous Fab/I domain complex, the protein samples of the Efalizumab Fab and the αL I domain were mixed, concentrated and then loaded onto a Superdex 200 column (GE Healthcare). The protein complex was eluted in 20 mM Tris-HCl (pH 7.9) and concentrated to ≈15 mg/mL for crystallization. The concentrations of the protein samples were measured using a Bio-Rad protein assay kit and the purity and homogeneity were confirmed by SDS/PAGE and dynamic light scattering analyses.
Crystallization and Diffraction Data Collection.
Crystallization was carried out using the hanging drop vapor diffusion method. Crystals of the Efalizumab Fab alone grew in the drop consisting of 1.2 μL of the protein solution (10 mg/mL) and 2.1 μL of the reservoir solution (15 mM citric acid, pH 5.0, and 12% wt/vol PEG 3350). Crystals of the Fab/I domain complex grew in 2 forms. Crystals of form I grew in the drop consisting of 1 μL of the protein complex solution (15 mg/mL) and 1 μL of the reservoir solution (0.1 M sodium citrate, pH 5.3, 0.2 M sodium potassium tartrate, and 1.6 M ammonium sulfate). Crystals of form II grew in the drop containing equal volume (2 μL) of the protein complex solution and the reservoir solution (0.1 M sodium cacodylate, pH 7.3, 0.2 M zinc acetate, and 11% wt/vol PEG 8000).
Crystal of the Fab alone was cryoprotected using the reservoir solution supplemented with 20% glycerol and then flash cooled into the liquid N2 stream (−170 °C). Diffraction data were collected using a Rigaku R-AXIS IV++ diffractometer and processed with the CrystalClear suite (53). Crystal of the Fab/I domain complex of form I was cryoprotected using Paratone-N (Hampton Research) and crystal of form II was dehydrated against 50% wt/vol PEG 8000, and then flash cooled into the liquid N2 stream (−170 °C). Diffraction data of the protein complex of form I were collected at beamline NW12 and that of form II at beamline 17A of Photon Factory, Japan. Both data sets were processed with the HKL2000 suite (54). The statistics of the diffraction data are summarized in Table 1 and Table S1.
Structure Determination and Refinement.
Both structures of the Efalizumab Fab alone and in complex with the LFA-1 I domain were solved with the MR method using a combination of the programs CNS (55) and Phaser (56). For the structure of the free Fab fragment, the structure of the antibody AF2 Fab (PDB code 1B2W) was used as the search model. For the structure of the protein complex of form I, the structures of the antibody 29G11 Fab (PDB code 1A0Q) and the αL I domain (PDB code 1ZOP) were used as the search models. For the structure of the protein complex of form II, the complex structure of form I was used as the search model. Structure refinement was carried out using CNS and model building was facilitated with the program Coot (57). The free Fab structure contains one Fab molecule and both complex structures contain 2 Fab/I domain complexes in the asymmetric unit. In the complex structure of form I, there was no evident electron density at the MIDAS of the I domain in both complexes. In the complex structure of form II, there was strong electron density at the MIDAS of both I domains which was putatively modeled as a Zn2+ ion because only Zn2+ was added in the crystallization solution. Electron density peaks at a height of at least 2.5 σ in difference Fourier maps were assigned as water molecules if they had reasonable geometry in relation to hydrogen bond donors or acceptors and had B factor of less than 60 Å2 after subsequent refinement.
Accession Codes.
Coordinates of the structures of the Efalizumab Fab alone and in complex with the LFA-1 αL I domain in 2 different crystal forms have been deposited in the RCSB Protein Data Bank with accession codes 3EO9, 3EOA, and 3EOB, respectively.
Supplementary Material
Acknowledgments.
We are grateful to the staff members at Photon Factory, Japan for technical support in diffraction data collection. This work was supported by grants from the Ministry of Science and Technology of China (2004CB720102, 2006AA02Z112, 2006AA02A313, 2006CB806501, and 2007CB914302), the National Natural Science Foundation of China (30570379, 30730028, 30623002, and 90713046), the Chinese Academy of Sciences (KSCX2-YW-R-107 and SIBS2008002), and the Science and Technology Commission of Shanghai Municipality (07XD14032).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EO9, 3EOA, and 3EOB).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810844106/DCSupplemental.
