Abstract
The insulin and insulin-like growth factor 1 receptors activate overlapping signalling pathways that are critical for growth, metabolism, survival and longevity. Their mechanism of ligand binding and activation displays complex allosteric properties, which no mathematical model has been able to account for. Modelling these receptors' binding and activation in terms of interactions between the molecular components is problematical due to many unknown biochemical and structural details. Moreover, substantial combinatorial complexity originating from multivalent ligand binding further complicates the problem. On the basis of the available structural and biochemical information, we develop a physically plausible model of the receptor binding and activation, which is based on the concept of a harmonic oscillator. Modelling a network of interactions among all possible receptor intermediaries arising in the context of the model (35, for the insulin receptor) accurately reproduces for the first time all the kinetic properties of the receptor, and provides unique and robust estimates of the kinetic parameters. The harmonic oscillator model may be adaptable for many other dimeric/dimerizing receptor tyrosine kinases, cytokine receptors and G-protein-coupled receptors where ligand crosslinking occurs.
Keywords: activation mechanism, binding kinetics, harmonic oscillator model, insulin receptor, mathematical modelling
Introduction
Insulin and insulin-like growth factors (IGF) 1 and 2 have similar structures and exert their action by activating two closely related receptor tyrosine kinases—the insulin receptor and the IGF1 type I receptor (IGF1 receptor), which share largely overlapping signalling pathways (Adams et al, 2000; De Meyts and Whittaker, 2002; De Meyts, 2004; Denley et al, 2005). Despite this similarity, the two hormones produce different responses: mostly metabolic for insulin and mitogenic for IGF1 (Kim and Accili, 2002). Dysregulation of their signalling may lead to two different life-threatening diseases: type II diabetes and cancer, which are among the largest global health challenges in the world. So far, there is poor understanding of how these hormones produce such different biological effects using similar signalling networks (Kim and Accili, 2002). It has become clear that a systems biology approach is required to understand the combinatorial nature of signalling specificity (Shymko et al, 1997; Kholodenko, 2007). Insulin analogues with altered kinetic properties show enhanced mitogenic potencies, although they bind to the same insulin receptor; they also cross-react to a variable extent with the IGF-I receptor (Shymko et al, 1997; Kurtzhals et al, 2000). Differences in kinetics of ligand binding and receptor activation by the insulin and IGF1 receptors may be one of the factors determining their specificity (Shymko et al, 1997). The two receptors' mechanism of ligand binding and activation displays complex allosteric properties (i.e. negative cooperativity and ligand dependence of the receptor dissociation rate), which no mathematical model has been able to fully account for (Jeffrey, 1982; Kohanski and Lane, 1983; Hammond et al, 1997; Wanant and Quon, 2000; Sedaghat et al, 2002). Thus, the development of a reliable mathematical model describing the two receptors' binding kinetics and activation is a critical first step in a systems biology approach to understand the function and specificity of these receptors.
The insulin and IGF1 receptors exist in the membrane as pre-formed covalent dimers of two identical moieties. Their extracellular domains comprise two leucine-rich repeat-containing large domains (L1 and L2) separated by a cystein-rich (CR) domain, followed by three fibronectin type III (Fn1–3) domains (Adams et al, 2000; De Meyts and Whittaker, 2002; De Meyts, 2004). A crystal structure of this extracellular (unliganded) insulin receptor dimer has recently been solved (McKern et al, 2006; Lawrence et al, 2007; Ward et al, 2007, 2008). The intracellular portion of the two receptors consists of a kinase domain flanked by regulatory regions (Hubbard and Miller, 2007).
The insulin and IGF1 receptors exhibit complex binding properties. The Scatchard plots for both receptors are concave up, indicating the presence of high- and low-affinity binding sites and/or negative cooperativity (De Meyts et al, 1973, 1976; De Meyts, 1994). The receptors bind only one ligand molecule with high affinity and at least another one with lower affinity. The ligand dissociation rate is dependent on its concentration (De Meyts et al, 1973, 1976; De Meyts, 1994). Furthermore, this dependence is bell-shaped for the insulin receptor, whereas for the IGF1 receptor, it is sigmoid (Christoffersen et al, 1994). When the insulin receptor is in a monomeric form, its affinity is reduced 30-fold, the Scatchard plot becomes linear and the dissociation rate of the ligand becomes independent of its concentration (De Meyts, 1994, 2004; De Meyts and Whittaker, 2002).
Insulin has two receptor-binding surfaces on the ‘opposite' sides of the molecule. The first ‘classical' binding surface, also involved in insulin dimerization, was defined in the early 1970s (Pullen et al, 1976; De Meyts et al, 1978), and later validated by alanine-scanning mutagenesis (Kristensen et al, 1997), whereas the second surface, also involved in insulin hexamerization, was mapped by alanine-scanning mutagenesis more recently (De Meyts, 2004; Gauguin et al, 2008). The dimerization surface interacts with a site located in the L1 module of the insulin receptor, as well as a 12 amino-acid peptide from the insert in Fn2, which combine to form ‘site 1' (Wedekind et al, 1989; Kurose et al, 1994; Williams et al, 1995; Mynarcik et al, 1996; De Meyts and Whittaker, 2002; Kristensen et al, 2002; Huang et al, 2004), whereas the hexamerization surface interacts with a site consisting of residues located in the C-terminal portion of L2 and in the Fn1 and Fn2 modules (site 2) (Fabry et al, 1992; De Meyts and Whittaker, 2002; Hao et al, 2006; Benyoucef et al, 2007; Whittaker et al, 2008). Schäffer (1994) suggested that the high-affinity binding could result from insulin crosslinking site 1 of one receptor half and site 2 of the other half of the receptor dimer, thus leaving the other two sites free for interaction with the ligand. However, this model had difficulty in explaining the ligand dependence of the ligand dissociation rate. De Meyts (1994) suggested that this problem could be solved by assuming that the four sites of the receptor dimer are arranged in a symmetrical antiparallel way, a postulate that was supported by the recent structure determination of the insulin receptor extracellular domain (McKern et al, 2006). Despite its seeming simplicity, this model turned out to be notoriously difficult for a quantitative analysis and some researchers even concluded that it did not explain the ligand dependence of the dissociation rate (Hammond et al, 1997). The problem is that the qualitative crosslinking models suggested by Schäffer (1994) and De Meyts (1994) are not detailed enough from a biochemical and structural viewpoint for mathematical modelling (especially concerning the precise mechanism that leads to receptor crosslinking). The modelling problem is further aggravated by combinatorial complexity arising from multivalent ligand binding to the receptor in multiple possible conformations. We have now solved this problem.
Here, we present the first mathematical model that accurately reproduces all the kinetic properties of the insulin receptor such as negative cooperativity and the bell-shaped ligand dependence of the receptor dissociation rate. On the basis of the available structural information, we develop a physically plausible model of the receptor activation, which is based on the concept of a harmonic oscillator. We justify thermodynamically that the symmetrically arranged subunits of the insulin receptor dimer experience harmonic oscillatory movements. Analysis of the behaviour of an ensemble of such harmonic oscillators in thermal equilibrium with the surrounding milieu allows to model the receptor activation in a simple way and to substantially reduce the combinatorial complexity. This model is the first one that gives a description of the insulin receptor binding and activation mechanism in terms of interactions between the molecular components and fully takes into account the combinatorial binding complexity (employing 35 insulin receptor intermediaries). Fitting of the model to experimental data provides unique and robust estimates of the kinetic parameters. With a small modification, the model can also be used for the IGF1 receptor. Furthermore, the harmonic oscillator model may be adaptable for many other dimeric or dimerizing receptor tyrosine kinases, cytokine receptors and G-protein-coupled receptors where ligand crosslinking occurs (De Meyts, 2008).
