Abstract
Addition of glucose to yeast cells increases their growth rate and results in a massive restructuring of their transcriptional output. We have used microarray analysis in conjunction with conditional mutations to obtain a systems view of the signaling network responsible for glucose-induced transcriptional changes. We found that several well-studied signaling pathways—such as Snf1 and Rgt—are responsible for specialized but limited responses to glucose. However, 90% of the glucose-induced changes can be recapitulated by the activation of protein kinase A (PKA) or by the induction of PKB (Sch9). Blocking signaling through Sch9 does not interfere with the glucose response, whereas blocking signaling through PKA does. We conclude that both Sch9 and PKA regulate a massive, nutrient-responsive transcriptional program promoting growth, but that they do so in response to different nutritional inputs. Moreover, activating PKA completely recapitulates the transcriptional growth program in the absence of any increase in growth or metabolism, demonstrating that activation of the growth program results solely from the cell's perception of its nutritional status.
Keywords: glucose signaling, protein kinase A, Saccharomyces, Sch9, Snf1
Introduction
To generate energy and central carbon metabolites, yeast cells ferment glucose or fructose in preference to other sugars and preferentially use any fermentable carbon source to any carbon source that has to be metabolized by oxidation (Johnston and Carlson, 1992). This hierarchical arrangement is established by allosteric regulation of various key enzymes in metabolic processes, such as glycolysis and gluconeogenesis, and by an extensive transcriptional regulatory network. Accordingly, addition of glucose to cells growing on a non-fermentable carbon source results in a rapid change in the pattern of protein phosphorylation as well as massive restructuring of the transcriptional state of the genome (Wang et al, 2004). These transcriptional and metabolic changes attendant on the transition to or from growth on glucose are mediated by several interlocking regulatory pathways whose relative contributions to overall regulation have not been fully clarified (Santangelo, 2006; Zaman et al, 2008). We have attempted to comprehensively define the glucose signaling network by determining how much of the glucose signal is mediated through each of the known glucose-responsive pathways.
Five systems are known to participate in glucose signaling in yeast, Ras/PKA, Gpr1/Gpa2, Sch9, Snf1 and Rgt2/Snf3 (Santangelo, 2006; Zaman et al, 2008). The Ras/PKA pathway plays the central role in responding to changes in glucose concentration and initiating the signaling processes that lead to cellular growth and division (Broach and Deschenes, 1990; Wang et al, 2004; Santangelo, 2006). Ras is a guanine nucleotide-binding protein that in its GTP-bound state activates adenylyl cyclase. Glucose addition to cells increases the level of GTP-bound Ras, yielding an increase in intracellular cAMP and subsequent activation of protein kinase A (PKA). The PKA catalytic subunits, encoded by TPK1, 2 and 3, phosphorylate a variety of proteins involved in metabolism and transcription to reorganize the metabolic and transcription landscape of the cell.
Gpr1 and Gpa2 define a nutrient sensing pathway that works in parallel with Ras to activate PKA. The GPCR-like protein, Gpr1 stimulates adenylyl cyclase in response to glucose through its associated GTP-binding protein, Gpa2 (Kubler et al, 1997; Xue et al, 1998). However, although the Gpr1/Gpa2 pathway clearly affects the developmental consequences of glucose availability (Kubler et al, 1997; Tamaki et al, 2000), it may play only a minor role in the acute transcriptional response of cells to glucose (Wang et al, 2004).
Sch9 is an AGC family kinase and the closest yeast homolog to the mammalian pro-survival Akt/PKB as well as to the TOR-regulated S6 kinase. SCH9 overexpression suppresses lethality caused by the loss of PKA signaling (Toda et al, 1988). Selective inhibition of an ATP analog-sensitive allele of SCH9 (sch9as) in cells growing in glucose media inhibits the expression of genes involved in ribosome biogenesis (Jorgensen et al, 2002), suggesting that it might play a similar role in ribosomal biogenesis as does PKA. Furthermore, Sch9 also mediates the regulation of ribosomal biogenesis by the TORC1 complex (Urban et al, 2007). We do not know whether the ability of Sch9 to suppress mutations in the Ras/PKA pathway reflects a convergence of Sch9 and PKA activities from different signaling paths or a direct participation of Sch9 in glucose signaling.
Snf1, the yeast homolog of mammalian AMP-activated PK, modulates gene expression upon glucose depletion through activation of the transcriptional activators Cat8 and Adr1 and inactivation of the Mig1 transcriptional repressor. Growth of cells in limiting glucose or in the presence of alternative carbon sources, such as glycerol and ethanol, activates the Snf1 PK through phosphorylation by one of three upstream kinases (Woods et al, 1994; Wilson et al, 1996; Hong et al, 2003; Nath et al, 2003). In the presence of glucose, the Reg1/Glc7 protein phosphatase 1 complex dephosphorylates and inactivates Snf1 (Sanz et al, 2000). Previous comparisons of global transcriptional changes following glucose downshift in SNF1 versus snf1Δ strains indicated that as many as 500 genes are regulated either directly or indirectly by Snf1 (Tachibana et al, 2005). The extent to which these genes are directly regulated by Snf1 and the extent of the overlap of the Snf1 regulatory network with other glucose-sensitive pathways are not known.
A final glucose-sensitive regulatory pathway couples expression of the multiple hexose transporter (HXT) genes to the level of glucose in the media through modulation of the Rgt1 repressor and the Mth1 and Std1 corepressors. Glucose-induced degradation of Mth1 and Std1 renders the Rgt1 repressor inactive, resulting in the induction of the HXT genes. Rgt1 activity is further influenced by the Snf1 and PKA pathways, and components of the Rgt system are subject to various positive and negative feedback loops (Kaniak et al, 2004; Palomino et al, 2006).
