Abstract
Nonribosomal peptide synthetases (NRPSs) catalyze the biosynthesis of many biologically active peptides and typically are modular, with each extension module minimally consisting of a condensation, an adenylation, and a peptidyl carrier protein domain responsible for incorporation of an amino acid into the growing peptide chain. C-1027 is a chromoprotein antitumor antibiotic whose enediyne chromophore consists of an enediyne core, a deoxy aminosugar, a benzoxazolinate, and a β-amino acid moiety. Bioinformatics analysis suggested that the activation and incorporation of the β-amino acid moiety into C-1027 follows an NRPS mechanism whereby biosynthetic intermediates are tethered to the peptidyl carrier protein SgcC2. Here, we report the biochemical characterization of SgcC5, an NRPS condensation enzyme that catalyzes ester bond formation between the SgcC2-tethered (S)-3-chloro-5-hydroxy-β-tyrosine and (R)-1-phenyl-1,2-ethanediol, a mimic of the enediyne core. SgcC5 uses (S)-3-chloro-5-hydroxy-β-tyrosyl-SgcC2 as the donor substrate and exhibits regiospecificity for the C-2 hydroxyl group of the enediyne core mimic as the acceptor substrate. Remarkably, SgcC5 is also capable of catalyzing amide bond formation, albeit with significantly reduced efficiency, between (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and (R)-2-amino-1-phenyl-1-ethanol, an alternative enediyne core mimic bearing an amine at its C-2 position. Thus, SgcC5 is capable of catalyzing both ester and amide bond formation, providing an evolutionary link between amide- and ester-forming condensation enzymes.
Keywords: enediyne, nonribosomal peptide synthetase
Nonribosomal peptide synthetases (NRPSs) are large multifunctional proteins that catalyze the synthesis of many pharmaceutically important peptides, including the anticancer drugs bleomycin and actinomycin and antibacterial antibiotics vancomycin and daptomycin. The prototypical NRPS is composed of loading, extension, and termination modules, with each extension module consisting of minimally 3 domains—an adenylation (A) domain, a peptidyl carrier protein (PCP) domain, and a condensation (C) domain (1–4). The A domain specifically selects an amino acid, activates it by formation of an aminoacyl adenylate, and transfers the activated substrate to the thiol group of the 4′-phosphopantetheinyl arm of the PCP domain to yield a thioester-linked amino acid. The C domain then catalyzes nucleophilic condensation between upstream and downstream PCP-tethered amino acids (also known as donor and acceptor substrates, respectively; ref. 5) to form a new amide bond, thus extending the growing peptide chain by 1 amino acid.
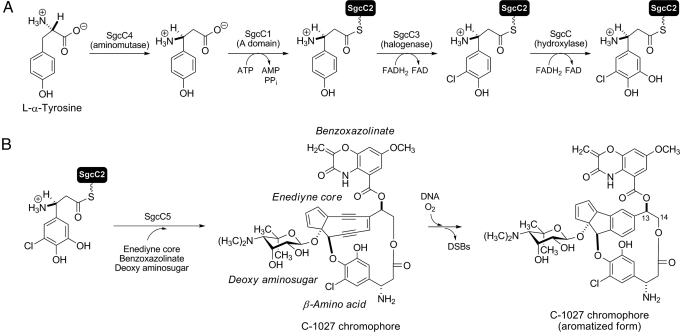
Although NRPS catalysis is often described to follow assembly-line enzymology, wherein the amino acid sequence of the peptide product can be directly predicted from the molecular architecture of NRPS domains and modules (3, 4), recent studies have revealed numerous NRPSs that deviate from this assembly-line molecular logic (6–8). For example, variations to the standard C-A-PCP module were found in the kutzneride (9) and bleomycin NRPSs (10), and iteratively acting domains have been observed for capreomycin (11), saframycin (12), and enterobactin (13) biosynthesis. Moreover, freestanding C, A, or PCP proteins may function in trans to form an NRPS module (8), and such freestanding proteins or didomain proteins have also been identified in biosynthetic machinery for natural products that do not even contain a peptide moiety. One such case was identified upon characterization of the biosynthetic gene cluster for the enediyne antitumor antibiotic C-1027, a cytotoxic secondary metabolite isolated from Streptomyces globisporus that undergoes an electronic rearrangement that, when in the presence of molecular oxygen, can lead to double-stranded DNA breaks (Fig. 1B) (14). In the C-1027 gene cluster, a freestanding C domain (SgcC5), A domain (SgcC1), and PCP (SgcC2) were identified, and together they constitute a minimal NRPS extension module that was predicted to be involved in the biosynthesis of the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of C-1027 (Fig. 1) (15).