Theory and results
Our goal is to develop a physically plausible mathematical model of the insulin receptor-binding and activation mechanism in terms of interactions between the molecular components. This, however, requires the knowledge of the precise mechanism of interaction between the components involved, for which some biochemical and structural details are still missing. Thus, some assumptions are unavoidable. All of the assumptions made in this article are justified from the physicochemical point of view and are therefore physically plausible.
Binding of insulin to the inactive conformation of the insulin receptor
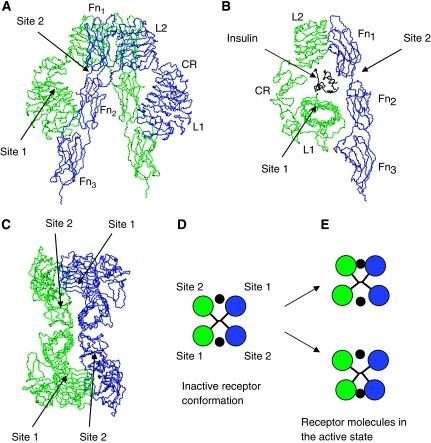
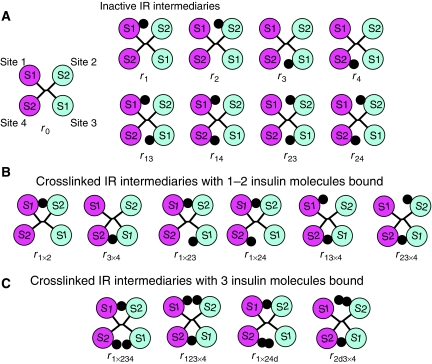
The structure of the insulin receptor dimer in the unliganded conformation displays a symmetrical antiparallel arrangement of the receptor's two binding sites for insulin (McKern et al, 2006) (see Figure 1A and C). From now on, a pair of sites 1 and 2 from the different receptor subunits will be referred to as a ‘crosslink'. The structure shows that the distance between sites 1 and 2 (within the same crosslink) is rather small (see Figure 1B), indicating that if an insulin molecule binds to either of these sites, there is not enough room for binding of a second insulin molecule. Thus, it is reasonable to assume that only one insulin molecule can bind to the same crosslink (either to site 1 or 2), when the receptor dimer is in the inactive conformation. Mathematical modelling of insulin binding to the inactive conformation of the insulin receptor is straightforward and requires only four parameters: association rate constants for sites 1 and 2 (designated a1 and a2, respectively) and dissociation rate constants for sites 1 and 2 (designated d1 and d2, respectively). The four sites will from now on be designated as sites 1, 2, 3 and 4, where sites 3 and 4 are identical to sites 1 and 2, respectively, but from a different crosslink (see Figure 2A). Binding of insulin to the inactive conformation of the insulin receptor gives rise to nine intermediaries, designated r0, r1, r2, r3, r4, r13, r14, r23 and r24 (where indices i and j in ri and rij designate the number of the site to which insulin is bound) (see Figure 2A).
Figure 1.
Insulin receptor structure. In all of the panels, the individual monomers of the insulin receptor dimer are coloured in green and blue, respectively, and location of the binding sites is shown approximately, as the residues involved in binding to insulin are not known precisely. (A) Crystal structure of the ecto-domain of the insulin receptor dimer (PDB code: 2DTG). Labelling of the modules is shown only for the blue-coloured monomer. (B) Half of the insulin receptor dimer is shown. The placement of insulin in the binding cavity is shown approximately. (C) A view of the insulin receptor dimer (shown in (A)) as seen from the ‘top'. (D) Simplified representation of the insulin receptor dimer, in which the insulin-binding subunits are represented as rigid bodies. (E) Crosslinked (tilted) conformations of the rigid-body representation of the insulin receptor dimer. Insulin is depicted as a black dot.
Figure 2.
Inactive (A) and crosslinked (B, C) insulin receptor intermediaries used in the model. S1 and S2 stand for sites 1 and 2, respectively. Insulin is depicted as a black dot.
Conformational change in the insulin receptor upon insulin binding
Currently, the precise mechanism of the insulin receptor crosslinking reaction required for mathematical modelling is not known. Therefore, we will analyse the available structural information from a physicochemical point of view to derive a plausible model.
In the absence of insulin, the insulin receptor adopts an inactive, symmetrical conformation (McKern et al, 2006) (see Figure 1A and C). Binding of insulin to both sites of a crosslink is thought to produce a conformational change in the insulin receptor, which is necessary for its activation. The crystal structure of the insulin receptor (McKern et al, 2006) is consistent with an assumption (as previously suggested by De Meyts, 1994) that this conformational change is produced by a tilt of the receptor subunits, leading to a movement of sites 1 and 2 towards each other and to a corresponding movement of sites from the second crosslink away from each other. This structural rearrangement is schematically shown in Figure 1D and E, in which the symmetrically arranged subunits of the receptor dimer are approximated by rigid bodies. Obviously, the tilted (activated) conformation has higher free energy than the symmetrical, inactive conformation (otherwise there would be a substantial receptor activation in the absence of insulin), probably due to slight distortion of bond angles and lengths. Thus, it is reasonable to assume that receptor molecules can exist in both inactive and active conformations, with equilibrium being strongly shifted towards the inactive (energetically more favourable) conformation in the absence of the ligand. However, when the ligand is present, it may bind simultaneously to sites 1 and 2 of the same crosslink when the receptor molecules are in the tilted conformation and thus stabilize the active conformation.
Insulin receptor as a harmonic oscillator: a physically plausible model of the receptor conformational change
As discussed above, the tilted (active) conformation of the insulin receptor has higher free energy compared with the non-tilted (inactive) conformation. It is reasonable to assume that larger tilt angles result in larger free energy values, and that the insulin receptor free energy has a local minimum at the zero tilt angle. Therefore, in a sufficiently small vicinity around the zero tilt angle, the system behaviour can be approximated by harmonic oscillations (Bloch, 1997). Let us consider the energy of the insulin receptor oscillations when the insulin receptor is in thermal equilibrium with the buffer. Then, the receptor molecules will have various energies (received from accidental collisions with the buffer molecules), and their distribution will be according to the Maxwell–Boltzmann formula (Bloch, 1997), which for a one-dimensional harmonic oscillator takes the form:
![]()
where P, E, k and T stand for probability, energy, Boltzmann constant and absolute temperature, respectively. As appears from Figure 3A, the most likely state of the receptor is that with the zero energy of oscillations (corresponding to the zero tilt angle, or an inactive conformation), with the probability density of finding receptor molecules with higher energies decreasing exponentially as a function of energy. The Maxwell–Boltzmann distribution implies that a small fraction of receptor molecules will have sufficient energy to reach the tilt angle of the activated receptor in the absence of insulin (Figure 3A). This fraction can be easily derived from the Maxwell–Boltzmann distribution if the activation energy (designated Eact), corresponding to the tilt angle of the activated receptor, is known:
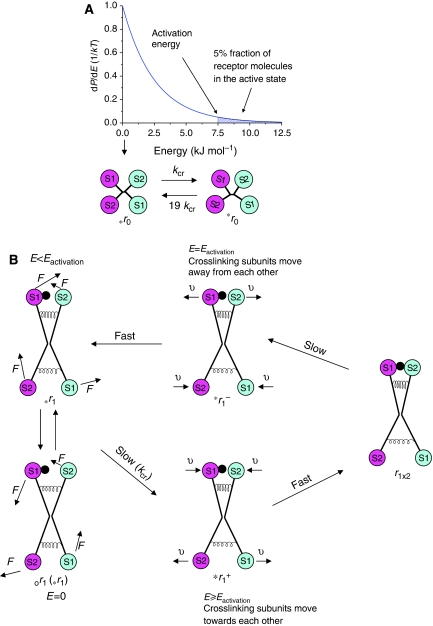
Figure 3.