Nutrient availability influences not only the transcriptional pattern of the cell but also its growth rate. Cells have the remarkable ability to maintain balanced growth over a wide range of growth rates (Regenberg et al, 2006; Castrillo et al, 2007; Brauer et al, 2008). Cells adapt to reduced nutritional availability, in part by altering the pattern of genes they transcribe. Although limitation for any one nutrient induces a nutrient-specific transcriptional response, much of the transcriptional change upon nutrient limitation is independent of which nutrient is limiting. This set of genes includes a group whose magnitude of expression change exhibits a strict dependence on growth rate (Regenberg et al, 2006; Brauer et al, 2008). As a result, Brauer et al (2008) could identify a collection of genes, the transcriptional levels of which provided a ‘growth rate signature' that was highly predictive of the growth rate of the cells from which the sample was taken. Thus, nutrient availability establishes both the growth rate of the cell and a corresponding highly stereotypic transcriptional pattern. However, causality in this correlation is unclear: does metabolism of available nutrients determine growth rate, which then specifies the transcriptional pattern, or does the cell's perception of nutrient availability set the transcriptional pattern, which in turn allows the cell to metabolize the available nutrients and grow at a particular rate? We have addressed this issue of causality in our studies of glucose sensing in yeast.
We previously examined how Saccharomyces cerevisiae regulates its transcriptional response to glucose by analyzing the Ras/PKA and Gpr1/Gpa2 pathways and using microarray analysis to capture the entire transcriptional response (Wang et al, 2004). That study not only identified the Ras/PKA pathway as the predominant locus through which glucose regulates transcription but also suggested that additional pathways played redundant, overlapping roles in this process. A recent report implicates signaling through TORC1 as a major parallel pathway to PKA (Slattery et al, 2008). In this present report, we provide a comprehensive view of glucose signaling in the cell by identifying the contributions of the five interlocking pathways to the transcriptional response. Our observations posit Ras/PKA as the primary mediators of the growth response of cells to glucose, whereas the Snf1 and Rgt pathways regulate a small set of genes specialized in alternate carbon metabolism and glucose uptake, respectively. Furthermore, although Sch9 activation can elicit essentially the same transcriptional response as does PKA, Sch9 does not participate significantly in the normal glucose response. Finally, our results indicate that yeast cells set their transcriptional growth pattern—and their growth rate—solely on their perception of the nutritional state of the cell, rather than on the products and activity attendant on metabolism of those nutrients.
Results
Glucose regulates transcription predominantly through the Ras/PKA pathway
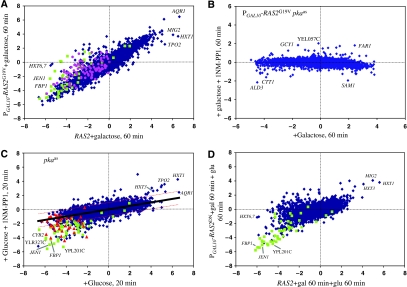
The Ras/PKA pathway likely plays a major role in the massive transcriptional restructuring attendant on glucose addition. Induction of an activated version of the RAS2 gene, by the addition of galactose to PGAL10-RAS2G19V strain growing in glycerol, recapitulates in direction and magnitude approximately 90% of the transcriptional changes resulting from glucose addition to wild-type yeast (Figure 1A). To confirm that Ras2 affects transcription solely though PKA, we constructed a strain carrying single mutations in each of TPK1, TPK2 and TPK3 that rendered each of the encoded kinases sensitive to inhibition by a modified kinase inhibitor, 1NM-PP1, which carries a bulky side group that precludes it from inhibiting wild-type PKA or any other kinase in the cell (Bishop et al, 2001). In the absence of the inhibitor, the mutant kinases function essentially normally in the cell and the strains exhibit normal growth and responses. We refer to the combination of tpk1as, tpk2as and tpk3as mutations as pkaas. Addition of 1NM-PP1 concurrently with galactose to a PGAL10-RAS2G19V pkaas strain growing in glycerol almost completely eliminates the Ras-induced transcriptional responses (Figure 1B). This shows that all of the Ras-induced signaling is mediated entirely through PKA. Thus, we conclude that Ras2 activation extensively recapitulates glucose-induced transcriptional changes solely through the activation of PKA.
Figure 1.
PKA mediates the primary transcriptional response of cells to glucose. Scatter plots of microarray data for ca. 5600 yeast genes, obtained from two different strains or conditions. Each point represents the log(2) change in levels of the mRNA for a single gene for one strain under one condition in the horizontal dimension and the log(2) change in mRNA levels for that gene in a different strain or condition in the vertical dimension. (A) Expression changes of all genes in a PGAL10-RAS2G19V strain (Y2866) pregrown on SC+3% glycerol, 60 min after the addition of 2% galactose relative to that at 0 min (vertical axis), versus a RAS2 strain (Y2864) pregrown on SC+3% glycerol, 20 min after glucose addition relative to 0 min (horizontal axis). (B) Both x- and y-axes show expression changes in PGAL10-RAS2G19V tpk1astpk2astpk3as (strain Y3621) pregrown on SC+3% glycerol, 60 min following the addition of galactose relative to 0 min. For the experiment in the vertical axis, 100 nM 1NM-PP1 was added concurrently with galactose. (C) Both x- and y-axes show expression changes in tpk1astpk2astpk3as (strain Y3561) grown on SC+3% glycerol 20 min following the addition of 2% glucose relative to 0 min. For the experiment in the vertical axis, 100 nM 1NM-PP1 was added concurrently with glucose. Solid black line shows the linear regression (y=0.26x), with the dotted red lines the two-fold limits from the regression line. (D) PGAL10-RAS2S24N (strain Y3168), pregrown on SC-glycerol and then preinduced with 2% galactose, 60 min after glucose addition relative to 0 min (vertical axis), versus RAS2 (strain Y2864) under the same conditions (horizontal axis). In all plots, genes whose expression is repressed more than two-fold by inactivation of Snf1 (see Figure 4) are shown in green, genes reported to be regulated by the Hap2/3/4/5 complex are shown in pink and those repressed more than two-fold by 125 μM glucose are shown in red.