Fig. 1.
Biosynthesis of (S)-3-chloro-5-hydroxy-β-tyrosine and its incorporation into the C-1027 chromophore. (A) Biosynthesis of the β-amino acid moiety from l-α-tyrosine featuring 4 functionally assigned enzymes. (B) Attachment of (S)-3-chloro-5-hydroxy-β-tyrosine onto the enediyne core by SgcC5, leading to the C-1027 chromophore. The timing for each of the coupling steps is unknown. C-1027 undergoes an electronic rearrangement that generates a benzenoid biradical species that can abstract hydrogen atoms from DNA, leading to DNA breaks and an aromatized C-1027 chromophore. DSBs indicates double-strand breaks.
Although NRPS C domains are known to catalyze amide bond formation between 2 PCP-tethered amino acids (3, 4), examination of the C-1027 chromophore structure suggests that SgcC5 instead catalyzes condensation to generate an ester bond by using a single PCP-tethered amino acid (Fig. 1B). C domains have been recently proposed to catalyze ester bond formation with carrier protein-tethered substrates, although they are homologous to the typical amide-forming C enzymes. For instance, the C domains located at the C terminus of the RapP and FkbP proteins from the rapamycin (16) and FK506 (17) NRPS biosynthetic machinery, respectively, were proposed to catalyze intramolecular cyclization via ester bond formation, hence releasing the final products from the carrier proteins. These C domains therefore act in a functional analogy to thioesterase domains, despite having no sequence homology between them. Some C domains embedded in elongation modules have also been proposed to catalyze chain extension via ester bond formation rather than amide bond formation on the basis of comparing the NRPS architecture with the expected product. This includes the nonribosomal peptides kutzneride (9), valinomycin (18), and cereulide (19), and the nonribosomal peptide-polyketide hybrid cryptophycin 1 (20). Recently, the C domain of a PCP-C didomain protein Fum14 in the biosynthesis of the fungal metabolite fumonisin was shown to generate 2 tricarballylic esters by using a thioester of N-acetylcysteamine as an acyl donor mimicking the phosphopantetheinyl group of the carrier protein, and in this example, the acceptor substrate was the free alcohol of the fumonisin precursor (21). This study provided the first biochemical evidence for a C domain to catalyze ester bond formation. The results suggested, as observed for the freestanding C enzyme VibH (22), that C domains can also use a free substrate as the acceptor nucleophile to couple with a carrier protein-tethered acyl donor for ester (as with Fum14) or amide (as with VibH) bond formation.
We have previously characterized the first 4 enzymatic steps of the biosynthetic pathway for the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of C-1027 leading to (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 (Fig. 1A), which includes generation of (S)-β-tyrosine by the aminomutase SgcC4 (23, 24), activation and loading of (S)-β-tyrosine to SgcC2 by the A enzyme SgcC1 (25, 26), and halogenation and hydroxylation of (S)-β-tyrosyl-S-SgcC2 by the FAD-dependent SgcC3 halogenase and SgcC hydroxylase, respectively (27, 28). We now report that SgcC5 does indeed catalyze ester bond formation between the SgcC2-tethered (S)-3-chloro-5-hydroxy-β-tyrosine and (R)-1-phenyl-1,2-ethanediol, an enediyne core analog that mimics the aromatized C-1027 structure (structure 1a; Figs. 1B and 2A), thereby supporting its proposed role in C-1027 biosynthesis (15). SgcC5 absolutely requires carrier protein-tethered molecules as the donor substrates and is regiospecific for the C-2 hydroxyl group of the enediyne core mimic as the acceptor substrate (C-2 of 1a corresponds to C-14 of the enediyne core; Figs. 1B and 2A). Remarkably, SgcC5 is also competent and regiospecific in catalyzing amide bond formation between the SgcC2-tethered (S)-3-chloro-5-hydroxy-β-tyrosine and (R)-2-amino-1-phenyl-1-ethanol, an enediyne core mimic bearing an amine as a nucleophile at its C-2 position (2a; Fig. 2A), as an alternative acceptor substrate. The broad acceptor tolerance of SgcC5 contrasts with the strict substrate specificity of other previously characterized C domains (3, 5, 29), although SgcC5 does exhibit a strong regiospecificity for the C-2 group as a nucleophile (i.e., C-2 hydroxyl or amino group) of the acceptor substrate. SgcC5 is an example of a C enzyme that catalyzes ester bond formation but retains the canonical amide-forming activity, albeit with significantly reduced efficiency. These findings provide biochemical evidence supporting an evolutionary link between amide- and ester-forming C enzymes that has been implied from structural similarities between C domains and chloramphenicol acetyltransferases (CATs) (30). Further mechanistic and structural characterization of SgcC5 should therefore allow us to engineer the ester-forming activity into other NRPS C domains or enzymes, thereby expanding the size and diversity of the novel peptide library by combinatorial biosynthesis or chemoenzymatic methods.
