Abstract
When a cell's environment changes, a large transcriptional response often takes place. The exquisite sensitivity and specificity of these responses are controlled in large part by the combinations of cis-regulatory elements that reside in gene promoters and adjacent control regions. Here, we present a study aimed at accurately modeling the relationship between combinations of cis-regulatory elements and the expression levels they drive in different environments. We constructed four libraries of synthetic promoters in yeast, consisting of combinations of transcription factor binding sites and assayed their expression in four different environments. Thermodynamic models relating promoter sequences to their corresponding four expression levels explained at least 56% of the variation in expression in each library through the different conditions. Analyses of these models suggested that a large fraction of regulated gene expression is explained by changes in the effective concentration of sequence-specific transcription factors, and we show that in most cases, the corresponding transcription factors are expressed in a pattern that is predicted by the thermodynamic models. Our analysis uncovered two binding sites that switch from activators to repressors in different environmental conditions. In both the cases, the switch was not the result of a single transcription factor changing regulatory modes, but most likely due to competition between multiple factors binding to the same site. Our analysis suggests that this mode of regulation allows for large and steep changes in expression in response to changing transcription factor concentrations. Our results demonstrate that many complex changes in gene expression are accurately explained by simple changes in the effective concentrations of transcription factors.
Keywords: competition, gene regulation, synthetic promoters, thermodynamic models, transcription factors
Introduction
Changes in a cell's environment often induce complex cascades of molecular events that result in a large-scale transcriptional response. These responses facilitate cellular processes such as differentiation (Gardner and Barald, 1991), proliferation (Radinsky, 1995), cellular defense (Owuor and Kong, 2002), and apoptosis (Matikainen et al, 2001). Quantitative models that describe how combinations of transcription factor (TF) binding sites dictate changes in expression will be an important part of understanding the transcriptional response of individual genes to environmental perturbations.
Many complex molecular events take place during regulated changes in transcription, but it is unclear how many of these events must be explicitly modeled to accurately capture the quantitative consequences of environmental changes on gene expression. Previous work suggests that in some prokaryotic and eukaryotic systems, changes in gene regulation can be accurately captured by modeling only changes in effective TF levels (Setty et al, 2003; Rosenfeld et al, 2005; Zinzen et al, 2006; Segal et al, 2008) (the concentration of a TF that can bind to its DNA site and regulate transcription); however, these studies rely on relatively few examples of promoters to make this claim. It is therefore unclear to what extent changes in TF concentrations can explain observed differences in expression levels between conditions. We showed earlier that expression levels driven by combinations of binding sites, in both synthetic and genomic promoters, are accurately captured by simple thermodynamic models that only account for protein–DNA and protein–protein interactions (Gertz et al, 2009); however, these models were only applied to expression in one steady state condition. Here, we extend this approach to model gene regulation changes in response to environmental perturbations to determine how well changes in effective TF concentrations capture environmental expression changes.
We present a thermodynamic analysis of four synthetic promoter libraries assayed for expression in each of the four environments. In each library, a single thermodynamic model that allows for fluctuations in effective TF concentrations captures over half of the variance in expression. Even though effective TF concentrations are influenced by post-translation modifications and localization, actual expression patterns matched the effective concentrations predicted by the models for the majority of TFs tested. Two of the sites that we analyzed exhibited switch-like behavior in which the site changed from an activating to a repressing site in different environments. Further analyses pointed toward competition between activators and repressors for the same site as the mechanism of switching. We show that this mode of regulation has important consequences on the dynamics of environment-specific regulation. Our results show that a substantial fraction of the transcriptional response of combinatorial promoters to changing environments can be captured by accounting for changes in TF concentrations.