Harmonic oscillator model of the insulin receptor activation mechanism. (A) Plot of the Maxwell–Boltzmann distribution for a one-dimensional harmonic oscillator and reaction scheme for the spontaneous receptor activation. The 5% fraction of the activated receptor molecules (with energy of oscillations ranging from the activation energy to ‘infinity') is indicated by the hatched region. (B) Reaction scheme for the insulin receptor activation. The forces acting to restore the inactive/symmetrical conformation of the receptor subunits are represented by elastic springs, which can be compressed and stretched during movement of the receptor subunits. E stands for energy of oscillations, Eactivation—activation energy, F—random force vector acting on the receptor subunits, υ—vector showing direction of movement of the crosslinking subunits. S1 and S2 stand for sites 1 and 2, respectively. Insulin is depicted as a black dot.
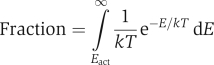 |
To estimate the fraction of the activated insulin receptor molecules in the absence of insulin, we note that the insulin receptor basal autophosphorylation rate is increased 10- to 20-fold in the presence of insulin (Flores-Riveros et al, 1989; Kohanski, 1993). It is reasonable to assume that the autophosphorylation rate is proportional to the fraction of the activated receptor molecules and that the basal rate is proportional to this fraction in the absence of insulin. Thus, around 5–10% of the receptor molecules are estimated to be activated in the absence of insulin. From this, the activation energy of the insulin receptor can be roughly estimated from the above equation as 5–8 kJ mol−1. Interestingly, the conformational change of the EGF receptor also requires about 4–8 kJ mol−1 (Ozcan et al, 2006), indicating that the above estimation is reasonable.
It should be pointed out that large-scale collective motions in a macromolecule are determined mostly by the macromolecule's global topology and are insensitive to structural details, and that the results obtained by assuming subunits to be rigid bodies connected by elastic springs provide a good description of the allosteric dynamics (for review, see Bahar and Rader, 2005). From this perspective, the rigid body model of the symmetrical insulin receptor dimer with elastic springs attached as shown in Figure 3B (compare it with Figure 1C) is expected to provide a decent description of the insulin receptor dynamics. Indeed, such a model will be capable of the conformational changes shown in Figure 1E and the movement of its subunits will be described by harmonic oscillations. In the following, this rigid body model of the insulin receptor dimer will serve as a basis for modelling of the crosslinking reaction.
Definition of the crosslinking constant
Let us consider the behaviour of the r0 intermediary in the absence of insulin. The harmonic oscillator model implies that in the absence of insulin there are no intermediaries with a certain stable tilt angle, although the zero tilt angle is most likely (see Figure 3A). The tilt angle will be fluctuating (due to collisions of the solvent molecules with the receptor subunits) until an activation state is achieved, which will only be transient because the oscillation will continue its swing in the reverse direction and quickly lose its energy due to interaction with the solvent. The r0 receptor molecules with energy of oscillations less than the energy of activation, designated *r0 (and therefore in the inactive state), will reach an active state, *r0, on average after a certain time period, τ. Then, transition from *r0 to *r0 can be modelled as a first-order reaction with a rate constant equal to 1/τ, designated from now on as the crosslinking constant, kcr (see Figure 3A). As approximately 5% of receptor molecules are in the active state (see above), the transition from *r0 to *r0 can be modelled as a first-order reaction with a rate constant equal to 19 kcr (Figure 3A).
Activation of the insulin receptor according to the harmonic oscillator model
Let us consider the activation of an inactive r1 intermediary (activation of all other inactive intermediaries can be considered in a similar way). The most likely state of this intermediary (with zero energy of oscillations) is designated as or1 and those states with energy of oscillations less than activation energy (including or1)—as *r1 (see Figure 3B), which represent the r1 receptor species in an inactive state. Subunits of the *r1 intermediaries will be experiencing random forces acting on them (shown by vectors F in Figure 3B) due to thermal motion of the solvent molecules. These forces will be varying in magnitude, direction and act for various periods of time. When these forces act synergistically for a sufficient period of time, an active state will be achieved, which (as mentioned above) will happen on average after a time period, τ, and transition to the active state, designated as *r1+ (see Figure 3B), from the inactive can be modelled as a first-order reaction with a rate constant, kcr, equal to 1/τ. As in the active state, the distance between insulin and site 2 is zero, the local concentration of insulin in the vicinity of site 2 becomes ‘infinite', which means that the subsequent binding of insulin to site 2, and thus formation of r1 × 2, is ‘instant' (see Figure 3B). To discriminate between *r1+ and r1 × 2, the *r1+ intermediary is shown in Figure 3B with a small distance between insulin and site 2, which can also be interpreted as a snapshot of *r1+ just before it reaches the active state. The r1 × 2 intermediary will exist until one of the sites of the crosslink dissociates. Let us assume that it is site 2 that dissociates. This leads to the formation of *r1− (see Figure 3B). Similar to the *r1+ intermediary, *r1− is shown with a small distance between site 2 and insulin, which can be interpreted as a snapshot of *r1− just after dissociation of site 2. There is an important difference between *r1− and *r1+. The two intermediaries have identical tilt angles, the same kinetic and potential energies of oscillations. But in *r1+ the crosslinking subunits move towards each other (see Figure 3B, the direction of movement is indicated by vectors υ), meaning that r1 × 2 will be formed in an instant, whereas in *r1− the crosslinking subunits move away from each other (see Figure 3B), meaning that r1 × 2 will not be formed because as soon as *r1− loses a small fraction of its energy due to interaction with the solvent, its energy becomes less than energy of activation, and thus by definition it becomes *r1. Thus, transition from *r1− to *r1 (see Figure 3B) can be considered as almost instant.
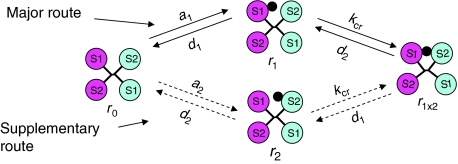
Reaction scheme for the insulin receptor activation in the harmonic oscillator model
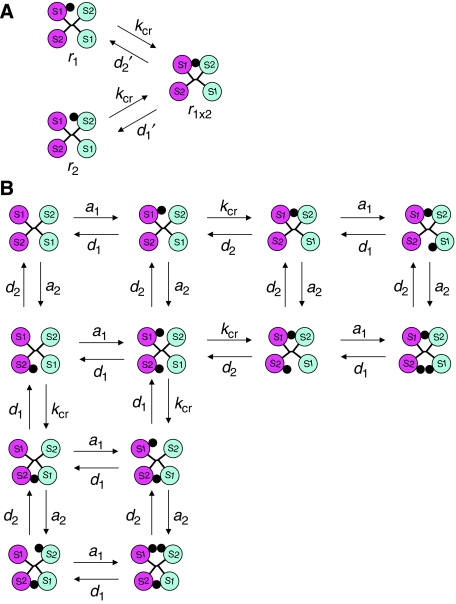
As can be seen from Figure 3B, formation of r1 × 2 from *r1 is rate limited by the first reaction (*r1 → *r1+) with a rate constant, kcr, and dissociation of r1 × 2 into *r1 is also rate limited by the first reaction (r1 × 2 → *r1−), which is a rate constant for dissociation of site 2 in r1 × 2, designated d2′. As the concentration of *r1 is practically the same as that of r1 (approximately 95% of that), the activation scheme shown in Figure 3B can be reduced to a very simple scheme: r1⇄r1 × 2 (see Figure 4A), where the forward reaction has a rate constant equal to kcr, and the reverse—d2′. By analogy, formation of r1 × 2 from r2 is described by a similar scheme: r2⇄r1 × 2 (see Figure 4A), where the rate constant of the reverse reaction (dissociation of site 1 in r1 × 2) is designated as d1′.