Fig. 2.
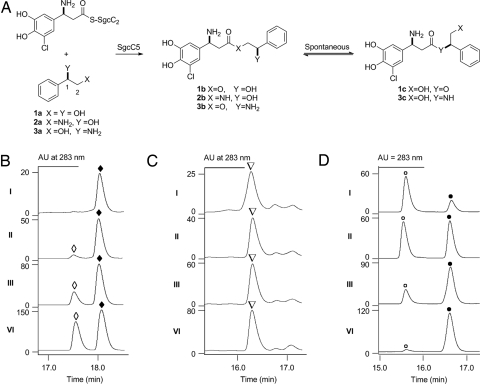
Characterization of SgcC5 as a condensation enzyme catalyzing both ester bond and amide bond formation. (A) SgcC5-catalyzed condensation reaction between SgcC2-tethered (S)-3-chloro-5-hydroxy-β-tyrosine and enediyne core mimics (R)-1-phenyl-1,2-ethanediol (1a), (R)-2-amino-1-phenyl-1-ethanol (2a), or (R)-2-phenylglycinol (3a) and the products established by spectroscopic analysis. HPLC profiles of time course analysis for SgcC5-catalzyed condensation between (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 incubated with (B) 1a and 1 μM SgcC5 for 1 min (i), 4 min (ii), 15 min (iii), and 90 min (iv), (C) 2a and 100 μM SgcC5 for 5 min (i), 15 min (ii), 60 min (iii), and 180 min (iv), and (D) 3a and 20 μM SgcC5 for 1 min (i), 2.5 min (ii), 5 min (iii), and 10 min (iv). 1b (◆), 1c (◇), 2b (▿), 3b (○), and 3c (●).
Results and Discussion
Bioinformatics Analysis of SgcC5.
The biosynthesis of the β-amino acid moiety and its incorporation into C-1027 are catalyzed by 6 proteins: SgcC, SgcC1, SgcC2, SgcC3, SgcC4, and SgcC5 (Fig. 1) (15). Among them, SgcC1 and SgcC2 have been identified as homologs to A domains and PCP domains in NRPSs, respectively, and the activity of SgcC1 to activate and load (S)-β-tyrosine to SgcC2 has been demonstrated (25, 26). SgcC5 features the highly conserved HHXXXDX14Y motif known to all modular NRPS C domains [supporting information (SI) Fig. S1] (22, 31–34), clearly supporting its functional assignment as a C enzyme but putatively catalyzing an ester bond formation (Fig. 1B). However, the overall sequence homology between SgcC5 and other well-studied NRPS C domains is low—SgcC5 shows only 19% identity/36% similarity to the SrfA-C C domain, 20% identity/38% similarity to the TycC C domain, and 16% identity/27% similarity to the freestanding C enzyme VibH, whose structures have all been solved by X-ray crystallography (22, 33, 34).
Overproduction of SgcC5 and Chemoenzymatic Preparation of Donor Substrate.
SgcC5 was overproduced in Escherichia coli BL21 (DE3), purified to homogeneity as an N-terminal, His6-tagged fusion protein (≈100 mg/L), and showed a single protein band upon SDS/PAGE analysis consistent with the predicted molecular weight (55 kDa) (Fig. S2).
Three recombinant proteins—SgcC1 (25), SgcC2 (27), and Svp, a promiscuous 4-phosphopantetheinyl transferase from the bleomycin biosynthetic pathway (35)—were produced as described previously to enzymatically prepare the SgcC2-tethered donor substrate. Apo-SgcC2 was first converted to holo-SgcC2 by Svp (35). The free substrate 3-chloro-5-hydroxy-β-tyrosine was chemically synthesized (27), activated, and loaded onto holo-SgcC2 by SgcC1 to yield (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2. SgcC1 activated and loaded only the (S)-3-chloro-5-hydroxy-β-tyrosine enantiomer to holo-SgcC2, despite using racemic 3-chloro-5-hydroxy-β-tyrosine, as determined by a chiral HPLC analysis in comparison with authentic (S)-3-chloro-5-hydroxyl-β-tyrosine and (R)-3-chloro-5-hydroxyl-β-tyrosine, respectively (Fig. S3 and SI Methods).
In Vitro Characterization of SgcC5 as an Ester-Forming Condensation Enzyme.