Results
Promoter libraries and expression analysis
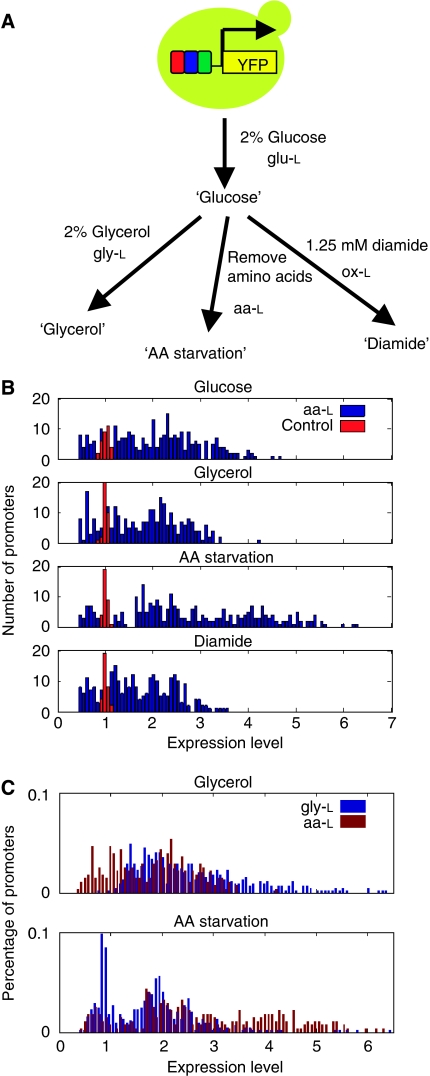
We constructed four synthetic promoter libraries in yeast, as described previously (Gertz et al, 2009), comprised of TF binding sites for both activators and repressors that should be responsive in specific environments. In the first library glu-L, made up of 376 promoters representing 183 unique combinations, we picked four sites that should be active in the presence of glucose: a Mig1/Mig2 (Lundin et al, 1994) site, a Gcr1 (Matys et al, 2003) site, a Rap1 (Matys et al, 2003) site, and a Reb1 (Liaw and Brandl, 1994) site. The second library gly-L, made up of 448 promoters representing 242 unique combinations, consisted of sites that should be active in the presence of glycerol: an Adr1 (Cheng et al, 1994) site, a carbon source response element (Roth et al, 2004) (CSRE; bound by Cat8 and Sip4), a Hap2/Hap3/Hap4/Hap5 (Chodosh et al, 1988) site, and an Rgt1 (Kim et al, 2003) site. In the third library aa-L, made up of 278 promoters representing 130 unique combinations, we picked four sites that should respond to amino acid starvation: a Cbf1 (Zhu and Zhang, 1999) site, a Gcn4 (Matys et al, 2003) site, a Met31/Met32 (Blaiseau et al, 1997) site, and an Nrg1 (Park et al, 1999) site. The final library ox-L, made up of 442 promoters representing 75 unique combinations, consists of three sites that respond to oxidative stress: an Msn2/Msn4 (Martinez-Pastor et al, 1996) site, an Smp1 (Dodou and Treisman, 1997) site, and an Xbp1 (Mai and Breeden, 1997) site. Each promoter is a random combination of the corresponding library's sites inserted upstream of yellow fluorescent protein driven by a moderately active basal promoter and integrated at the TRP1 locus in the yeast genome.
To study the relationship between combinations of TF binding sites and expression levels in different environments, each yeast strain in the synthetic promoter libraries was grown in four environments (outlined in Figure 1A): high glucose, glycerol (lone carbon source), amino acid starvation, and in the presence of the oxidative stress agent diamide (see Materials and methods for specific media and growth protocols). After being grown in the environment for a specified amount of time, each strain was then analyzed for expression by flow cytometry. The overall expression distribution for library aa-L is shown in Figure 1B (see Supplementary Figure S1 for expression distribution of other libraries). The expression levels for promoters in aa-L on the whole are higher in amino acid starvation compared with the other three conditions. A comparison of expression distributions for gly-L and aa-L in glycerol and amino acid starvation is shown in Figure 1C. The overall distribution of gly-L expression values is higher in glycerol, whereas aa-L expression values are higher in amino acid starvation. These results suggest that the binding sites chosen for the aa-L library do indeed have larger effects during amino acid starvation than in other conditions. Promoters containing only a basal promoter without any binding sites, which are used to calculate the technical variance, are shown in red in Figure 1B. In aa-L, the technical variance is 0.25% of the total variance. The biological replicate variance, or the disparity between expression levels driven by promoters with the same sequence in the same environment, is 7.17% of the total variance (see Supplementary Table S1 for error levels in each library). Overall, we see reproducible variation in expression created by combinations of TF binding sites (see Supplementary Datasets A–D for promoter sequences and expression values).
Figure 1.
Experimental setup and expression analysis in synthetic promoter libraries. (A) Four combinatorial synthetic promoter libraries (glu-L, gly-L, aa-L, and ox-L) were constructed, and each promoter was grown in four cellular environments. Cells were then analyzed for fluorescence using flow cytometry. (B) Expression distribution for library aa-L (blue) along with basal promoter only controls (red) in each of the four environments. (C) Comparison of expression distributions of libraries gly-L (blue) and aa-L (red) in glycerol and AA starvation.
Gene expression model
To model the relationship between promoter sequence and expression levels in different environments, we used a thermodynamic framework, first proposed by Shea and Ackers (1985) and described previously (Gertz et al, 2009). The main feature of the thermodynamic framework is the assumption that gene regulation is dictated entirely by the binding of proteins to DNA and proteins to other proteins. Each thermodynamic model is specified by the changes in free energies associated with different binding events and the relative concentrations of the TFs in the different conditions, while ignoring any possible kinetic events such as enzymatic modifications of RNA polymerase (RNAP) or histones. We assume that the free energies of the molecular interactions do not change in response to the environment. Therefore, the only way to achieve differential expression is through changes in the TF concentrations and thus the frequency of TF–DNA binding.