Although most genes repressed by glucose are repressed to the same extent by RAS2G19V induction, about 160 genes repressed by glucose are not repressed, or are not repressed as substantially, following the induction of RAS2G19V (Figure 1A). These genes (Supplementary Table S1) are highly enriched for those involved in oxidative phosphorylation and the citric acid cycle. Moreover, a majority of these genes are highly enriched in those regulated by the Snf1 pathway (P=1.8 × 10−36, see below) or are listed in YEASTRACT (www.yeastract.com) as being subject to regulation by the Hap2/3/4/5 transcription factor complex (P=4.3 × 10−57) (Supplementary Table S1). Genes regulated by Snf1 do not significantly overlap those regulated by Hap2/3/4/5. Thus, our analysis reveals that at least three distinct pathways, Ras/PKA, Snf1 and Hap, participate in glucose repression.
Although our results above indicate that Ras/PKA activation is sufficient to recapitulate glucose signaling, these results do not establish that glucose signaling actually proceeds through Ras/PKA. To address this issue, we measured glucose-induced transcriptional changes under conditions of compromised Ras/PKA signaling. Specifically, we examined transcriptional changes following concurrent addition of glucose and 1NM-PP1 to a pkaas strain relative to the addition of glucose alone (Figure 1C). The glucose effect on the transcription of most genes was uniformly reduced to approximately one-fourth that seen in the absence of drug. We obtained the same result using a dominant-negative RAS2 allele (Figure 1D). These results indicate that PKA is required for most of the glucose-induced transcriptional changes.
Although the absence of a functional PKA pathway attenuated glucose-induced transcriptional changes in most genes, 75–100 genes exhibited essentially normal repression and approximately 40 genes showed essentially normally induction (Figure 1C and D; Supplementary Table S1). The set of genes exhibiting essentially normal repression in these two experiments strongly overlap the set of genes unregulated by RAS2G19V induction (P=10−60), as well as the set of genes regulated by the Snf1 pathway (P<10−300; see Figure 1C and below). In sum, these data clearly indicate that glucose regulation of most genes proceeds predominantly through the Ras/PKA pathway and that a small but significant number of genes are regulated by glucose independently of the Ras/PKA pathway.
Sch9 plays a limited role in glucose signaling
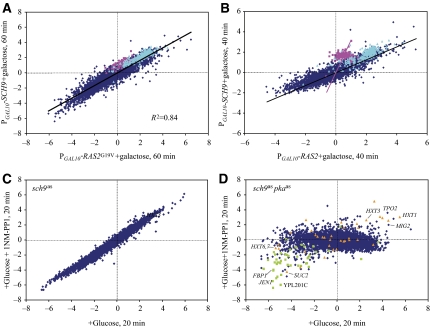
Sch9 was isolated as a high-copy suppressor of loss-of-function mutations of Ras and PKA. Moreover, global transcriptional studies of Sch9 indicated that many of the genes stimulated by PKA activation required Sch9 for expression (Jorgensen et al, 2002). Accordingly, we examined the effect on global transcription of the yeast genome of overexpressing Sch9 in a PGAL1-SCH9 strain grown in glycerol and compared that to the effect of glucose addition or RAS2G19V induction. We found that overexpression of Sch9 recapitulated in direction and magnitude 90% of the transcriptional effects of glucose addition to cells (data not shown) as well as the effects of RAS2G19V induction (Figure 2A). This shows that overexpression of Sch9 leads to the induction and repression of the same set of genes as does activation of Ras2 and suggests that SCH9 is a high-copy suppressor of the Ras/PKA pathway because it can regulate similar functional processes as Ras. Interestingly, the pattern of expression resulting from Sch9 overexpression differed from that of Ras2 activation during the early phase of induction. In particular, the changes in expression of most genes following Sch9 activation occurred more slowly than those promoted by Ras activation. However, ribosomal biogenesis genes and, more dramatically, ribosomal protein gene induction occurred significantly more rapidly, as judged by the extent of deviation of these genes from a Gaussian fit to the average kinetic behavior of all other genes (P<10−20) (Figure 2B). Thus, although the final result of Sch9 activation mimics that of Ras2/PKA activation, the mechanism by which these changes occur may differ in the two cases.
Figure 2.
Sch9 plays a minor role in glucose signaling. Microarray expression data presented as in Figure 1. (A) Expression changes of all genes in a PGAL1-SCH9 strain (Y3506) pregrown on SC+3% glycerol, 40 min after the addition of 2% galactose relative to that at 0 min (y-axis), versus a PGAL10-RAS2G19V strain (Y2866) pregrown on SC+3% glycerol, 40 min after galactose addition relative to 0 min (x-axis). Pink dots: cytoplasmic ribosomal protein genes; cyan dots: ribosomal biogenesis genes. Black line is the linear regression for the entire set of genes (slope=0.57) and the pink line passes through the origin and the centroid of ribosomal protein genes (slope=3.14). (B) Same as in (A) except at 60 min post-induction for both strains. (C) Both x- and y-axes show expression changes in sch9as (strain Y3561) grown on SC+3% glycerol 20 min following the addition of 2% glucose relative to 0 min. For the experiment in the y-axis, 100 nM 1NM-PP1 was added concurrently with glucose. Linear regression line (not shown) has a slope of 1.03 with an R2 value of 0.97. (D) Both x- and y-axes show expression changes in tpk1astpk2astpk3as sch9as (strain Y3508) grown on SC+3% glycerol 20 min following the addition of 2% glucose relative to 0 min. For the experiment in the vertical axis, 100 nM 1NM-PP1 was added concurrently with glucose. Green dots: genes whose expression is repressed more than 2 × by inactivation of Snf1; orange dots: genes induced more than 2 × by activation of Rgt2 (Figure 5).