SgcC5 was initially assayed with 3-chloro-5-hydroxy-β-tyrosine as the donor substrate and (R)-1-phenyl-1,2-ethanediol (1a; Fig. 2A) as the acceptor substrate, the latter of which serves as an enediyne core mimic representing the aromatized version of the C-1027 enediyne chromophore (the exact structure of the native substrate for SgcC5 remains unknown; hence, it is unavailable) (Fig. 1B). HPLC analysis of the assay solution, however, showed no product formation, suggesting that SgcC5 may require carrier protein-tethered substrate. SgcC5 was next assayed with (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and (R)-1-phenyl-1,2-ethanediol as the donor and acceptor substrates, respectively, and the reaction was followed by HPLC analysis (Fig. 2B). After 15 min of incubation, 2 products formed, with the major one at retention time (rt) 18.2 min and the minor one at rt 17.6 min, and the 2 products reached a 4:5 ratio after 90 min of incubation (Fig. 2B iii and iv). Each of the 2 products was isolated and subjected to electrospray ionization mass spectroscopy (ESI-MS) analysis, yielding the same pair of [M + H]+ ions at 352.1 and 354.1 in a 3:1 ratio, characteristic of monochlorinated species, which was consistent with the molecular weight of the predicted ester products 1b and 1c (calculated [M + H]+ ions at m/z = 352.1 and 354.1; Fig. 2A). A large-scale reaction was then carried out to produce both products (Fig. S4 and SI Methods), and 1H, 13C, and 2D NMR spectroscopic analyses (Table S1) confirmed them as the ester products arising from 3-chloro-5-hydroxy-β-tyrosine coupled to the C-2 and C-1 hydroxyl groups of (R)-1-phenyl-1,2-ethanediol, respectively (1b and 1c; Fig. 2A).
Kinetic Analysis of Esterification by SgcC5.
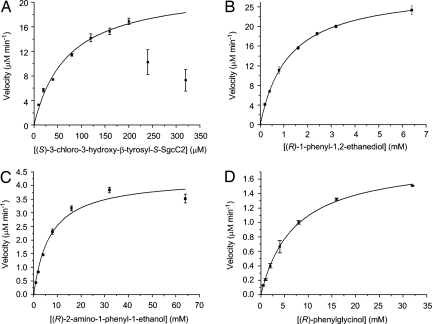
HPLC analysis following the SgcC5-catalyzed ester formation between (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and (R)-1-phenyl-1,2-ethanediol showed that the enzymatic reaction was time-dependent, enabling the determination of single-substrate kinetic constants (Fig. 2B). (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 showed substrate inhibition at concentrations >200 μM, and the kinetics constants obtained therefore are only estimates. The formation of product with constant (R)-1-phenyl-1,2-ethanediol (5 mM) and variable (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 exhibited Michaelis–Menten kinetics, with a Km = 71 ± 9 μM and a kcat = 22 ± 2 min−1 (Fig. 3A), and assays with 200 μM (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and variable (R)-1-phenyl-1,2-ethanediol yielded a Km = 1.2 ± 0.1 mM and kcat = 27 ± 2 min−1 (Fig. 3B).
Fig. 3.
Single-substrate kinetic analysis of SgcC5-catalyzed ester or amide formation. Reactions with variable donor substrate (A) (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and with variable acceptor substrates (B) (R)-1-phenyl-1,2-ethanediol (1a), (C) (R)-2-amino-1-phenyl-1-ethanol (2a), and (D) (R)-2-phenylglycinol (3a), respectively. See Fig. 2A for 1a, 2a, and 3a structures.
In Vitro Characterization of SgcC5 as an Amide-Forming Condensation Enzyme.
Inspired by the canonical amide bond-forming activity of NRPS C domains and enzymes, SgcC5 was next assayed as an amide-forming C enzyme with (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 as the donor substrate and (R)-2-amino-1-phenyl-1-ethanol (2a; Fig. 2A) as the acceptor substrate. Remarkably, HPLC analysis of the assay solution showed time-dependent formation of a new product with an rt of 16.3 min (Fig. 2C). This new peak was collected and subjected to HR-MALDI-MS analysis, yielding a pair of [M + Na]+ ions at m/z = 373.0926 and 375.0900 and with a 3:1 ratio characteristic of the monochlorinated amide product (2b; Fig. 2A) (calculated [M + Na]+ ions at m/z = 373.0931 and 375.0897 in a 3:1 ratio). Production and purification of 2b from a scale-up reaction and subsequent 1H, 13C, and 2D NMR analyses unambiguously established its amide structure (Table S2). The apparent kinetic parameters were determined with 200 μM (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and variable (R)-2-amino-1-phenyl-1-ethanol to give Km = 7.0 ± 1.3 mM and kcat = 0.043 ± 0.002 min−1 (Fig. 3C). Under the conditions examined, SgcC5 was ≈3,700-fold more efficient at catalyzing ester bond formation with 1a than catalyzing amide bond formation with 2a as the acceptor substrate on the basis of kcat/Km value comparison (Fig. 2A).