We fit a full thermodynamic model for each library separately. We also fit models in which the TF concentrations were not allowed to fluctuate. In each library, models that allow TF concentrations to change in response to the environment fit the data significantly better compared with models that maintain constant TF concentrations across all environments (Table I). In every case, cross-validation of the models on 20% of each library resulted in fits that were within 2% of those obtained by fitting on all data. These models are therefore not over fit to the data, which is expected because in the worst case we fit 20 parameters to 1112 observations.
Table 1.
R2 values (fraction of variance explained) for thermodynamic models and cross-validation experiments
| Library | Model without varying TF concentrations | Model with varying TF concentrations | Cross- validation |
|---|---|---|---|
| glu-L | 0.47 | 0.61 | 0.61 |
| gly-L | 0.37 | 0.56 | 0.56 |
| aa-L | 0.43 | 0.6 | 0.59 |
| ox-L | 0.54 | 0.63 | 0.61 |
In each library, at least 56% of the variance in expression in every environment is captured with a completely thermodynamic model. By simply changing the concentrations of TFs, we capture the majority of gene regulation in our system. This relatively simple approach worked equally well for all libraries in all conditions. The results suggest that simple protein–protein and protein–DNA interactions underlie much of combinatorial cis-regulation and that a majority of the response to environmental perturbation is accurately captured by simple changes in the effective concentrations of TFs.
The parameter values for aa-L are shown in Table II as an example (see Supplementary Table S2 for all parameter values). According to the model for aa-L, Nrg1 represses by having an unfavorable interaction with RNAP. It is present at similar concentrations in all four environments. The three activators, Cbf1, Gcn4, and Met31/Met32, have favorable interactions with RNAP and all are at higher concentrations when cells are starved for amino acids. As the activators are present at higher concentrations and the repressor remains unchanged when faced with amino acid starvation, the overall distribution of expression for the entire library is shifted up (Figure 1B). With this simple model for the aa-L library of combinatorial promoters, we can explain 60% of the variance in expression for all environments.
Table 2.
Parameter values and 95% confidence intervals for the aa-L library
| Parameter | Value (±95% CI) |
|---|---|
| [Cbf1Glucose]/[Cbf1AA starvation] | 0.04±0.02 |
| [Gcn4Glucose]/[Gcn4AA starvation] | 0.04±0.03 |
| [Met31/Met32Glucose]/[Met31/Met32AA starvation] | 0.01±0.01 |
| [Nrg1Glucose]/[Nrg1AA starvation] | 0.93±0.49 |
| [Cbf1Glycerol]/[Cbf1AA starvation] | 0.02±0.01 |
| [Gcn4Glycerol]/[Gcn4AA starvation] | 0.01±0.01 |
| [Met31/Met32Glycerol]/[Met31/Met32AA starvation] | 0.002±0.002 |
| [Nrg1Glycerol]/[Nrg1AA starvation] | 0.6±0.3 |
| [Cbf1Diamide]/[Cbf1AA starvation] | 0.04±0.02 |
| [Gcn4Diamide]/[Gcn4AA starvation] | 0.0003±0.003 |
| [Met31/Met32Diamide]/[Met31/Met32AA starvation] | 0.01±0.005 |
| [Nrg1Diamide]/[Nrg1AA starvation] | 0.65±0.33 |
| ΔGCbf1−RNAP | −2.84±0.72 |
| ΔGCbf1'−RNAP | −2.54±0.73 |
| ΔGGcn4−RNAP | −3.85±1.41 |
| ΔGGcn4'−RNAP | −4.23±1.51 |
| ΔGMet31/Met32−RNAP | −2.31±0.9 |
| ΔGMet31/Met32'−RNAP | −0.82±0.55 |
| ΔGNrg1−RNAP | 1.58±0.46 |
| ΔGNrg1'−RNAP | 2.33±0.84 |
The symbol ' denotes reverse orientation. ΔG <0 indicates a favorable interaction.
Overall, 11 of the 15 sites exhibited their expected effect on gene expression. In two cases, Gcr1 and Xbp1, the sites had no significant effect on gene regulation in our system. The other two outliers were present in gly-L. The Adr1 site behaved as a repressor, whereas Adr1 is known to be an activator (Bemis and Denis, 1988). The Rgt1 site behaved as an activator, although it is primarily thought to be a repressor (Kim et al, 2003); however, there is some evidence to suggest that Rgt1 is both an activator and a repressor (Ozcan et al, 1996; Mosley et al, 2003). To test whether Rgt1 was behaving as an activator, we placed promoters with only Rgt1 sites into an Δrgt1 deletion strain. The Rgt1 sites still activated expression in the absence of Rgt1 (Supplementary Figure S2), showing that the activation of gene expression by these sites is not through Rgt1 but most likely another TF binding the Rgt1 site. The Adr1 site is discussed below.