Figure 4.
Reaction scheme for the insulin receptor binding. (A) Scheme of the crosslinking reaction. (B) Simplified scheme of the insulin receptor kinetic network. S1 and S2 stand for sites 1 and 2, respectively. Insulin is depicted as a black dot.
Binding of insulin to the crosslinked insulin receptor intermediaries
The crosslinking reaction leads to the formation of another six crosslinked/activated intermediaries, designated r1 × 2, r3 × 4, r1 × 23, r1 × 24, r13 × 4 and r23 × 4 (where sign ‘ × ' in 1 × 2 (or 3 × 4) means that insulin crosslinks sites 1 and 2 (or sites 3 and 4) from the inactive intermediaries (r1, r2, r3, r4, r13, r14, r23 and r24) (Figure 2B). Everything described so far for the insulin receptor can also be applied to the IGF1 receptor. Because of the tilt of the insulin receptor subunits in r1 × 2, the distance between sites 3 and 4 is increased (compared with the inactive conformation) (see Figure 1D and E), indicating that the insulin receptor may now bind two more insulin molecules (to sites 3 and 4), in contrast to inactive r1 (which can only bind one more insulin molecule, either to site 3 or 4). Thus, we make an assumption that the insulin receptor can bind up to three insulin molecules, only when it is in the active state. As will be shown later, this assumption leads to a bell-shaped ligand dependence of the dissociation rate. Without this assumption, this dependence becomes sigmoid. Thus, for the IGF1 receptor it will be assumed that a third IGF1 molecule cannot bind to the receptor, which is reasonable because IGF1 is larger in size compared with insulin. In addition, the geometry of the binding sites in the IGF1 receptor is somewhat different from that of the insulin receptor, which could also be the reason why the third IGF1 molecule cannot bind to the receptor. Site 1 is more extensive in the IGF1 receptor complex because in addition to the ‘classical' binding surface binding to L1, the C-peptide of IGF1 (absent in insulin) binds to the CR region (Keyhanfar et al, 2007). Other structural differences between site 1 of the insulin and IGF1 receptors in the L1 domain are reviewed in Lou et al (2006). Binding of the third insulin molecule to the insulin receptor leads to two more intermediaries: r1 × 234 and r123 × 4 (Figure 2C). The r1 × 2 intermediary will exist until one of the sites of the crosslink dissociates, leading to either r1 (dissociation of site 2) or r2 (dissociation of site 1), with the corresponding dissociation rate constants, d2′ for site 2, and d1′ for site 1. It is tempting to assume that d1′=d1 and d2′=d2. This is not necessarily so. For dissociation of insulin from r1 to take place, some energy should be supplied to r1 for breaking the binding of insulin from site 1 (the energy comes from random collisions with the solvent molecules). However, it is obvious that slightly less energy is required when the insulin receptor is in r1 × 2 conformation, because this conformation is ‘strained' (in contrast to the ‘relaxed' r1 conformation), and the pull of the ‘strained' receptor subunits contributes to breaking the binding of insulin from site 1. Thus, d1′ is expected to be larger than d1. Using the same reasoning, d2′ is expected to be larger than d2. Because the energy of the ‘strained' r1 × 2 conformation is expected to be rather small (7.5 kJ mol−1), one may expect that d1′ (d2′) is not much larger than d1 (d2). But as it is not possible to predict precisely how much larger, two cases will be considered. In the first, it will be assumed that the increase is insignificant, or d1′=d1 and d2′=d2. In the second case, there will be no such assumption.
Effect of insulin dimerization
Insulin dimerizes in solution with a Kd value of approximately 7 μM (Pekar and Frank, 1972). After insulin dimerization, only the hexamerization site is available for interaction of the insulin dimer with sites 2 and 4. It is not known whether the insulin dimer can bind to the receptor. However, if we make an assumption that the dimer can bind to the receptor when it is in the active state, then simulations show that this binding is negligible. Thus, we can use our modelling results as evidence that the dimer does not bind to the receptor. However, as it is not possible to predict this result in advance, binding of the dimer is considered in the model. The insulin dimer could potentially bind only to site 2 of r3 × 4 and site 4 of r1 × 2, leading to another two intermediaries r2d3 × 4 and r1 × 24d (Figure 2C).
Modelling the insulin receptor binding kinetics
Binding of insulin to the insulin receptor leads to the formation of 19 intermediaries (see Figure 2) through a network of 72 individual interactions (if one counts all possible interactions between these 19 intermediaries). The essential features of this network (with 12 intermediaries and 32 interactions) are demonstrated in Figure 4B. The complete network is shown in Supplementary Figure S3 and the corresponding differential equations in Supplementary Figure S6. It should be noted that the model does not ‘allow' direct dissociation of the crosslinking insulin (binding sites 1 and 2 simultaneously) from r1 × 234 until one of the insulin molecule dissociates from sites 3 or 4. This is a consequence of the assumption that the third molecule can bind only when the receptor is in the active state (see above). This means that as long as the third molecule is bound, the receptor is in the active state (if not, the third molecule would be bound to the receptor in the inactive state, contradicting the assumption). If one of the receptor's sites binding to the crosslinking insulin dissociates, the active conformation will still be maintained (as the third insulin molecule is bound). However, as the local concentration of the crosslinking insulin in the vicinity of the dissociated site is ‘infinite' (because the active state is still maintained), the crosslinking insulin is expected to ‘instantly' rebind. Another reason is that the crosslinking insulin could be entrapped between the receptor subunits when the receptor is in the active state (see Figure 1B), and as long as the active state is maintained, insulin may not be able to escape from the binding cavity due to steric hindrances. However, the crosslinking insulin is ‘allowed' to dissociate indirectly, for example, as follows: r1 × 234 → r1 × 23 → r13 → r3 (see Figure 2), leading to dissociation of the insulin molecule, which was initially crosslinking sites 1 and 2 in r1 × 234.
Modelling the ligand dependence of the receptor dissociation rate requires explicit consideration of the binding of two types of ligand: unlabelled (‘cold') and radioactively labelled (‘hot'), which leads to the formation of 77 distinct receptor intermediaries. The number of intermediaries can be reduced to 35 if the initial conditions of the used experimental set-up are imposed. Mathematical modelling of the interactions between these 35 intermediaries (employing a system of the corresponding 35 differential equations) is described in the Supplementary information.
Modelling the IGF1 receptor binding kinetics
The IGF1 receptor kinetics can be modelled in essentially the same way as described for the insulin receptor except that (1) the third IGF1 molecule (as justified before) is not ‘allowed' to bind to the receptor and (2) IGF1 does not dimerize (Brzozowski et al, 2002).