Regiospecificity of SgcC5 Toward Acceptor Substrates.
The regiospecificity of SgcC5 was first examined by following the time course of 1b and 1c formation from (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and (R)-1-phenyl-1,2-ethanediol (1a) as substrates (Fig. 2A). The formation of 1b is SgcC5-dependent; 1b is produced exclusively in the early stage, but 1c appears upon longer incubation (Fig. 2B), indicating that 1c originates from 1b. Both products were subsequently purified, and incubation of each of them under the assay conditions in the absence of SgcC5 indeed confirmed the spontaneous conversion between 1b and 1c, with an equilibrium constant of 1c/1b = 0.80 ± 0.10 (Fig. 2A and SI Methods). These observations support that the SgcC5-catalyzed ester bond formation with (R)-1-phenyl-1,2-ethanediol as the acceptor substrate is regiospecific for the C-2 hydroxyl group.
(R)-2-amino-1-phenyl-1-ethanol and (R)-2-phenylglycinol (2a and 3a; Fig. 2A) were next used as acceptor substrates to further confirm the regiospecificity of SgcC5 with (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 as the donor substrate by following their reaction time courses, respectively. (R)-2-amino-1-phenyl-1-ethanol (2a) afforded the corresponding 2b exclusively, as described earlier (Fig. 2C). (R)-2-phenylglycinol (3a) yielded 2 products initially, but the first product at rt of 15.5 min completely converted to the second product at rt 16.6 min after prolonged incubation (Fig. 2D). The second product at rt of 16.6 min was collected and analyzed by HR-ESI-MS, giving a pair of [M + H]+ ions at m/z = 351.1106 and 353.1089 in the 3:1 ratio expected for the monochlorinated coupling product (3c; Fig. 2A) (calculated M + H]+ ions at m/z = 351.1112:353.1082 ≈ 3:1). Scale-up reactions afforded purified 3c, whose amide structure was confirmed by 1H NMR analysis (SI Methods). The first product at rt 15.5 min, predicted to be the ester product (3b; Fig. 2A), was unstable, and thus no spectroscopic analysis was performed. Steady-state kinetic analysis using 200 μM (S)-3-choro-5-hydroxy-β-tyrosyl-(S)-SgcC2 and variable (R)-2-phenylglycinol was also carried out, yielding a Km = 7.1 ± 0.3 mM and kcat = 1.87 ± 0.03 min−1 (Fig. 3D). SgcC5 is ≈100-fold less efficient using 3a compared with 1a as the acceptor substrate on the basis of their kcat/Km value comparison. Taken together, these observations support that SgcC5 catalyzes either ester bond formation with (R)-1-phenyl-1,2-ethanediol (1a) or (R)-2-phenylglycinol (3a) or amide bond formation with (R)-2-amino-1-phenyl-1-ethanol (2a) as the acceptor substrate and is regiospecific for the C-2 hydroxyl or amino group, respectively. Of the 3 substrates examined, the preferred substrate for SgcC5 is a 1,2-diol enediyne mimic 1a; the nascent product 1b, however, readily undergoes a 1,2-migration to afford 1c with an equilibrium constant of 1c/1b = 0.80 ± 0.10 (Fig. 2A).
SgcC5 Is an Ester Bond-Forming and Amide Bond-Forming Condensation Enzyme.
We have experimentally demonstrated that SgcC5 is indeed a C enzyme-catalyzing ester bond formation to incorporate the SgcC2-tethered (S)-3-chloro-5-hydroxy-β-tyrosine moiety into C-1027, concluding functional assignment of the minimal NRPS module, composed of 3 freestanding SgcC1, SgcC2, and SgcC5 proteins, in C-1027 biosynthesis (Fig. 1). This was accomplished by taking advantage of the chemoenzymatically prepared (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 as the donor substrate and the enediyne core mimic (R)-1-phenyl-1,2-ethanediol as the acceptor substrate for SgcC5. SgcC5 absolutely requires an SgcC2-tethered donor substrate and efficiently and regiospecifically couples it, in an ester linkage, with the C-2 hydroxy of the free acceptor substrate that mimics the C-14 hydroxy of the enediyne core (Figs. 1B and 2A). The fact that SgcC5 recognizes the enediyne core mimics, such as 1a, 2a, and 3a (Fig. 2) is indicative of relaxed specificity for the acceptor substrate, and this is in contrast to previous investigations that suggest C domains in general have strict specificity for acceptor substrates (3, 5, 29).