Transcription factor expression patterns are accurately predicted by models
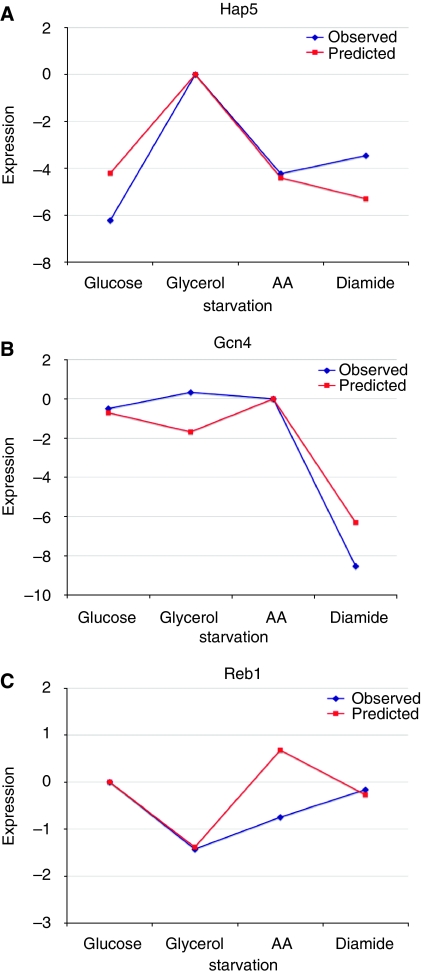
As each thermodynamic model fits parameters that correspond to the relative effective TF concentrations in each condition, we tested whether the patterns of TF levels predicted by the model match the expression levels of the TFs. To measure the expression levels of each TF, we used green fluorescent protein (GFP) tagged TFs (Ghaemmaghami et al, 2003; Huh et al, 2003). We grew each strain in the same four environments as the promoter libraries and measured GFP levels using flow cytometry. The results of representative TFs are shown in Figure 2. The majority of TFs, 6 out of 10 (Smp1 was not in the collection; Gcr1, Xbp1, Rgt1 and Adr1, discussed earlier, were excluded) showed expression patterns that significantly correlated (P<0.05) with predicted expression patterns based on the model. Of the TFs that do not significantly match the predicted expression patterns, Reb1 showed a similar pattern, but the correlation coefficient did not meet the significance threshold (ρ=0.38). The expression patterns of Mig1/Mig2, Nrg1, and Cbf1 were not similar to the predictions made by the thermodynamic models. In each of these cases, it is likely that regulation by these factors involves more than simple changes in TF concentration. Mig1 changes its localization in response to carbon source (De Vit et al, 1997). The DNA binding efficiency of Cbf1 is regulated by interactions with other proteins (Kuras et al, 1997). It has been postulated that Nrg2, a protein similar to Nrg1, binds to the same site as Nrg1. Nrg1 and Nrg2 are expressed in inverse patterns (Berkey et al, 2004) and both may be phosphorylated by Snf1 (Vyas et al, 2001). The thermodynamic model accurately predicts expression patterns for the majority of TFs that we tested. When the mode of regulation of a TF involves mechanisms other than changes in concentration, the actual expression pattern deviates from the effective concentrations predicted by the models; however, the predicted changes in effective TF concentrations allow us to accurately capture the expression patterns of the corresponding sites.
Figure 2.
General agreement between observed and predicted expression patterns of transcription factors. The observed expression patterns of Cat8 (A) and Gcn4 (B) are significantly correlated to predictions based on the models. The observed expression pattern for Reb1 (C) shows some correlation with predictions based on the model; however, it is not significant.
Two sites exhibit switch-like behavior
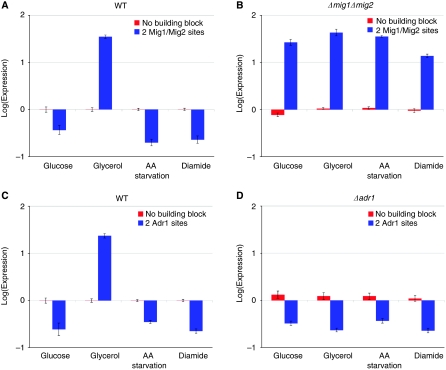
Out of the 15 sites analyzed, two—Adr1 and Mig1/Mig2—showed switch-like behavior, where they behave as activators in one condition and repressors in the other conditions. Mig1 and Mig2 are repressors that bind the same site in the presence of high concentrations of glucose (2%) (Lutfiyya et al, 1998). There is also some evidence that Mig1 can activate genes in certain genetic backgrounds (Treitel and Carlson, 1995). The Mig1/Mig2 site in the glu-L library represses transcription in the presence of glucose, but strongly activates expression when glucose is replaced by glycerol (Figure 3A). Adr1 is an activator in the presence of alternative carbon sources, such as glycerol (Bemis and Denis, 1988). The Adr1 site in the gly-L library activates in the presence of glycerol, as expected, but represses when glucose is present (Figure 3C).