Experimental data used for modelling
Two sets of data were used for modelling. In the first one (a competition experiment), IM9 and 293 EBN cells were incubated for 2.5 h with hot insulin and 4 h with hot IGF1, respectively, in the presence of various concentrations of the respective cold ligand. Concentration of the hot ligand was 7 pM in both cases. Thereafter, the amount of bound radioactivity was measured (Figure 5A and B). In the second set (quantification of the ligand dependence of the dissociation rate), IM9 and 293 EBN cells were pre-incubated for 2 h with 24 pM hot insulin and IGF1, respectively. Thereafter, the hot ligand was removed and cells were incubated in a 40-fold dilution for various periods of time with various concentrations of the respective cold ligand (De Meyts et al, 1973). The amount of the remaining pre-bound radioactivity was then measured (Figure 5C and D). To minimize the effect of receptor recycling on the apparent kinetics, the data were obtained at 15°C. The recycling rate at 15°C is expected to be reduced 50- to 100-fold compared with that at 37°C. The reduced rate is still too high to be totally ignored because of long incubation times (up to 4 h). Modelling receptor recycling is described in the Supplementary information.
Figure 5.
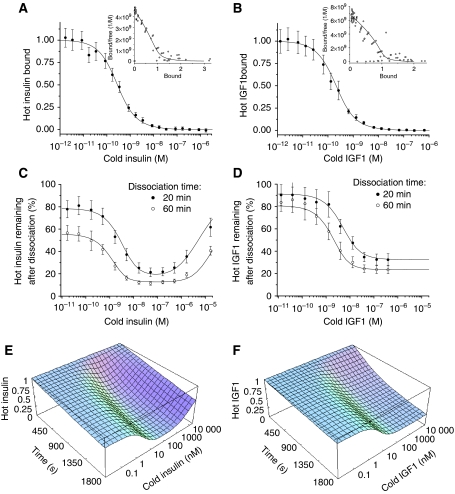
Fitting of the model to the experimental data for the insulin/IGF1 binding. (A, B) Competition experiments for the insulin and IGF1 binding, respectively (shown also as Scatchard plots in the insets). (C, D) Percentages of hot insulin and IGF1, respectively, remaining after dissociation for 20 and 60 min in the presence of various concentrations of the respective cold ligand. (E, F) Simulation of the ligand dependence of the receptor dissociation for insulin and IGF1, respectively, using the estimated parameter values.
Fitting of the insulin receptor model to the experimental data
Fitting of the insulin receptor kinetics was carried out in Mathematica 5.2 (Wolfram Research, 2005) using a genetic algorithm (Mitchell, 1996) (with random initial values of all of the parameters) in combination with gradient minimization (see Supplementary information). The fitting procedure was repeated 25 times (every time with a different set of randomly selected initial parameters) and the average values of the fitted parameters and their standard deviations are shown in Table I. As appears from Table I, the genetic algorithm converges to very similar solutions, regardless of the initial choice of parameters, which indicates that all of the used parameters can be identified uniquely. As can be seen from Figure 5A and C, there is a very good fit between the simulated and experimental data points for the competition experiment (Figure 5A) (shown also as a Scatchard plot in Figure 5A) and for the ligand dependence of the dissociation rate (Figure 5C), which reflects the fact that on average the simulated data points are within ±0.53 standard deviations from the experimental data points. If one does not assume that d1′=d1 and d2′=d2, then the best fit is produced when d1′ and d2′ are approximately three times greater than d1 and d2, respectively. However, the fit is improved only marginally, and the values of the other parameters are not changed significantly, which means that for practical purposes the assumption that d1′=d1 and d2′=d2 is reasonable. The Kd values for the two sites are approximately 6.4 and 400 nM. As the experimentally determined Kd for site 1 is approximately 10 nM, and for site 2 in the sub-micromolar range (Schäffer, 1994), it is reasonable to assign the 6.4 nM Kd to site 1, and the 400 nM Kd to site 2. Crosslinking of sites 1 and 2 leads to an apparent high-affinity binding with a 0.19 nM Kd (determined from the initial slope of the simulated Scatchard plot), which is in good agreement with the experimentally determined value for these cells (approximately 0.2 nM). It is obvious that simulation of insulin binding to the monomeric insulin receptor would result in approximately 34-fold (6.41/0.19) reduction of the apparent affinity (in good agreement with a 30-fold reduction observed experimentally), and the monomeric form would show no ligand dependence for the dissociation rate (also in accordance with the experiments). However, the simulated Scatchard plot of the monomeric receptor would be curvilinear (because sites 1 and 2 have different affinity), whereas the experimental one seems to be linear (De Meyts, 1994). It should be noted that site 2 consists of residues located in three insulin receptor domains (L2, Fn1 and Fn2) and disruption of the insulin receptor quaternary structure (due to reduction of the covalent S–S bonds, necessary for production of the monomeric form) is quite likely to disrupt this site.
Table 1.
Fitted parameter values for the insulin and IGF1 receptor kinetics
| Parameters | Insulin kinetics | IGF1 kinetics |
|---|---|---|
| Apparent Kd (nM) | 0.19±0.005 | 0.12±0.006 |
| Kcr (s−1) | 0.33±0.099 | 0.33 |
| Site 1 Kd (nM) | 6.41±0.78 | 9.09±0.40 |
| Site 2 Kd (nM) | 399±76 | 490±460 |
| a1 (s−1 M−1) | 490 000±26 000 | 347 000±7600 |
| d1 (s−1) | 0.0031±0.00031 | 0.0032±0.000091 |
| a2 (s−1 M−1) | 27 900±12 100 | 8700±8200 |
| d2 (s−1) | 0.010±0.0024 | 0.0042±0.00014 |
| Endocytosis rate (% per h) | 12.1±3.2 | 20.6±0.1 |
| Exocytosis rate (% per h) | 8.3±12.1 | 0.2±0.3 |
Fitting of the IGF1 receptor model to the experimental data
Initial fitting of the IGF1 receptor kinetics in the same way as for the insulin receptor produced a good data fit, but converged to similar solutions only for the a1 and d1 parameters (data not shown). It should be noted that the a2 and d2 parameters in the insulin receptor fitting are constrained by the upturn of the curve for the ligand dependence of the receptor dissociation rate (see Figure 5C), which happens at insulin concentrations of higher than 100 nM (this corresponds to binding of the third insulin molecule at site 2 with 400 nM Kd). However, this upturn is absent in the curve for the IGF1 receptor (see Figure 5D), and as a result, the site 2 parameters are not constrained sufficiently to produce a unique determination from the binding data alone.
To estimate the site 2 parameters, we made an assumption that the crosslinking constant, kcr, of the IGF1 receptor is similar to that of the insulin receptor. The plausibility of this assumption is based on the fact that kcr depends on the dynamics of the large-scale collective motions in the receptor, which are essentially determined by the macromolecule's global topology and are insensitive to structural details (for review, see Bahar and Rader, 2005). The structure of the IGF1 receptor dimer has not yet been determined, but the structure of the IGF1 receptor L1-CR-L2 fragment is very similar to the equivalent insulin receptor fragment (Lou et al, 2006). Taking also into account high sequence similarity between the two receptors and the fact that the hybrid insulin–IGF1 receptors bind IGF-I with high affinity, there is no doubt that the global topology of the two receptors is very similar. Thus, the assumption that the crosslinking constants of the two receptors are similar is quite reasonable.
Fitting data with the fixed kcr parameter (equal to the kcr for the insulin receptor) results in similar solutions regardless of the initial choice of the other parameters (see Table I) and a good data fit (see Figure 5B and D). The only parameter that is not well defined is a2. Compared with the insulin receptor, the IGF1 receptor appears to have 1.6 higher apparent affinity, but slower association and dissociation kinetics: a1 is lower 1.4-fold, d1 is unchanged, a2 appears to be lower 3.2-fold (but this result is not very reliable due to a large uncertainty in a2) and d2 is lower 2.4-fold. A comparison in the ligand dependence of the receptor dissociation for the insulin and IGF1 receptors, based on the estimated parameter values (see Table I), is shown in Figure 5E and F.