Kinetic analysis was performed with (R)-1-phenyl-1,2-ethanediol (1a) as a surrogate acceptor substrate, yielding a Km = 1.2 mM and kcat = 27 min−1. Despite using a nonnative substrate, the Km is comparable to VibH with respect to norspermidine as an acceptor substrate, but the turnover of SgcC5 is substantially lower when compared with VibH (kcat of 6,010 min−1) (22) or when compared with the ester bond-forming CAT with chloramphenicol as an acceptor (kcat of 3,600 min−1) (36). In contrast, dihydrolipoamide acetyltransferases, which also belong to the same superfamily as condensation enzymes and CAT, have similar Km with respect to acceptor substrates but significantly lower kcat compared with SgcC5, as exemplified with yeast dihydrolipoamide acetyltransferase (Km of 4.0 mM and kcat = 0.13 min−1) (37). The significance of these kinetic constants—particularly in the absence of kinetic data with natural substrate—is currently uncertain. It is apparent, however, that SgcC5-catalyzed amide bond formation occurs at a substantially lower efficiency, manifested by a large decrease in kcat. Thus, amide bond formation clearly has no physiological relevance, but instead may present the opportunity to apply combinatorial biosynthetic approaches for structural diversification.
NRPS C domains, including SgcC5, consist of ≈450 amino acids and contain an HHXXXDX14Y motif, with the Asp and second His essential for activity (excluding only VibH, where catalysis was unaffected by mutation to the His residue) (3) (SI Methods and Fig. S1). Recent structural investigations with VibH (22), SrfA-C C domain (34), and TycC C domain (33) have revealed that C domains are composed of 2 subdomains, each of which has a similar fold to a monomer of CATs. CATs also contain an essential HHXXXDX14Y motif and are catalytically active as trimers, forming 3 active sites at the monomer interface (30). The structural similarities of C domains and CATs suggest that a similar strategy is used in amide bond and ester bond formation, respectively, and alludes to the possibility that C domains may be able to catalyze ester bond formation.
Here, we have shown that SgcC5 not only catalyzes ester bond formation, as was predicted from the structure of C-1027 (15), but also retains the canonical amide-forming activity typical of NRPS condensation enzymes, albeit with significantly reduced efficiency. SgcC5 now joins Fum14 (21) in the category of C domains that have been biochemically demonstrated to catalyze ester bond formation, but to our knowledge, SgcC5 is the first C domain to be shown to catalyze both ester and amide bond formation. SgcC5, therefore, may represent a rare event of evolution caught in action, providing an opportunity to investigate how to evolve an amide-forming C enzyme to an ester-forming enzyme or vice versa. Interestingly, the biosynthetic gene cluster for the enediyne maduropeptin contains an SgcC5 homolog, MdpC5, that has high sequence homology (45/58% identity/similarity), and hence likely similar function; however, in contrast to SgcC5, MdpC5 is predicted to catalyze amide bond formation based on examination of the maduropeptin structure (38). The relative simplicity of SgcC5 with respect to architecture and substrate specificity of typical C domains makes SgcC5 an excellent candidate to further explore the mechanistic details of C domains in general, and structural analysis of SgcC5 should provide critical insights into the selectivity for a hydroxy or amine nucleophile of the acceptor substrate. In view of the central role of C domains in nonribosomal peptide biosynthesis, and given the technical difficulties associated with studying the prototypical C domains, such studies should pave the way for new opportunities to generate novel natural products by combinatorial biosynthesis or chemoenzymatic methods.
Materials and Methods
General.
Details for chemicals and instruments used; bioinformatics analysis of SgcC5; preparation of SgcC, SgcC1, SgcC2, SgcC3, SgcC5, SgcE6, and Svp; enzymatic synthesis of the donor substrate (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2; purification of the acceptor substrate (R)-2-amino-1-phenyl-1-ethanol (2a); time courses of SgcC5-catalyzed ester and amide bond formation and HPLC analyses; and the large-scale preparation of the ester and amide products 1b, 2b, 1c, and 3c and their structural characterization of these products by MS and NMR analyses are provided in the SI Methods.
Activity Assay for SgcC5.