Figure 3.
Adr1 and Mig1/Mig2 sites exhibit a switch-like behavior. Average log base 2 expression levels for promoters with only the basal promoter and with two Mig1/Mig2 sites are shown in the wild-type strain (A) and a Δmig1Δmig2 strain (B). The deletion of Mig1 and Mig2 abolishes repression, but uncovers activation in all conditions. Average log base 2 expression levels for promoters with only the basal promoter and with two Adr1 sites are shown in the wild-type strain (C) and an Δadr1 strain (D). The deletion of Adr1 abolishes activation in glycerol but does not remove repression in the other environments. Error bars ±1 s.e.m.
We attempted to determine the general mechanism by which these sites switch from behaving as activators to repressors. When promoters with two Mig1/Mig2 sites were placed in a Δmig1Δmig2 double-deletion strain, we observed activation, expression significantly above the basal level, in all four environments (Figure 3B). This clearly shows that Mig1 and Mig2 are not responsible for the activation observed in glycerol through the Mig1/Mig2 sites, but Mig1 and Mig2 are responsible for the repression observed in the other three environments. These results suggest that Mig1 and Mig2 successfully compete with an unknown activator, which is present in all four environments, for the Mig1/Mig2 site in the presence of glucose. In the glycerol environment, the balance is shifted such that the unknown activator binds to and activates the promoters. Other Mig1/Mig2 binding sites have also been indicated as harboring an unknown activator-binding site (Wu and Trumbly, 1998).
When promoters with two Adr1 sites were placed in an Δadr1 deletion strain, we no longer observed activation in the presence of glycerol and observed repression, expression significantly below the basal level, in all four environments (Figure 3D). This shows that Adr1 is not responsible for the repression observed in glucose through the Adr1 sites, but that Adr1 is responsible for activation in glycerol. These results indicate that Adr1 successfully competes with an unknown repressor, which is present in all four environments, for the Adr1 site in the presence of glycerol. When glucose is present in the environment, the balance is shifted such that the unknown repressor binds to and represses the promoters. Neither the Mig1/Mig2 site nor the Adr1 site matches with any other known TF binding sites in yeast. Known sites are also not created by the ligation junctions between TF binding sites. The competition between activators and repressors for the same sites may be an underappreciated and efficient mode of the transcriptional response to different environments.
Model of competition
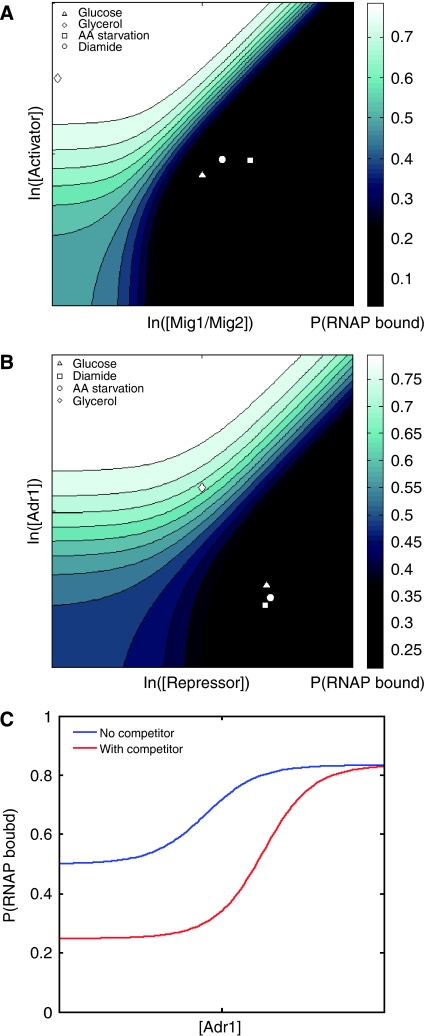
The thermodynamic models discussed earlier do not allow for activator–repressor switching and, therefore, cannot capture fully switch-like sites. Within the thermodynamic framework, the simplest method of explaining these switch-like sites is to introduce unknown competing TFs into the model. We do not have direct evidence of competition; although, it is congruent with the data described earlier. When a factor competing with Mig1/Mig2 for its site is added to the model of the glu-L library, the R2 increases from 0.61 to 0.65 (P<0.001, F-test). By adding an unknown factor that competes with Adr1 for its site in the model for gly-L, the R2 value increases from 0.56 to 0.62 (P<0.001, F-test). In each case, thermodynamic models that introduce competing factors are significantly better at capturing expression patterns. For sites that did not exhibit a switch-like behavior, adding an unknown competing factor did not create a significantly better model.