As Sch9 activation recapitulates much of the glucose signaling, we asked how much of the glucose-induced changes in gene expression depended on Sch9. To do so, we inactivated Sch9, by adding 1NM-PP1 to a strain carrying a previously described analog-sensitive allele of SCH9 (Jorgensen et al, 2002), at the same time that we added glucose to the glycerol-grown culture. Loss of Sch9 activity did not in any way attenuate the glucose-induced transcriptional changes to the cell (Figure 2C). This suggests that very little of the glucose-induced changes are mediated by Sch9. Nonetheless, inactivating Sch9 concomitantly with inactivating PKA, using a strain carrying both sch9as and pkaas, completely eliminated the ability of the cell to mount a major transcriptional response to glucose addition (Figure 2D). From these experiments and those above, we conclude that Sch9 impinges on the same transcriptional circuit as does PKA, thus accounting for the ability of Sch9 at high levels to compensate for the loss of PKA activity, but that Sch9 provides only a minor conduit for glucose-induced transcriptional responses.
Interaction of the Gpr1/Gpa2 and the Sch9 signaling pathways
Several previous studies have suggested that the G-protein-coupled receptor-like protein Gpr1 participates in glucose signaling, through modulation of a heterotrimeric protein that includes the Gα protein Gpa2, in response to binding of various sugar ligands. We confirmed our previous results (Wang et al, 2004) showing that the induction of an activated allele of GPA2 weakly recapitulates the glucose-induced transcriptional changes and that these effects are mediated solely by PKA (Figure 3A). Moreover, we confirmed that GPR1 is not required for sensing or responding to glucose addition (Figure 3B).
Figure 3.
Interaction of Gpr1/Gpa2 and Sch9. Microarray expression data presented as in Figure 1. (A) Both x- and y-axes show expression changes in a PGAL10-GPA2Q300L tpk1astpk2astpk3as strain (Y3581) pregrown on SC+3% glycerol, 60 min following the addition of galactose relative to 0 min. For the experiment in the y-axis, 100 nM 1NM-PP1 was added concurrently with galactose. (B) Expression changes in a gpr1Δ strain (Y3573) pregrown on SC+3% glycerol, 20 min after the addition of 2% glucose relative to that at 0 min (y-axis), versus a GPR1 strain (Y2864) pregrown on SC+3% glycerol, 20 min after glucose addition relative to 0 min (x-axis). (C) Expression changes of all genes in a PGAL10-GPA2Q300L tpk1astpk2astpk3as strain (Y3581) pregrown on SC+3% glycerol, 60 min after the addition of 2% galactose relative to that at 0 min (y-axis), versus a PGAL10-GPA2Q300L tpk1astpk2astpk3as sch9as strain (Y3578) pregrown on SC+3% glycerol, 60 min after galactose addition relative to 0 min (x-axis). (D) Expression of genes in a gpr1Δ strain (Y3573) relative to wild type (Y2864) during exponential growth in SC+3% glycerol (y-axis) versus that in a sch9as strain (Y3507) relative to wild type (Y2864) under the same conditions. Genes previously determined to be induced by treatment with ketoconazole (Agarwal et al, 2003) are shown in pink and those encoding the pauperin family of cell wall proteins are shown in cyan.
However, further studies revealed unexpected interactions between Gpr1/Gpa2 and Sch9. First, we observed that gene expression changes upon activation of GPA2 exhibited a significantly greater dynamic range in an sch9as background than in a wild-type background (Figure 3C). In particular, several hundred genes are repressed by the activation of GPA2 at least four-fold greater in an sch9as strain than in the isogenic SCH9 strain. These genes are substantially enriched for those required for alcohol metabolism and gluconeogenesis (P<10−12), genes that are normally repressed by glucose. sch9as is a hypomorphic allele, which would suggest that Sch9 normally attenuates the transcriptional response elicited by Gpa2. The increased dynamic range in the transcriptional response to GPA2 induction in our sch9as strain could be due either to a more robust response following Gpa2 activation or to a difference in the initial transcriptional state of sch9as versus wild-type cells. To examine this, we assessed the transcriptional pattern of our PGAL10-GPA2 sch9as strain growing on glycerol, the initial state in the GPA2 induction experiment, relative to that of the isogenic SCH9 strain. Except as noted below, most genes showed the same level of expression during growth on glycerol in the sch9as strain relative to an isogenic wild-type strain (data not shown). Accordingly, we conclude that attenuation of Sch9 activity augments the ability of cells to respond to activation by Gpa2.
Our studies revealed a second connection between Sch9 and Gpr1/Gpa2. Although most genes were expressed normally during growth on glycerol in sch9as strains, approximately 90 genes were induced by more than two-fold in an sch9as strain relative to an isogenic SCH9 strain (Supplementary Table S1). Remarkably, gpr1Δ strains overexpressed essentially the same set of genes during growth on glycerol (Figure 3D; P=0). We observed no differences in expression between wild-type and sch9as or gpr1 strains during growth on glucose. Thus, loss of Gpr1 or reduction in Sch9 activity only affects steady-state gene expression during growth on a non-fermentable carbon source, a condition under which at least Gpr1 is thought to be inactive. The genes overexpressed under these conditions included members of the ergosterol biosynthesis pathway as well as the TIP1 gene family (DAN and TIR genes) and all but one member of a family of 22 genes encoding serine-poor proteins (PAU genes). The TIP1 and PAU gene families are involved in cell wall biogenesis or modification and are induced both by chronic anaerobiasis and by sterol deprivation (Rachidi et al, 2000; Agarwal et al, 2003; Lai et al, 2005). Induction of these genes is likely not in response to anaerobiasis, as classic anaerobic response genes, such as ANB1 and COX5b, are not induced in gpr1Δ or sch9as strains. Rather, the induced set of genes strongly match those induced by ketoconazole treatment (P<10−245), which induces ERG and TIP genes, through the activation of the Upc2 and Ecm22 transcription factors as a consequence of sterol deprivation (Agarwal et al, 2003; Davies and Rine, 2006). Thus, we conclude that deletion of GPR1 or attenuation of Sch9 activity induces the same small subset of genes during growth on a non-fermentable carbon source, in part by the activation of Upc2/Ecm22 either directly, or indirectly through depletion of sterols.