Discussion
In this article, we present the first mathematical model that accurately reproduces all the kinetic properties of the insulin and IGF1 receptors. In previous models of the insulin receptor binding kinetics (kinetics of the IGF1 receptor has not been modelled before) (Hammond et al, 1997; Wanant and Quon, 2000; Sedaghat et al, 2002), a two-site binding model was used to generate a curvilinear Scatchard plot resembling that of the insulin receptor, but modelling of the ligand dependence of the dissociation rate was not even attempted. Our model is different from the previous ones not only because it is capable of accurately reproducing all the kinetic properties of the insulin and IGF1 receptors but also because it is the first model that gives a physically plausible description of the two receptors' binding and activation in terms of interactions between the molecular components and takes fully into account the combinatorial complexity arising from multivalent binding to multiple receptor conformations.
The presented mathematical model builds on a structurally and thermodynamically justified assumption that the insulin receptor conformational change (required for the receptor activation) can be described by harmonic oscillations of the receptor subunits. By analysis of the behaviour of an ensemble of the insulin receptor ‘harmonic oscillators' in thermal equilibrium with the surrounding milieu, we developed a physically plausible model of the receptor crosslinking (activation), which allows to model the latter in a simple way, using only one parameter. As description of insulin binding to two different sites requires four parameters, the harmonic oscillator model requires only five parameters. In spite of this, the model gives a very accurate description of the insulin receptor binding kinetics, and with a minor modification—of the IGF1 receptor as well.
The presented model uses a rather complex network of interactions for description of the insulin kinetics. However, now that the parameters have been determined, this network may be reduced substantially depending on the experimental set-up. Simulations show that at physiological concentrations of the two ligands (i.e. lower than 0.1 nM), the kinetic network shown in Figure 4B simplifies to a very simple scheme, shown in Figure 6, in which the major route (accounting for 77% at equilibrium) is shown by solid lines, and the supplementary (accounting for 23%)—by dashed lines. If the supplementary route is ignored, the equilibrium level of r1 × 2 can be calculated as follows:
Figure 6.
Scheme of the insulin receptor binding at physiological insulin concentrations.
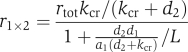 |
where L is the insulin concentration and rtot is the total receptor level. By comparing this expression with the formula for a single site binding:
![]()
we obtain that the insulin receptor Kd can be calculated from the values of the individual parameters as
Using the determined parameter values (see Table I), we obtain from this formula that the insulin receptor Kd can be estimated as 0.186 nM (in good agreement with experiments). The scheme shown in Figure 6 is also valid for the physiological concentrations of IGF1, and applying the above formula to the estimated IGF1 parameters (see Table I), the IGF1 receptor Kd can be estimated as 0.114 nM (also in good agreement with experiments).
Our model represents an essential first step in building a systems biology analysis of the insulin/IGF-I signalling networks to explain the combinatorial nature of their biological specificity. It has been demonstrated previously that mitogenic potencies of insulin analogues with altered kinetic properties correlate well with the analogues' dissociation rate from the receptor: the slower the dissociation, the higher the mitogenic potency (Shymko et al, 1997; Kurtzhals et al, 2000). In this study, we have shown that IGF1 dissociates from its receptor much slower than insulin (compare dissociations shown in Figure 5C with D at physiological concentrations of the two ligands, i.e. below 0.1 nM). The apparent dissociation rates for IGF1 and insulin (in the presence of the two ligands at physiological concentrations) can be estimated from Figure 5C and D as 0.16 and 0.54 h−1, respectively. Thus, IGF1 also complies with the above-mentioned correlation of mitogenicity versus dissociation rate, as IGF1 dissociates slower and it is more mitogenic than insulin. This suggests that kinetic proofreading could be one of the factors determining the signalling specificity of insulin, IGF1 and the insulin analogues (at least as far as mitogenicity is concerned). In the kinetic proofreading model of signalling specificity (McKeithan, 1995; Hlavacek et al, 2001), the activated receptor must complete a cascade of reversible modifications before a cellular response can occur. If the ligand dissociates before the full set of modifications is complete, the receptor reverts to its basal unmodified state. Thus, the average lifetime of the receptor in the activated state (which is equal to the reciprocal of the ligand dissociation rate constant) determines whether or not a particular biological response occurs in a signalling cascade that is under a strict kinetic proofreading control. However, application of the kinetic proofreading concept to the insulin/IGF1 signalling appears to be problematic, because the average lifetime of the insulin and IGF1 receptors in the ligand-bound state is approximately 2 and 6 h (calculated from the rate constants estimated above), respectively, whereas all of the insulin/IGF1 receptor effectors require at most 20 min for maximum activation in brown adipocytes (Krüger et al, 2007) and 45 min in white adipocytes (Schmelzle et al, 2006), with the majority of the effectors requiring only from 1 to 5 min for the maximum activation.
However, according to the presented model, the active state is maintained continuously for a much shorter period of time than the ligand-bound state. If we consider an insulin receptor intermediary with one insulin molecule bound (which corresponds to the binding under physiological insulin concentrations), then the insulin receptor molecule is expected to shuttle between the active r1 × 2 state and the inactive r1 and r2 states (see Figure 6). On the basis of the determined rate constants (see Table I), it is easy to calculate that the insulin receptor is expected to be continuously in the active state on average for approximately 76 s, whereas the IGF1 receptor for approximately 135 s. Thus, the difference in the average lifetime of the activated states of the two receptors is approximately two-fold. This two-fold difference, as demonstrated by Hlavacek et al (2001), can result in high signalling specificity depending on the kinetic parameters of a particular signalling pathway. As the majority of the insulin/IGF1 receptor's effectors reach maximum activation with a delay of 1–5 min after stimulation with the ligand (Krüger et al, 2007), the predicted lifetimes of the active states of the two receptors are sufficiently short to have a potential effect on some of the effectors due to kinetic proofreading. Whether or not a particular effector is under control of kinetic proofreading requires a careful kinetic analysis of the signalling pathway involved, which is not the aim of this study. However, it is interesting to note that activation of the Ras–MAP kinase pathway, responsible for the control of cell growth, results in a maximum activation of the ERK1 and ERK2 kinases with a delay of approximately 5 min (Krüger et al, 2007). If we hypothesize that this pathway is under control of kinetic proofreading, and use the 5 min delay as a rough estimate of the time period necessary for activation of the two MAP kinases, then IGF1 is expected to activate this pathway more efficiently than insulin, as the average lifetime of the activated state of the IGF1 receptor is two-fold smaller than the time period required for the MAP kinase activation, whereas in the case of the insulin receptor it is four-fold smaller. The above observation provides a possible explanation for the fact that IGF1 is more mitogenic than insulin, and in general for the correlation between the mitogenicity of insulin analogues and the kinetics of their dissociation. Whether or not it is true requires further investigation.
The soluble insulin receptor has a 10-fold higher affinity than the receptor expressed in the cell membrane (Schäffer, 1994). The reason for this interesting phenomenon has until now been lacking. However, the presented model provides a natural explanation for it. As discussed before, the crosslinking constant depends on the dynamics of the receptor conformational change. As the conformational change is coupled with some movement of the receptor transmembrane subunits in the membrane, the dynamics in solution are expected to be faster due to lower viscosity of the buffer solution (used in the binding experiments) in comparison with the cell membrane. The faster dynamics are expected to result in a larger value for the crosslinking constant, and thus higher affinity. For more discussion of this topic as well as an analysis of the receptor dissociation routes leading to the ligand dependence of the receptor dissociation, see Supplementary information.