To determine the landscape of expression levels in the presence of competing TFs, we used the thermodynamic models to simulate the influence of varying TF concentrations on expression. We examined the response of two Mig1/Mig2 sites and two Adr1 sites to different levels of their corresponding TF and an unknown competing TF. The model predicts that a promoter with two Mig1/Mig2 sites is repressed in any environment with glucose and fully activated in glycerol. The predicted TF concentrations in the repressed environments are placed at the foot of a steep gradient (Figure 4A). The same pattern is predicted for a promoter with two Adr1 sites, except that in glycerol, the unknown repressor keeps Adr1 from fully activating at the promoter (Figure 4B). In each case, the TF concentrations in glucose indicate a promoter in a repressed state that is poised to dramatically change expression levels in response to slight changes in TF concentrations.
Figure 4.
Competition between activators and repressors. The probability of RNA polymerase (RNAP) being bound to a promoter with two Mig1/Mig2 sites (A) and two Adr1 sites (B) based on the thermodynamic models involving competition are shown. The predicted concentrations of the competing TFs in glucose (▵), glycerol (◊), AA starvation (□), and diamide (○) are also presented. (C) The effect on Adr1 concentrations on the probability of RNAP bound with and without a competing repressor.
The presence of a competitive factor causes a more dramatic response of expression to changes in TF concentrations. In the case of Adr1, the model predicts that the dynamic range of expression levels is over twice as large with a competitive repressor at the glucose concentration than without a competitive repressor present (Figure 4C). The maximum gradient is also twice as steep with a competitive repressor. When competing TFs are present, the model predicts that promoters not only display a larger dynamic range in expression but are also more sensitive to TF concentrations.
Discussion
Using large libraries of synthetic combinatorial promoters, we were able to accurately and quantitatively model how combinations of regulatory elements impact expression levels in different environments. We found that four separate thermodynamic models, each based solely on binding events between DNA and proteins, accounted for approximately 60% of the variance in expression in all four environments. These models capture the majority of gene regulation in our system, while only allowing effective TF concentrations to vary between environments. In many cases, we showed that changes in TF concentrations match closely with those predicted by the model. In these cases, changes in TF concentrations are the likely primary mode of regulation between conditions. In other cases, although the model accurately predicted the expression of library members containing the TF binding sites, the expression of the TFs themselves did not match the predicted levels. These sites were bound by TFs that are known to have a significant post-translational mode of regulation. The discrepancies between the predictions and observed TF levels may be indicators of significant post-translation regulation.
In analyzing 15 binding sites, we found that two behaved in a switch-like manner, acting as both an activating and a repressing site depending on the environment. A likely mechanism of switching is the existence of competing TFs. In some expression systems, competing TFs are known to impact regulation (Gregori et al, 1993; Kwon et al, 1999; Pierce et al, 2003); however, the number of known competing TFs is small, possibly due to difficulties in finding switch-like binding sites. The regulatory roles of Adr1 and Mig1/Mig2 have been thoroughly studied using gene knockouts (Hu et al, 2007; Westergaard et al, 2007); however, more information is needed to find switch-like binding sites. Only because we isolate and analyze individual TF binding sites, as opposed to entire promoters, and we are quantitatively aware of our basal promoter activity, we are able to observe the phenomenon of binding site switching. In each case, Adr1 and Mig1/Mig2 sites, we found that competition between different factors was the most likely mechanism of switching. Having two factors compete for the same site is an efficient way to tune expression to fluctuations in the environment, creating larger dynamic ranges of expression and steeper responses to TF concentrations. It also makes for an interesting evolutionary landscape for non-coding DNA. If competition is common in promoters, then the evolution of regulatory elements is a multidimensional optimization problem. For example, mutations in a switch-like regulatory element may influence the binding of an activator, the binding of a repressor, or both. Therefore, the particular sequence of a switch-like regulatory element will strongly influence its corresponding transcriptional response to an environment through the relative binding of an activator and a repressor.
Materials and methods
Strains and plasmids
The strain harboring the synthetic promoter library was derived from the haploid strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) as described in Brachmann et al (1998). The library of promoters was constructed in plasmid pJG102 (Gertz et al, 2009). TF-GFP fusions were obtained from Invitrogen (Carlsbad, CA) and are described in Ghaemmaghami et al (2003) and Huh et al (2003). To measure the activity of Adr1 and Rgt1 sites, we used BY4742 and MATα deletion strains derived from BY4742 described in Brachmann et al (1998). To look at the activity of Mig1/Mig2 sites, we used BY4742 and Δmig1Δmig2 strain YM6682. YM6682, which was provided by Mark Johnston, Washington University, was created by mating Δmig1 and Δmig2 haploid strains, sporulating them, and selecting double-deletion spores.