Snf1 regulates only a limited set of glucose-repressed genes
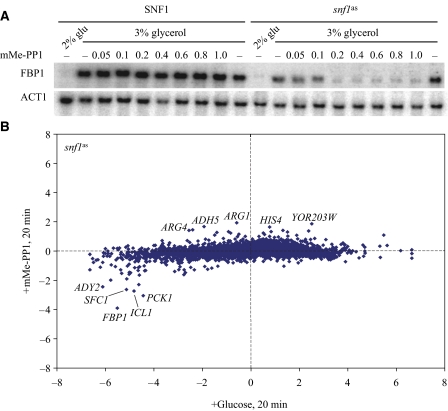
Snf1, the yeast homolog of mammalian AMP kinase, activates a transcriptional response during growth in low glucose or in alternative carbon sources such as sucrose and glycerol but is less active during growth in high glucose concentration. Accordingly, we assessed the contribution of Snf1 to the glucose transcriptional response by constructing a strain in which we substituted an analog-sensitive mutant of Snf1 (snf1as) for the wild-type gene. Addition of the ATP analog mMe-PP1 to this strain mimics the effect of glucose addition, as witnessed by suppression in the expression of the Snf1-regulated gene, FBP1, following addition of the analog (Figure 4A). Thus, to identify the genes that are regulated by glucose in an Snf1-dependent manner, we compared the extent of transcriptional changes that occurred after inhibition of Snf1as activity to the changes that occurred in the same strain after the addition of glucose. Inhibition of Snf1as activity altered the expression of only a few genes: only 66 genes showed a change of greater than two-fold, 47 repressed and 19 induced (Figure 4B; Supplementary Table S1). The repressed genes were in general not as affected by inhibition of Snf1as as they were by the addition of glucose. This was true using a 2.5-fold higher concentration of mMe-PP1 or a 25-fold higher concentration of 23DM-PP1, another inhibitor of Snf1as (data not shown). This suggests either that the inhibitors do not completely inhibit Snf1as or that other signaling systems cooperate with Snf1 to repress target genes. In this context, 27 of the 47 genes repressed upon Snf1 inactivation were also repressed more than four-fold by Ras activation (see Figure 1A). From these data, we conclude that the Snf1 pathway mediates only a small portion of the glucose signal in yeast and does so in cooperation with other signaling pathways.
Figure 4.
Snf1 controls a small set of glucose-regulated genes. (A) Strains Y2864 (SNF1) and Y3504 (snf1as) were grown to a density of A600=0.25 in SC+3% glycerol, at which point glucose was added to 2% or mMe-PP1 was added to the indicated concentration (μM). The northern blot of RNA samples from cells harvested 1 h after drug treatment, probed for FBP1 and ACT1 is shown. (B) Microarray data presented as in Figure 1 for strain Y3504 (snf1as) pregrown in SC+3% glycerol 20 min after the addition of 0.4 μM mMe-PP1 relative to 0 min (y-axis) or 20 min after the addition of glucose to 2% relative to 0 min (x-axis).
Genes repressed by inactivation of Snf1as cells were substantially enriched in those involved in carboxylic acid metabolism (P=1.6 × 10−20) and fatty acid β-oxidation (P=10−8) (Supplementary Figure S1). Cat8 and Adr1 are transcription activators, the activity of which depends on phosphorylation by Snf1. In total, 16 of 28 genes bound by Cat8 in vivo during growth on a non-fermentable carbon source and 11 of 32 genes bound by Adr1 in vivo under these conditions (Tachibana et al, 2005) were repressed upon inactivation of Snf1 (Supplementary Figure S1) (P=0 for both associations by Gene Set Enrichment Analysis (www.broad.mit.edu/gsea/)). In all, our study showed that 25 of 47 genes repressed by inactivation of Snf1 were bound in vivo by Adr1 or Cat8. This was in contrast to genes showing differential expression in steady-state cultures of SNF1 versus snf1Δ, where only 10% of the genes were found to be bound by Adr1 or Cat8. However, the genes induced by Snf1 inactivation are not normally induced by glucose, whereas the genes repressed by inactivation of Snf1 are normally repressed by glucose and a substantial fraction of these are repressed by glucose even in the absence of PKA and Sch9 function (P=0; Figures 1 and 2). Thus, although Snf1 regulates a limited number of genes, it mediates a significant branch of the glucose repression mechanism not subject to PKA regulation.
Rgt1 regulates a limited set of target genes for glucose induction
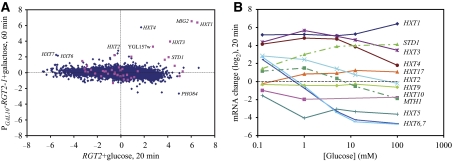
Glucose binding to either of two membrane sensors, Rgt2 and Snf3, results in proteolysis of two corepressors, Mth1 and Std1, that function with Rgt1 to repress a number of genes, many of which encode hexose transporters. To assess the extent to which this network contributes to the overall transcriptional response of cells to glucose, we placed RGT2-1, a hyperactive allele (Ozcan et al, 1998), under control of the GAL10 promoter and examined the transcriptional effect of induction of this allele in cells growing on glycerol. A small number of genes—predominantly those encoding hexose transporters—are induced upon activation of RGT2-1. Our results are consistent with those from a previous study providing a comprehensive identification of the targets of Rgt1 (Kaniak et al, 2004). Moreover, a significant proportion of the genes induced two-fold or more by RGT2-1 overexpression are induced normally by glucose in the absence of Sch9 and PKA activity (P=10−54) (Figure 5A). Thus, the Rgt1 pathway appears to work in parallel with PKA to regulate a subset of glucose-inducible genes.