Standard assays were carried out in 200 μL of reaction mixture containing 200 μM (S)-3-chloro-5-hydroxy-β-tyrosyl-(S)-SgcC2 as the donor substrate, 10 mM acceptor substrates [(R)-1-phenyl-1, 2-ethanediol (1a) or (R)-2-amino-1-phenyl-1-ethanol (2a) or (R)-2-phenylglycinol (3a)], 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 50 mM phosphate buffer (pH 7.5), and SgcC5 (1.0 μM for 1a, 100 μM for 2a, and 20 μM for 3a). HPLC was used to monitor the reaction, with detection at 254 nm (for acceptor substrates) and 283 nm (for products). The reactions were quenched by addition of trifluoroacetic acid to a final concentration of 16%, and after centrifugation the clarified supernatant was subjected to HPLC analysis (SI Methods).
Kinetics of Activity of SgcC5.
To determine the kinetics for the donor substrate (S)-3-chloro-5-hydroxy-β-tyrosyl-SgcC2 in the production of 1b and 1c, 195 μL of reaction mixtures contained 100 mM Tris·HCl (pH 7.5), 5 mM ATP, 2 mM TCEP, 12.5 mM MgCl2, 5 mM 3-chloro-5-hydroxy-β-tyrosine, 10 μM Svp, 10 μM SgcC1, and varying concentrations of apo-SgcC2 (10–320 μM) and CoA (50 μM to 1.6 mM; final concentration for 200 μL of total assay solution upon addition of SgcC5). The reactions were incubated at 25 °C for 45 min to allow for the loading of 3-chloro-5-hydroxy-β-tyrosine to holo-SgcC2 in situ before the addition of 5 mM 1a. This loading was determined to be complete based on HPLC analysis, and the reaction components were shown not to inhibit SgcC5 activity. The reaction was initiated by the addition of 1 μM SgcC5 and carried out in triplicate. The reactions were quenched with 16% trifluoroacetic acid after being incubated at 25 °C for 5 min. After centrifugation, the resulting clarified supernatant was subjected to HPLC analysis. To quantify the amount of products formed from each reaction, a calibration curve based on the HPLC peak area with UV detection at 283 nm was generated with a known amount of synthetic 3-chloro-5-hydroxy-β-tyrosine, and all of these compounds exhibited identical UV absorption at this wavelength. The Michaelis–Menten equation was fit to plots of initial rate of product formation versus substrate concentration to extract values for the Km and kcat parameters.
To determine the kinetic parameters for acceptor substrates (R)-1-phenyl-1,2-ethanediol (1a), (R)-2-amino-1-phenyl-1-ethanol (2a), and (R)-2-phenylglycinol (3a), 180 μL of reactions contained 100 mM Tris·HCl (pH 7.5), 200 μM apo-SgcC2, 5 mM ATP, 1 mM CoA, 2 mM TCEP, 12.5 mM MgCl2, 5 mM 3-chloro-5-hydroxy-β-tyrosine, 10 μM Svp, and 10 μM SgcC1 (final concentration for 200 μL of total assay solution upon addition of SgcC5). The reactions were similarly incubated at 25 °C for 45 min to allow for complete loading of 3-chloro-5-hydroxy-β-tyrosine to holo-SgcC2 in situ, and the reaction components were shown not to inhibit SgcC5 activity. The reactions were initiated by the addition of varied concentrations of acceptor substrates (0.2–6.4 mM for 1a, 1–64 mM for 2a, and 0.5–32 mM for 3a), followed by the addition of SgcC5 (1 μM SgcC5 for 1a and 3a, and 100 μM SgcC5 for 2a), and each reaction was performed under initial velocity conditions and carried out in triplicate. The reactions were quenched with 16% trifluoroacetic acid after being incubated at 25 °C for different times (5 min for 1a, 15 min for 2a, and 30 min for 3a). Product formation was quantified by HPLC peak area to obtain velocity data for single-substrate kinetic analysis. Data were fit to the Michaelis–Menton equation to determine the Km and kcat..
Supplementary Material
Acknowledgments.
We thank Dr. Y. Li, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences (Beijing, China), for the wild-type S. globisporus strain and the Analytical Instrumentation Center of the School of Pharmacy, University of Wisconsin, Madison for support in obtaining MS and NMR data. This work is supported in part by National Institutes of Health (NIH) Grants CA78747 and CA113297. S.V.L. is the recipient of an NIH postdoctoral fellowship (CA1059845).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808880106/DCSupplemental.