Library construction
To create the building blocks that make up the synthetic promoters, we used the procedure and oligonucleotide pairs described in Gertz et al (2009). The Gcr1 site we used was:
5′-GATCGTACAGCTTCCTCTAC-3′
3′-CATGTCGAAGGAGATGCTAG-5′
Expression analysis
All cultures were grown with shaking at 30°C. Cultures of synthetic promoter strains (including deletion experiments) were grown to log phase in 2 ml 96-well plates in 500 ul of synthetic complete media lacking uracil with 2% glucose. ‘Glucose' cultures were fixed at this point. For ‘diamide' cultures, diamide was added to a final concentration of 1.25 mM and grown for 7 h. ‘amino acid starvation' cultures were first spun down at 3000 g for 5 min. The supernatant was removed and 500 ul of minimal media containing 2% glucose and supplemented with histidine, leucine, lysine, and tryptophan was added, and the cultures were grown for 6 h. Glycerol cultures were first spun down at 3000 g for 5 min. The supernatant was removed and 500 ul of synthetic complete media lacking uracil with 2% glycerol was added and the cultures were grown for 16 h. TF-GFP fusions were grown in the same ways described earlier, except that uracil was added to each media.
Each culture was fixed at the corresponding time point by adding a 4% paraformaldehyde solution (4% paraformaldehyde, 100 mM sucrose) to a final concentration of 1% and incubating at room temperature for 15 min. The cells were then spun down at 3000 g for 5 min. The supernatant was removed, and the cells were resuspended in 250 ul of phosphate-buffer saline and stored at 4°C.
The fluorescence intensities and electronic volumes of 25 000 events from each well were measured on a Beckman Coulter Cell Lab Quanta SC with a multiplate loader. For each well, the mean of fluorescence divided by electronic volume for 25 000 events was taken as the expression value for that well. On each plate, the expression value of the four no insert controls were averaged to calculate a plate effect to account for changes in laser intensity or growth differences. Each expression value on the plate was then divided by the plate effect.
Sequencing
Synthetic promoters were sequenced and analyzed as described previously (Gertz et al, 2009).
Thermodynamic model
All calculations were performed using the Matlab package from The Mathworks, Inc. (Natick, MA). To model gene expression, we implemented a thermodynamic model of polymerase occupancy that was proposed by Shea and Ackers (1985). The model and implementation was described previously in Gertz et al (2009). In brief, the parameters that comprise the model are ϖ's that describe the changes in free energies from the binding of two proteins (TFs and/or RNAP) and q's for each protein that are confounded parameters. The q parameters represent the natural log of 1/Kd for the protein–DNA interaction plus the natural log of the active (meaning able to bind DNA) protein concentration. With these parameters, Boltzmann weights for each possible state of the promoter are calculated. Boltzmann weights are calculated by taking the sum of the q values for all DNA–protein interactions and ϖ values for all protein–protein interactions occurring in a particular state and exponentiating the negation of that sum. For instance, in a state where a TF and RNAP are bound to a promoter, the Boltzmann weight is equal to the exponentiation of −(qRNAP+qTF+ϖRNAP−TF). The probability of RNAP binding is then determined by dividing the sum of Boltzmann weights for the states with RNAP bound by the sum of Boltzmann weights for all states.
Different expression levels in different environments are modeled by changing the active TF concentrations and therefore the q values. This is because changes in free energies (ϖ and Kd) do not depend on environment. To fit expression levels, q values are allowed to change with the environments; however, the q values in the reference environment (e.g., glycerol for gly-L) are fixed at a neutral value of zero. As we are only measuring expression levels, q and ϖ values are dependent on each other. Therefore, q values can only describe relative changes in active TF concentrations.
To model competition between TFs for the same site, we allow the site to have three states: unbound, bound by the first TF, or bound by the second TF; compared with two states: bound or unbound by the TF. Therefore, the only difference is that there are more states where Boltzmann weights need to be calculated. Once the Boltzmann weights are calculated, the weights are partitioned in the same way as described previously (Gertz et al, 2009).
Parameter fitting
Parameters were fit for the thermodynamic models as described previously (Gertz et al, 2009). When fitting the models with competition, the initial guess for the parameter values was the final parameter fit for the model for the same library without competition and all zeros for parameters pertaining to the competing TF.
Cross-validation was performed by first partitioning the promoters randomly into 20% blocks. We removed one block at a time and fit parameters on the remaining 80%. The accuracy of the model was then measured on the block that was left out. This was repeated for all five blocks, and the results were combined to calculate the overall R2 of the cross-validation.
Supplementary Material
Contains supplementary figures and tables
Contains promoter sequences and expression values for glu-L.
Contains promoter sequences and expression values for gly-L.