Figure 5.
The Rgt network regulates a small number of glucose-induced genes. (A) Microarray data presented as in Figure 1 for a PGAL10-RGT2-1 strain pregrown in SC+3% glycerol 60 min after galactose addition relative to 0 min (y-axis) versus a RGT2 strain 20 min after glucose addition relative to 0 min. Genes that showed essentially normal induction by glucose in a tpk1astpk2astpk3as sch9as strain in the presence of 1NM-PP1 (Figure 2) are shown in pink. (B) Expression changes (log(2)) obtained from microarray experiments of the indicated HXT genes as well as STD1 and MTH1 as a function of concentration of glucose 20 min after its addition to wild-type strain Y2864 grown to mid log in SC+3% glycerol.
As several of the genes induced by overexpression of RGT2-1 were repressed by 2% glucose, we examined the expression of the hexose transporter genes by microarray analysis, as a function of the concentration of glucose added to the glycerol-grown culture. All hexose transporter genes induced by RGT2-1 show increased mRNA levels at 20 min following the addition of 0.125 mM glucose (Figure 5B). However, consistent with previous observations (Ozcan and Johnston, 1995; Boles and Hollenberg, 1997), higher glucose concentrations repress HXT2 and HXT6,7, which encode high-affinity glucose transporters (Reifenberger et al, 1997). Examining all our microarray data, we observe distinct, complex patterns of expression of all the HXT genes (Supplementary Table S2). For instance, HXT5 was induced only following transfer from glucose to glycerol medium, HXT9 is essentially unregulated by glucose and HXT17 responds to galactose addition. Finally, as previously noted, the Rgt1 co-repressor genes STD1 and MTH1 show opposite expression responses to increasing glucose levels, consistent with proposed distinction in their roles in induction versus maintenance of expression of the hexose transporter genes (Kim et al, 2006). Thus, we conclude that the Rgt pathway provides a glucose response pathway that is sensitive to lower concentrations of glucose than is the PKA pathway.
Specific 5′ and 3′ motifs mediate glucose-induced transcriptional responses
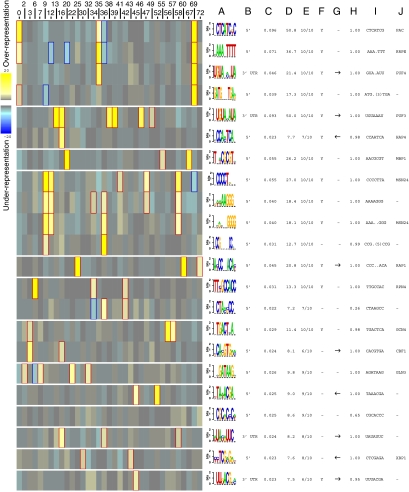
To identify putative regulatory motifs mediating glucose regulation of transcription in yeast, we used Iclust to cluster all genes based on the data resulting from our 200 expression experiments and then applied FIRE software to extract motifs enriched in the 5′ and 3′ regions of those genes that exhibit a similar transcriptional pattern. The Iclust algorithm extracts clusters of genes such that the expression profiles of genes within the same cluster are highly informative about one another, or in other words, have maximal dependency (Slonim et al, 2005; Elemento et al, 2007). Given the gene clusters, FIRE automatically discovers motifs, or putative regulatory elements, that are highly enriched or depleted across various clusters (Slonim et al, 2005; Elemento et al, 2007).
With default parameters, Iclust returned 75 clusters with at least 10 genes per cluster (Supplementary Table S4), half of which were enriched for specific functional groups of genes (Supplementary Table S5). Using FIRE to extract motifs enriched in individual clusters of co-regulated genes revealed a number of 5′ motifs previously associated with glucose regulation (Wang et al, 2004), including the RRPE and PAC motifs and binding sites for Hap4, Rap1, Mbp1, Cbf1, Msn2 and Msn4 (Figure 6). These observations highlight a role for PKA and Sch9 in regulating these transcription factors as well as the recently identified repressors Stb3, Dot6 and Ybl057c that act through RRPE and PAC (Liko et al, 2007). We also identified the sterol-regulatory element (SRE), binding site for Upc2 and Ecm22, associated with the cluster of genes, the expression of which showed increased expression in gpr1 or sch9 strains grown on glycerol. FIRE also returned the motif CCG(N)5CCG, but not the canonical Hap1-binding site CCG(N)6CCG, in a cluster of genes repressed by glucose in a PKA/Sch9-independent manner. This may suggest the involvement of a novel transcription factor in glucose signaling or that the Hap1-binding site may be more plastic with regard to spacing than previously reported (Ha et al, 1996). Finally, FIRE returned motifs not currently associated with known transcription factors, such as the GATCN3TGA motif enriched in a cluster containing Ribi genes that also carry RRPE/PAC sites. The identity of the corresponding factor and its role in glucose regulation remain to be determined.
Figure 6.