It should be pointed out that the scope of our model is far from being purely theoretical. It has in fact wide-ranging applications due to its ability to provide robust determination of experimental binding parameters. Thus, we have now evaluated the five constants that define the model for the two alternatively spliced isoforms of the insulin receptor and solved some kinetic issues that had been controversial for many years (Knudsen et al, manuscript in preparation). We have shown that the insulin receptor A isoform (lacking exon 11) has 1.5-fold higher affinity and slightly faster apparent association and dissociation kinetics compared with the insulin receptor B isoform (containing exon 11). The differences between the two isoforms appear to be entirely due to the parameters of the site 1: the a1 and d1 are larger three- and two-fold, respectively, in the A isoform compared with the B-isoform. We have also started a detailed evaluation of the kinetics of binding of a number of ‘supermitogenic' insulin analogues to both insulin receptor isoforms, to determine which parameters correlate with enhanced mitogenicity. This is a critical issue in the molecular toxicology of insulin analogues, which are increasingly replacing regular insulin in the therapy of diabetes mellitus; enhanced mitogenicity in vitro of some analogues has been an issue of great concern (Kurtzhals et al, 2000; Hansen, 2008).
In conclusion, we present the first model that allows to explain mathematically all the kinetic properties of the insulin and IGF1 receptors. The model may be adaptable to other RTKs as well as cytokine receptors. Also, there is growing evidence that many receptors of the GPCR family (containing more than 800 members in humans) function as pre-bound dimers, and some of them have been shown to display negative cooperativity in ligand binding similar to that of the insulin receptor/IGF1R (Springael et al, 2007). The present model may thus be used for modelling not only the insulin and IGF1 receptors' binding but also a large number of other receptors in which ligand crosslinking occurs.
Supplementary Material
1. General remarks 2. Rule based model of the insulin receptor binding 3. Rule based model of the IGF1 receptor binding 4. Reaction scheme for the insulin/IGF1 receptor binding 5. Modelling receptor recycling 6. Modelling of the competition experiment 7. Modelling of the ligand dependence of the dissociation rate 8. Fitting of the model to the experimental data 9. Supplementary Discussion 10. Supplementary figures
Acknowledgments
The Hagedorn Research Institute is an independent basic research component of Novo Nordisk A/S. Soetkin Versteyhe and Lisbeth Gauguin were supported by a joint PhD scholarship from the Danish Ministry of Science, Technology and Development, and Novo Nordisk A/S (CORA).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams TE, Epa VC, Garrett TP, Ward CW (2000) Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci 57: 1050–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I, Rader AJ (2005) Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol 15: 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyoucef S, Surinyia KH, Hadaschik D, Siddle K (2007) Characterization of insulin/IGF hybrid receptors: contributions of the insulin receptor L2 and Fn1 domains and the alternatively spliced exon 11 sequence to ligand binding and receptor activation. Biochem J 403: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch SC (1997) Introduction to Classical and Quantum Harmonic Oscillators. New York: John Wiley & Sons Inc. [Google Scholar]
- Brzozowski AM, Dodson EJ, Dodson GG, Murshudov GN, Verma C, Turkenburg JP, de Bree FM, Dauter Z (2002) Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry 41: 9389–9397 [DOI] [PubMed] [Google Scholar]
- Christoffersen CT, Bornfeldt KE, Rotella CM, Gonzales N, Vissing H, Shymko RM, ten Hoeve J, Groffen J, Heisterkamp N, De Meyts P (1994) Negative cooperativity in the insulin-like growth factor-I (IGF-I) receptor and a chimeric IGF-I/insulin receptor. Endocrinology 135: 472–475 [DOI] [PubMed] [Google Scholar]
- De Meyts P (1994) The structural basis of insulin and insulin-like growth factor-I (IGF-I) receptor binding and negative cooperativity, and its relevance to mitogenic versus metabolic signaling. Diabetologia 37 (Suppl 2): S135–S148 [DOI] [PubMed] [Google Scholar]
- De Meyts P (2004) Insulin and its receptor: structure, function and evolution. BioEssays 26: 1351–1362 [DOI] [PubMed] [Google Scholar]
- De Meyts P (2008) The insulin receptor: prototype of dimeric, allosteric membrane receptors. Trends Biochem Sci 33: 376–384 [DOI] [PubMed] [Google Scholar]
- De Meyts P, Bianco AR, Roth J (1976) Site–site interactions among insulin receptors: characterization of the negative cooperativity. J Biol Chem 251: 1877–1888 [PubMed] [Google Scholar]
- De Meyts P, Roth J, Neville J, Gavin JR III, Lesniak MA (1973) Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem Biophys Res Comm 55: 154–161 [DOI] [PubMed] [Google Scholar]
- De Meyts P, Van Obberghen E, Roth J, Wollmer A, Brandenburg D (1978) Mapping of the residues of the receptor binding region of insulin responsible for the negative cooperativity. Nature 273: 504–509 [DOI] [PubMed] [Google Scholar]
- De Meyts P, Whittaker J (2002) Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov 1: 769–783 [DOI] [PubMed] [Google Scholar]
- Denley A, Cosgrove LJ, Booker GW, Wallace JC, Forbes BE (2005) Molecular interactions of the IGF system. Cytokines Growth Factor Rev 16: 421–439 [DOI] [PubMed] [Google Scholar]
- Fabry M, Schaeffer E, Ellis L, Kojzo E, Fahrenholz F, Brandenburg D (1992) Detection of a new hormone contact site within the insulin receptor ectodomain by the use of a novel photoreactive insulin. J Biol Chem 267: 8950–8956 [PubMed] [Google Scholar]
- Flores-Riveros JR, Sibley E, Kastelic T, Lane MD (1989) Substrate phosphorylation catalyzed by the insulin receptor tyrosine kinase. Kinetic correlation to autophosphorylation of specific sites in the beta subunit. J Biol Chem 264: 21557–21572 [PubMed] [Google Scholar]
- Gauguin L, Delaine C, Alvino CL, McNeil KA, Wallace JC, Forbes BE, De Meyts P (2008) Alanine scanning of a putative receptor binding surface of insulin-like growth factor-I (IGF-I). J Biol Chem 283: 20821–20829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BF (2008) Insulin analogues with increased mitogenic potency—are they safe?. Horm Metab Res 93: 3618–3626 [DOI] [PubMed] [Google Scholar]
- Hao C, Whittaker L, Whittaker J (2006) Characterization of a second ligand binding site of the insulin receptor. Biochem Biophys Res Commun 347: 334–339 [DOI] [PubMed] [Google Scholar]
- Hammond BJ, Tikerpae J, Smith GD (1997) An evaluation of the cross-linking model for the interaction of insulin with its receptor. Am J Physiol Endocrinol Metab 272: E1136–E1144 [DOI] [PubMed] [Google Scholar]