Contains promoter sequences and expression values for aa-L.
Contains promoter sequences and expression values for ox-L.
Acknowledgments
The authors thank Eric Siggia, Gary Stormo, Robi Mitra, Mark Johnston, Katherine Varley, and members of the Cohen lab for helpful discussion and suggestions. BAC and JG were supported by NIH grant R01 GM078222. JG was also supported by NSF Graduate Research Fellowship DGE-0202737.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bemis LT, Denis CL (1988) Identification of functional regions in the yeast transcriptional activator ADR1. Mol Cell Biol 8: 2125–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey CD, Vyas VK, Carlson M (2004) Nrg1 and nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot Cell 3: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D (1997) Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol 17: 3640–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast (Chichester, England) 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Cheng C, Kacherovsky N, Dombek KM, Camier S, Thukral SK, Rhim E, Young ET (1994) Identification of potential target genes for Adr1p through characterization of essential nucleotides in UAS1. Mol Cell Biol 14: 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Olesen J, Hahn S, Baldwin AS, Guarente L, Sharp PA (1988) A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell 53: 25–35 [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M (1997) Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell 8: 1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Treisman R (1997) The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol 17: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner CA, Barald KF (1991) The cellular environment controls the expression of engrailed-like protein in the cranial neuroepithelium of quail-chick chimeric embryos. Development (Cambridge, England) 113: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA (2009) Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457: 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gregori C, Kahn A, Pichard AL (1993) Competition between transcription factors HNF1 and HNF3, and alternative cell-specific activation by DBP and C/EBP contribute to the regulation of the liver-specific aldolase B promoter. Nucleic Acids Res 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR (2007) Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kim JH, Polish J, Johnston M (2003) Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol 23: 5208–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Barbey R, Thomas D (1997) Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J 16: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Kim MS, Edenberg HJ, Hur MW (1999) Sp3 and Sp4 can repress transcription by competing with Sp1 for the core cis-elements on the human ADH5/FDH minimal promoter. J Biol Chem 274: 20–28 [DOI] [PubMed] [Google Scholar]
- Liaw PC, Brandl CJ (1994) Defining the sequence specificity of the Saccharomyces cerevisiae DNA binding protein REB1p by selecting binding sites from random-sequence oligonucleotides. Yeast (Chichester, England) 10: 771–787 [DOI] [PubMed] [Google Scholar]
- Lundin M, Nehlin JO, Ronne H (1994) Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol 14: 1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M (1998) Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150: 1377–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai B, Breeden L (1997) Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol 17: 6491–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15: 2227–2235 [PMC free article] [PubMed] [Google Scholar]
- Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, Sherr DH, Tilly JL (2001) Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat Genet 28: 355–360 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Lakshmanan J, Aryal BK, Ozcan S (2003) Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem 278: 10322–10327 [DOI] [PubMed] [Google Scholar]
- Owuor ED, Kong AN (2002) Antioxidants and oxidants regulated signal transduction pathways. Biochem Pharmacol 64: 765–770 [DOI] [PubMed] [Google Scholar]
- Ozcan S, Leong T, Johnston M (1996) Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol 16: 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Koh SS, Chun JH, Hwang HJ, Kang HS (1999) Nrg1is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol 19: 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK (2003) Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol 23: 4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radinsky R (1995) Modulation of tumor cell gene expression and phenotype by the organ-specific metastatic environment. Cancer Metastasis Rev 14: 323–338 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science (New York, NY) 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Roth S, Kumme J, Schuller HJ (2004) Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet 45: 121–128 [DOI] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U (2008) Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Setty Y, Mayo AE, Surette MG, Alon U (2003) Detailed map of a cis-regulatory input function. Proceedings of the National Academy of Sciences of the United States of America 100: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MA, Ackers GK (1985) The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol 181: 211–230 [DOI] [PubMed] [Google Scholar]
- Treitel MA, Carlson M (1995) Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proceedings of the National Academy of Sciences of the United States of America 92: 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Kuchin S, Carlson M (2001) Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard SL, Oliveira AP, Bro C, Olsson L, Nielsen J (2007) A systems biology approach to study glucose repression in the yeast Saccharomyces cerevisiae. Biotechnol Bioeng 96: 134–145 [DOI] [PubMed] [Google Scholar]
- Wu J, Trumbly RJ (1998) Multiple regulatory proteins mediate repression and activation by interaction with the yeast Mig1 binding site. Yeast (Chichester, England) 14: 985–1000 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang MQ (1999) SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics (Oxford, England) 15: 607–611 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D (2006) Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16: 1358–1365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains supplementary figures and tables
Contains promoter sequences and expression values for glu-L.
Contains promoter sequences and expression values for gly-L.
Contains promoter sequences and expression values for aa-L.
Contains promoter sequences and expression values for ox-L.