Identification of motifs associated with glucose-regulated genes. FIRE (finding informative regulatory elements) (Elemento et al, 2007) analysis of clustered data from Supplementary Figure S2 provided a heat map of the motifs (in rows) and the clusters in which they occur (in columns, numbers defined in Supplementary Tables S4 and S5). Only those clusters from which motifs were extracted are shown. Each identified motif is associated with (A) a predicted motif, (B) location of motif, either 5′ promoter region or 3′UTR, (C) mutual information (MI) score of the motif over the cluster indices (in bits), (D) z-score that measures the significance of the MI value as calculated using randomization tests, (E) robustness value calculated by doing 10 jack-knife trials after removing 1/3 of the data to test whether the motif repeatedly comes up as statistically significant, (F) position bias indicating whether the motif is concentrated at a specific location with respect to the translation start or stop site, (G) orientation bias, (H) conservation index as determined by comparison to Saccharomyces bayanus, (I) seed sequence and (J) motif name. The intensity of the yellow color in the heat map represents the degree to which the motif is over-represented in the specified cluster, whereas the intensity of the blue color represents the degree to which the motif is under-represented in the specified cluster. The genes in each cluster are listed in Supplementary Table S4 and statistically significant enrichments of functional categories in the different clusters are listed in Supplementary Table S5.
In addition to motifs in the promoter region of co-regulated genes, FIRE also highlighted a number of motifs in the 3′ untranslated regions (UTRs) of different groups of co-regulated genes. Several of these are variants of previously identified binding sites for various Puf proteins involved in regulating subcellular distribution, translation and decay of mRNAs from specific subsets of genes (Olivas and Parker, 2000; Gerber et al, 2004). FIRE analysis revealed a motif that matches the Puf3-binding site enriched in several different clusters, all of which were over-represented by genes with mitochondrial-related functions. However, these clusters exhibited different regulation by glucose. Thus, although Puf3 may affect the stability of a number of mRNAs, it does not appear to mediate glucose regulation of the levels of these mRNAs in a uniform manner. FIRE analysis also identified the motif UGUAN3UA, which likely corresponds to the binding site for Puf4 (Grigull et al, 2004; Foat et al, 2005), in 3′ UTRs of genes in clusters enriched for those involved in ribosome biogenesis. To assess whether this motif participated in glucose regulation of mRNA levels, we replaced the 3′ UTR of one such gene, UTP5, with that from ADH1. The resulting construct, UTP5-3′UTRADH1 was expressed two-fold lower in cells grown on glycerol than did the normal UTP5 gene but showed the same induction by glucose as did UTP5 itself (data not shown). Similarly, the motif UAUAUUC, related to the PRSE motif (Foat et al, 2005), was enriched in genes associated with carboxylic acid metabolism and oxidative phosphorylation, which are repressed upon glucose addition. Replacement of the 3′ UTR of two such genes, ICL1 and CAT8, with the 3′ UTR of ADH1 as above resulted in reduced expression of the two genes during growth on glycerol but did not affect the extent of repression observed upon glucose addition. From these studies, we conclude that specific RNA-binding motifs are associated with specific subsets of genes both upregulated and downregulated by glucose addition but these motifs, and their associated RNA-binding proteins, do not participate in this glucose regulation.
Growth-responsive genes are regulated by nutrient signaling, not nutrient metabolism
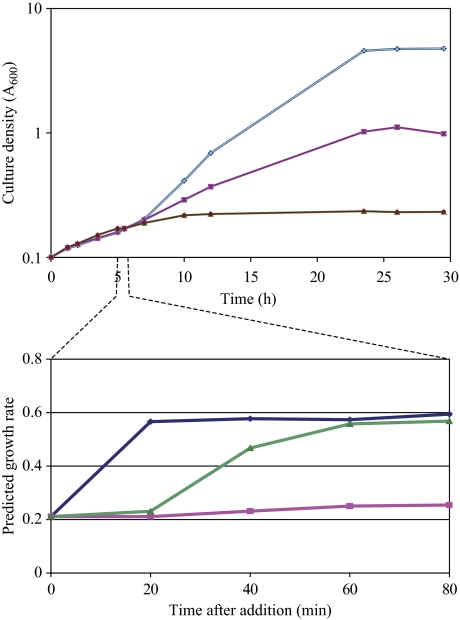
As noted in the introduction, nutrient availability establishes both the growth rate of the cell and a corresponding highly stereotypic transcriptional pattern. To address the causality of this correlation, we examined the growth rate of cells under conditions in which we could artificially induce a high growth rate transcriptional pattern in the absence of added nutrient. In particular, as noted above, induction of RAS2G19V in glycerol-grown cells results in a transcriptional pattern highly reminiscent of glucose addition. Applying the algorithm developed by Brauer et al (2008), a culture of the PGAL10-RAS2G19V strain growing on glycerol exhibits a growth rate signature of 0.2, which changes to 0.6 within 20 min of addition of glucose, consistent with the change in doubling time from 5.8 to 2.6 h (Figure 7). Similarly, the transcriptional pattern observed 60 min following the induction of RAS2G19V matched the growth rate signature of the glucose-growing culture. However, in contrast to the glucose addition, RAS2G19V induction did not yield an increase in growth but an immediate decrease in growth rate followed by complete cessation of growth within 4 h. From these observations, we conclude that the cell sets its growth-specific transcriptional pattern not on the basis of the energy or metabolites produced from available nutrients but rather on the basis of its perception of its nutritional environment. Moreover, a mismatch between the cell's actual nutritional state and its perception of its nutritional environment can result in growth attenuation and cell death (Fedor-Chaiken et al, 1990).
Figure 7.
Growth versus growth rate prediction. Upper panel: growth of strain Y2866 (gal1 PGAL10-RAS2G19V) at 30°C on SC+3% glycerol with no additions (pink squares), addition of glucose to 2% at 5 h (blue diamonds) or addition of galactose to 2% at 5 h (green triangles). Lower panels: samples were taken at 20-min intervals from the time of additions from the cultures depicted in the upper panel, RNA was extracted and analyzed by microarray hybridization. The predicted growth rate based on the gene expression pattern in each culture using the algorithm described in Brauer et al (2008) is plotted as a function of time post-addition.