- Hlavacek WS, Redondo A, Metzger H, Wofsy C, Goldstein B (2001) Kinetic proofreading models for cell signaling predict ways to escape kinetic proofreading. Proc Natl Acad Sci USA 98: 7295–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Xu B, Hu SQ, Chu YC, Hua QX, Qu Y, Li B, Wang S, Wang RY, Nakagawa SH, Theede AM, Whittaker J, De Meyts P, Katsoyannis PG, Weiss MA (2004) How insulin binds: the B-Chain alpha-helix contacts the L1 beta-helix of the insulin receptor. J Mol Biol 341: 529–550 [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT (2007) Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol 19: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD (1982) The interaction of insulin with its receptor: crosslinking via insulin association as the source of receptor clustering. Diabetologia 23: 381–385 [DOI] [PubMed] [Google Scholar]
- Keyhanfar M, Booker GW, Whittaker J, Wallace JC, Forbes BE (2007) Precise mapping of an IGF-I binding site on the IGF1-R. Biochem J 401: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN (2007) Untangling the signalling wires. Nat Cell Biol 9: 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Accili D (2002) Signalling through IGF-I and insulin receptors: where is the specificity? Growth Horm IGF Res 12: 84–90 [DOI] [PubMed] [Google Scholar]
- Kohanski RA (1993) Insulin receptor autophosphorylation. I. Autophosphorylation kinetics of the native receptor and its cytoplasmic kinase domain. Biochemistry 32: 5766–5772 [DOI] [PubMed] [Google Scholar]
- Kohanski RA, Lane MD (1983) Binding of insulin to solubilized insulin-receptor from human placenta––evidence for a single class of noninteracting binding sites. J Biol Chem 258: 7460–7468 [PubMed] [Google Scholar]
- Kristensen C, Andersen AS, Ostergaard S, Hansen PH, Brandt J (2002) Functional reconstitution of insulin receptor binding site from non-binding receptor fragments. J Biol Chem 277: 18340–18345 [DOI] [PubMed] [Google Scholar]
- Kristensen C, Kjeldsen T, Wiberg FC, Schäffer L, Hach M, havelund S, Bass J, Steiner DF, Andersen AS (1997) Alanine scanning mutagenesis of insulin. J Biol Chem 272: 12978–12983 [DOI] [PubMed] [Google Scholar]
- Krüger M, Kratchmarova I, Blagoev B, Tseng Y-H, Kahn CR, Mann M (2007) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc Natl Acad Sci USA 105: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurose T, Pashmforoush M, Yoshimasa Y, Carroll R, Schwartz GP, Burke GT, Katsoyannis PG, Steiner DF (1994) Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the α-subunit of the insulin receptor. J Biol Chem 269: 29190–29197 [PubMed] [Google Scholar]
- Kurtzhals P, Schäffer L, Sørensen A, Kristensen C, Jonassen I, Schmid C, Trüb T (2000) Correlations of receptor binding and metabolic and mitogenic potencies of insulin analogues designed for clinical use. Diabetes 49: 999–1005 [DOI] [PubMed] [Google Scholar]
- Lawrence MC, McKern NM, Ward CW (2007) Insulin receptor structure and its implications for the IGF-1 receptor. Curr Opin Struct Biol 17: 699–705 [DOI] [PubMed] [Google Scholar]
- Lou M, Garett TPJ, McKern NM, Hoyne PA, Epa VC, Bentley JD, Lovrecz GO, Cosgrove LJ, Frenkel MJ, Ward CW (2006) The first three domains of the insulin receptor differ structurally from the insulin-like growth factor-1 receptor in the regions governing ligand specificity. Proc Natl Acad Sci USA 103: 12429–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan TW (1995) Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA 92: 5042–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, Elleman TC, Richards Km, Bentley JD, Pilling PA, Hoyne PA, Cartledge KA, Pham TM, Lewis JL, Sankowich SE, Stoichevska V, Da Silva E, Robinson LP, Frenkel MJ, Sparrow LG et al. (2006) Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 443: 218–221 [DOI] [PubMed] [Google Scholar]
- Mitchell M (1996) An Introduction to Genetic Algorithms. Cambridge, MA and London, UK: The MIT Press [Google Scholar]
- Mynarcik DC, Yu GQ, Whittaker J (1996) Alanine-scanning mutagenesis of a C-terminal ligand binding domain of the insulin receptor α subunit. J Biol Chem 271: 2439–2442 [DOI] [PubMed] [Google Scholar]
- Ozcan F, Klein P, Lemmon MA, Lax I, Schlessinger J (2006) On the nature of low- and high-affinity EGF receptors on living cells. Proc Natl Acad Sci USA 103: 5735–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekar AH, Frank BH (1972) Conformation of proinsulin. A comparison of insulin and proinsulin self-association at neutral pH. Biochemistry 11: 4013–4016 [DOI] [PubMed] [Google Scholar]
- Pullen RA, Lindsay DG, Woos SP, Tickle IJ, Blundell TL, Wollmer A, Krail G, Brandenburg D, Zahn H, Gliemann J, Gammeltoft S (1976) Receptor-binding region of insulin. Nature 259: 369–373 [DOI] [PubMed] [Google Scholar]
- Schäffer L (1994) A model for insulin binding to the insulin receptor. Eur J Biochem 221: 1127–1132 [DOI] [PubMed] [Google Scholar]
- Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM (2006) Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55: 2171–2179 [DOI] [PubMed] [Google Scholar]
- Sedaghat AR, Sherman A, Quon MJ (2002) A mathematical model of metabolic insulin signaling pathways. Am J Physiol Endocrinol Metab 283: E1084–E1101 [DOI] [PubMed] [Google Scholar]
- Shymko RM, De Meyts P, Thomas R (1997) Logical analysis of timing-dependent receptor signalling specificity: application to the insulin receptor metabolic and mitogenic signalling pathways. Biochem J 326: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, Urizar E, Costagliola S, Vassart G, Parmentier M (2007) Allosteric properties of G protein-coupled receptor oligomers. Pharmacol Ther 115: 410–418 [DOI] [PubMed] [Google Scholar]
- Wanant S, Quon MJ (2000) Insulin receptor binding kinetics: modeling and simulation studies. J Theor Biol 205: 355–364 [DOI] [PubMed] [Google Scholar]
- Ward C, Lawrence M, Streltsov V, Garrett T, McKern N, Lou MZ, Lovrecz G, Adams T (2008) Structural insights into ligand-induced activation of the insulin receptor. Acta Physiol 192: 3–9 [DOI] [PubMed] [Google Scholar]
- Ward CW, Lawrence MC, Streltsov VA, Adams TE, McKern NM (2007) The insulin and EGF receptor structures: new insights into ligand-induced receptor activation. Trends Biochem Sci 32: 129–137 [DOI] [PubMed] [Google Scholar]
- Wedekind F, Baer-Pontzen K, Bala-Mohan S, Choli D, Zahn H, Brandenburg D (1989) Hormone binding site of the insulin receptor: an analysis using photoaffinity-mediated avidin complexing. Biol Chem Hoppe Seyler 370: 251–258 [DOI] [PubMed] [Google Scholar]
- Whittaker L, Hao C, Fu W, Whittaker J (2008) High-affinity insulin binding: insulin interacts with two ligand binding sites. Biochemistry 47: 12900–12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PF, Mynarcik DC, Qin YG, Whittaker J (1995) Mapping of an NH2-terminal ligand binding site of the insulin receptor by alanine scanning mutagenesis. J Biol Chem 270: 1–5 [DOI] [PubMed] [Google Scholar]
- Wolfram Research, Inc. (2005) Mathematica Edition: Version 5.2. Wolfram Research, Inc. Champaign, Illinois, USA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. General remarks 2. Rule based model of the insulin receptor binding 3. Rule based model of the IGF1 receptor binding 4. Reaction scheme for the insulin/IGF1 receptor binding 5. Modelling receptor recycling 6. Modelling of the competition experiment 7. Modelling of the ligand dependence of the dissociation rate 8. Fitting of the model to the experimental data 9. Supplementary Discussion 10. Supplementary figures