References
- 1.Kishimoto TK, et al. The leukocyte integrins. Adv Immunol. 1989;46:149–182. doi: 10.1016/s0065-2776(08)60653-7. [DOI] [PubMed] [Google Scholar]
- 2.Ruoslahti E. Integrins. J Clin Invest. 1991;87:1–5. doi: 10.1172/JCI114957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 4.Stefanidakis M, Koivunen E. Cell-surface association between matrix metalloproteinases and integrins: Role of the complexes in leukocyte migration and cancer progression. Blood. 2006;108:1441–1450. doi: 10.1182/blood-2006-02-005363. [DOI] [PubMed] [Google Scholar]
- 5.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Madrid F, Nagy JA, Robbins E, Simon P, Springer TA. A human leukocyte differentiation antigen family with distinct α-subunits and a common β-subunit: The lymphocyte function-associated antigen (LFA-1), the C3bi complement receptor (OKM1/Mac-1), and the p150,95 molecule. J Exp Med. 1983;158:1785–1803. doi: 10.1084/jem.158.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gahmberg CG, Tolvanen M, Kotovuori P. Leukocyte adhesion–structure and function of human leukocyte β2-integrins and their cellular ligands. Eur J Biochem. 1997;245:215–232. doi: 10.1111/j.1432-1033.1997.00215.x. [DOI] [PubMed] [Google Scholar]
- 8.Giblin PA, Lemieux RM. LFA-1 as a key regulator of immune function: Approaches toward the development of LFA-1-based therapeutics. Curr Pharm Des. 2006;12:2771–2795. doi: 10.2174/138161206777947731. [DOI] [PubMed] [Google Scholar]
- 9.Arnaout MA. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75:1037–1050. [PubMed] [Google Scholar]
- 10.Makgoba MW, et al. ICAM-1 a ligand for LFA-1-dependent adhesion of B, T and myeloid cells. Nature. 1988;331:86–88. doi: 10.1038/331086a0. [DOI] [PubMed] [Google Scholar]
- 11.Shimaoka M, et al. Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yalamanchili P, Lu C, Oxvig C, Springer TA. Folding and function of I domain-deleted Mac-1 and lymphocyte function-associated antigen-1. J Biol Chem. 2000;275:21877–21882. doi: 10.1074/jbc.M908868199. [DOI] [PubMed] [Google Scholar]
- 13.Leitinger B, Hogg N. Effects of I domain deletion on the function of the β2 integrin lymphocyte function-associated antigen-1. Mol Biol Cell. 2000;11:677–690. doi: 10.1091/mbc.11.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JO, Rieu P, Arnaout MA, Liddington RC. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 15.Qu A, Leahy DJ. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, αLβ2) integrin. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JO, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): A pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 17.Kallen J, et al. Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain. J Mol Biol. 1999;292:1–9. doi: 10.1006/jmbi.1999.3047. [DOI] [PubMed] [Google Scholar]
- 18.Qu A, Leahy DJ. The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin. Structure. 1996;4:931–942. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
- 19.Legge GB, et al. NMR solution structure of the inserted domain of human leukocyte function associated antigen-1. J Mol Biol. 2000;295:1251–1264. doi: 10.1006/jmbi.1999.3409. [DOI] [PubMed] [Google Scholar]
- 20.Nishida N, et al. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Hartgring SA, Bijlsma JW, Lafeber FP, van Roon JA. Interleukin-7 induced immunopathology in arthritis. Ann Rheum Dis. 2006;65(Suppl3):69–74. doi: 10.1136/ard.2006.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. Lancet. 2007;370:263–271. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 23.Ramsay AG, Marshall JF, Hart IR. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 2007;26:567–578. doi: 10.1007/s10555-007-9078-7. [DOI] [PubMed] [Google Scholar]
- 24.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8:604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidmaier R, Baumann P. Anti-adhesion evolves to a promising therapeutic concept in oncology. Curr Med Chem. 2008;15:978–990. doi: 10.2174/092986708784049667. [DOI] [PubMed] [Google Scholar]
- 26.Nicolls MR, Gill RG. LFA-1 (CD11a) as a therapeutic target. Am J Transplant. 2006;6:27–36. doi: 10.1111/j.1600-6143.2005.01158.x. [DOI] [PubMed] [Google Scholar]
- 27.Demierre MF, Higgins PD, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–942. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 28.Mehlis SL, Gordon KB. The immunology of psoriasis and biologic immunotherapy. J Am Acad Dermatol. 2003;49:S44–50. doi: 10.1016/s0190-9622(03)01134-4. [DOI] [PubMed] [Google Scholar]