References
- 1.Felnagle EA, et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 3.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 5.Belshaw PJ, Walsh CT, Stachelhaus T. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science. 1999;284:486–489. doi: 10.1126/science.284.5413.486. [DOI] [PubMed] [Google Scholar]
- 6.Wenzel SC, Müller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: Deviations from textbook biosynthetic logic. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Haynes SW, Challis GL. Non-linear enzymatic logic in natural product modular mega-synthases and -synthetases. Curr Opin Drug Disc Develop. 2007;10:203–218. [PubMed] [Google Scholar]
- 8.Donadio S, Monciardini P, Sosio M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 9.Fujimori DG, et al. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci USA. 2007;104:16498–16503. doi: 10.1073/pnas.0708242104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L, Sánchez C, Chen M, Edwards DJ, Shen B. The biosynthetic gene cluster for antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptides synthetase and a polyketide synthase. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 11.Felnagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG. Identification of the biosynthetic gene cluster and an additional resistance gene for the antituberculosis drug capreomycin. Appl Environ Microbiol. 2007;73:4162–4170. doi: 10.1128/AEM.00485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, et al. Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehmann DE, Shaw-Reid CA, Losey HC, Walsh CT. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: Sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc Natl Acad Sci USA. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao RG, Zhen JM. Enediyne anticancer antibiotic lidamycin: Chemistry, biology and pharmacy. Anticancer Agents Med Chem. 2008;8:292–304. doi: 10.2174/187152008783497055. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 16.Schwecke T, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motamedi H, Shafiee A. The biosyntheic gene cluster for macrolactone ring of the immunsuppressant FK506. Eur J Chem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- 18.Cheng YQ. Deciphering the biosynthetic codes for the potent Anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. Chembiochem. 2006;7:471–477. doi: 10.1002/cbic.200500425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehling-Schulz M, et al. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl Environ Microbiol. 2005;71:105–113. doi: 10.1128/AEM.71.1.105-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magarvey NA, et al. Biosynthetic characterization and chemoenzymatic assembly of the cryptophycins. Potent anticancer agents from Nostoc cyanobionts. ACS Chem Biol. 2006;1:766–779. doi: 10.1021/cb6004307. [DOI] [PubMed] [Google Scholar]
- 21.Zaleta-Rivera K, et al. A bidomain nonribosomal peptide synthetase encoded by FUM14 catalyzes the formation of tricarballylic esters in the biosynthesis of fumonisins. Biochemistry. 2006;45:2561–2569. doi: 10.1021/bi052085s. [DOI] [PubMed] [Google Scholar]
- 22.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Mol Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 23.Christenson SD, Wu W, Spies MA, Shen B, Toney MD. Kinetic analysis of the 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. Biochemistry. 2003;42:12708–12718. doi: 10.1021/bi035223r. [DOI] [PubMed] [Google Scholar]
- 24.Christianson CV, Montavon TJ, VanLanen SG, Shen B, Bruner SD. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry. 2007;46:7205–7214. doi: 10.1021/bi7003685. [DOI] [PubMed] [Google Scholar]
- 25.Van Lanen SG, et al. Biosynthesis of the β-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring β-amino acyl-S-carrier protein intermediates. J Am Chem Soc. 2005;127:11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- 26.Van Lanen SG, Lin S, Dorrestein PC, Kelleher NL, Shen B. Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Biol Chem. 2006;281:29633–29640. doi: 10.1074/jbc.M605887200. [DOI] [PubMed] [Google Scholar]
- 27.Lin S, VanLanen SG, Shen B. Regiospecific chlorination of (S)-β-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Am Chem Soc. 2007;129:12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin S, Van Lanen SG, Shen B. Characterization of the 2-component, FAD-dependent monooxygenase SgcC that requires carrier protein-tethered substrates for the biosynthesis of the enediyne antitumor antibiotic C-1027. J Am Chem Soc. 2008;130:6616–6623. doi: 10.1021/ja710601d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootz HD, Schwarzer D, Marahiel MA. Construction of hybrid peptide synthetases by module and domain fusions. Proc Natl Acad Sci USA. 2000;97:5848–5853. doi: 10.1073/pnas.100075897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw WV, Leslie AGW. Chloramphenicol acetyltransferase. Annu Rev Biophys Biophys Chem. 1991;20:363–386. doi: 10.1146/annurev.bb.20.060191.002051. [DOI] [PubMed] [Google Scholar]
- 31.Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 32.Rausch C, Hoof I, Weber T, Wohlleben W, Huson D. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez C, Du L, Edwards DJ, Toney MD, Shen B. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 36.Lewendon A, Murray IA, Shaw WV. Replacement of catalytic histidine-195 of chloramphenicol acetyltransferase: Evidence for a general base role for glutamate. Biochemistry. 1994;33:1944–1950. doi: 10.1021/bi00173a043. [DOI] [PubMed] [Google Scholar]
- 37.Niu X, Stoops JK, Reed LJ. Overexpression and mutagenesis of the catalytic domain of dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Biochemistry. 1990;29:8614–8619. doi: 10.1021/bi00489a017. [DOI] [PubMed] [Google Scholar]
- 38.Van Lanen SG, Oh T, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.