- 29.Champe M, McIntyre BW, Berman PW. Monoclonal antibodies that block the activity of leukocyte function-associated antigen 1 recognize three discrete epitopes in the inserted domain of CD11a. J Biol Chem. 1995;270:1388–1394. doi: 10.1074/jbc.270.3.1388. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Springer TA. A binding interface on the I domain of lymphocyte function-associated antigen-1 (LFA-1) required for specific interaction with intercellular adhesion molecule 1 (ICAM-1) J Biol Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- 31.Edwards CP, et al. Identification of amino acids in the CD11a I-domain important for binding of the leukocyte function-associated antigen-1 (LFA-1) to intercellular adhesion molecule-1 (ICAM-1) J Biol Chem. 1995;270:12635–12640. doi: 10.1074/jbc.270.21.12635. [DOI] [PubMed] [Google Scholar]
- 32.Binnerts ME, et al. Antibodies that selectively inhibit leukocyte function-associated antigen 1 binding to intercellular adhesion molecule-3 recognize a unique epitope within the CD11a I domain. J Biol Chem. 1996;271:9962–9968. doi: 10.1074/jbc.271.17.9962. [DOI] [PubMed] [Google Scholar]
- 33.Lu C, Shimaoka M, Zang Q, Takagi J, Springer TA. Locking in alternate conformations of the integrin αLβ2 I domain with disulfide bonds reveals functional relationships among integrin domains. Proc Natl Acad Sci USA. 2001;98:2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, Shimaoka M, Salas A, Springer TA. The binding sites for competitive antagonistic, allosteric antagonistic, and agonistic antibodies to the I domain of integrin LFA-1. J Immunol. 2004;173:3972–3978. doi: 10.4049/jimmunol.173.6.3972. [DOI] [PubMed] [Google Scholar]
- 35.Werther WA, et al. Humanization of an anti-lymphocyte function-associated antigen (LFA)-1 monoclonal antibody and reengineering of the humanized antibody for binding to rhesus LFA-1. J Immunol. 1996;157:4986–4995. [PubMed] [Google Scholar]
- 36.Gottlieb A, et al. Effects of administration of a single dose of a humanized monoclonal antibody to CD11a on the immunobiology and clinical activity of psoriasis. J Am Acad Dermatol. 2000;42:428–435. doi: 10.1016/s0190-9622(00)90214-7. [DOI] [PubMed] [Google Scholar]
- 37.Jullien D, et al. T-cell modulation for the treatment of chronic plaque psoriasis with efalizumab (Raptiva): Mechanisms of action. Dermatology. 2004;208:297–306. doi: 10.1159/000077660. [DOI] [PubMed] [Google Scholar]
- 38.Hildreth JE, August JT. The human lymphocyte function-associated (HLFA) antigen and a related macrophage differentiation antigen (HMac-1): Functional effects of subunit-specific monoclonal antibodies. J Immunol. 1985;134:3272–3280. [PubMed] [Google Scholar]
- 39.Dougherty GJ, Hogg N. The role of monocyte lymphocyte function-associated antigen 1 (LFA-1) in accessory cell function. Eur J Immunol. 1987;17:943–947. doi: 10.1002/eji.1830170708. [DOI] [PubMed] [Google Scholar]
- 40.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 43.Shimaoka M, et al. AL-57, a ligand-mimetic antibody to integrin LFA-1, reveals chemokine-induced affinity up-regulation in lymphocytes. Proc Natl Acad Sci USA. 2006;103:13991–13996. doi: 10.1073/pnas.0605716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su HP, Golden JW, Gittis AG, Hooper JW, Garboczi DN. Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein. Virology. 2007;368:331–341. doi: 10.1016/j.virol.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies DR, Padlan EA, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 47.Wilson IA, Stanfield RL. Antibody-antigen interactions: New structures and new conformational changes. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 48.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 49.Casasnovas JM, Stehle T, Liu JH, Wang JH, Springer TA. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bella J, Kolatkar PR, Marlor CW, Greve JM, Rossmann MG. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmiedel J, Blaukat A, Li S, Knochel T, Ferguson KM. Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer Cell. 2008;13:365–373. doi: 10.1016/j.ccr.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karpusas M, et al. Crystal structure of the α1β1 integrin I domain in complex with an antibody Fab fragment. J Mol Biol. 2003;327:1031–1041. doi: 10.1016/s0022-2836(03)00203-1. [DOI] [PubMed] [Google Scholar]
- 53.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W, Carter CW., Jr Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.DeLano WL. [Accessed on Jan 8, 2008];The PyMOL Molecular Graphics System. 2002 Available at http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.