Discussion
The glucose signaling network
As described in this study, we used global transcriptional analysis in combination with genetics, particularly relying on conditional alleles, to obtain a nearly complete view of glucose signaling in yeast. We can account for the vast majority of glucose-mediated transcriptional responses through the contributions of five distinct but interlocking pathways (Figure 8). PKA activation is both sufficient, as we showed previously, and necessary, as we show here, to induce the vast majority of transcriptional changes in response to glucose. Activation of PKA recapitulates both the magnitude and direction of transcriptional response of >90% of all genes regulated by glucose; inactivation of PKA concurrent with glucose addition eliminates most of the response of these genes.
Figure 8.
The glucose signaling network. Diagram of the regulatory wiring connecting the addition of glucose to the transcriptional responses of the cell. Dotted line indicates a limited or indirect connection. See text for details.
Several pathways account for PKA-independent regulation by glucose. Snf1 regulates a significant fraction of approximately 100 genes that show essentially normal glucose repression in the absence of PKA function and an overlapping set of approximately 160 glucose-repressed genes that are not fully repressed by activation of PKA. A significant fraction of the genes repressed independently of PKA are enriched in those previously identified as targets of the heme-activated transcriptional regulators Hap1 and the Hap2/3/4/5 complex. Previous work has shown that overexpression of Hap4 elicits activation of a number of glucose-repressible genes (Lascaris et al, 2003). Moreover, we observed that glucose represses the expression of HAP4 and does so independently of both PKA and Snf1. Thus, this pathway may be a direct regulatory circuit rather than an indirect consequence of heme metabolism. Finally, a few genes are induced by glucose in the absence of PKA function and a subset of those is not fully induced by PKA activation in the absence of glucose. These are almost completely accounted for by regulation through the Rgt1 pathway. In sum, PKA, Snf1, Rgt1 and heme-dependent transcriptional activators account for essentially all of glucose signaling of transcription in yeast (Figure 8).
Even for those genes whose glucose regulation is not mediated solely by PKA, PKA often influences the expression in conjunction with other signaling pathways. For instance, a significant fraction of Snf1-regulated genes also shows robust repression upon PKA activation. Similarly, genes subject to Rgt1 regulation also both respond to PKA activation and require PKA activity for full activation by glucose. These overlapping responses provide confirmation and define the extent of the transcriptional consequences of the interconnection between PKA signaling and both Snf1 and Rgt signaling, whose mechanistic underpinnings have been previously suggested (Kaniak et al, 2004; Palomino et al, 2006).
cis-Regulatory elements in glucose signaling
Assessing the mutual information between gene sequence and the gene clusters extracted from our expression measurements allowed us to identify a number of motifs that participate in glucose regulation. The observation that many of the 5′ motifs identified by FIRE analysis correspond to transcription factor-binding sites previously recognized as mediating regulation of glucose-responsive genes underscores the reliability of this motif search algorithm. Some of these factors likely couple changes in growth capacity with the change in nutrient quality, such as those involved in ribosome biogenesis (RRPE, PAC and Rap1), increased biosynthetic capacity (Gln3, Gcn4 and Cbf3) and cell cycle progression (Mbp1), whereas several other factors regulate stress response (Msn2/4, Rpn4 and Xbp1) or carboxylic acid metabolism (Hap4 and the Hap1-like sequence CCG(N)5CCG). Several other motifs identified using FIRE highlight novel targets of the glucose signaling network, such as sterol metabolism regulated by Ecm22/Upc1, and some that are not readily assignable to a known transcription factor. In most cases, the mechanism by which the transcription factor activity responds to upstream signaling pathways has not been clearly defined, although for reasons noted below, the factors likely respond directly to the signaling network, rather than to metabolic changes in the cell. Determining the mechanistic bases for these connections offers a rich area for further investigation.
FIRE also identified a number of previously recognized motifs within the 3′ UTRs that were associated with genes exhibiting characteristic expression patterns. These included Puf3- and Puf4-binding sites as well as the PRSE motif (Foat et al, 2005). Our analysis of these motifs suggests that, although these motifs contribute to the translation, stability and subcellular localization of mRNAs, they do not appear to mediate the immediate regulatory response of the cell to the addition of glucose.
Sch9 plays a limited role in glucose signaling
Previous genetic observations and our current results have suggested that Sch9 provides a parallel pathway to PKA for glucose-mediated transcriptional changes. Sch9 was originally isolated as a high-copy suppressor of loss of PKA signaling. Moreover, we find that overexpression of Sch9 elicits essentially the same transcriptional response as does activation of PKA signaling. However, our results suggest that although Sch9 and PKA regulate a common set of genes, they do so through different signaling pathways. First, although induction of Sch9 in glycerol-grown cells elicits a similar transcriptional response as PKA activation, loss of Sch9 activity does not detectably diminish glucose regulation of those genes. This suggests that Sch9 does not contribute significantly to glucose signaling as does PKA. Moreover, although the final transcriptional pattern achieved upon Sch9 induction matches that obtained by PKA activation, the kinetics of regulation are quite distinct. In particular, ribosomal protein genes are induced more rapidly by Sch9 activation than by PKA activation, whereas all other genes are induced or repressed more slowly. Thus, Sch9 and PKA impinge on the transcriptional machinery through distinct mechanisms. Both pathways regulate Rim15 to modulate quiescence but through distinct mechanisms (Swinnen et al, 2006) and both pathways control the nuclear localization of the Sfp1 protein (Jorgensen et al, 2004), which modulates the expression of Ribi and RP genes. Both of these pathways also influence the activity of Ribi gene repressors, Stb3, Dot6 and Ybl054w, which act through RRPE and PAC elements (Liko et al, 2007). Sch9 and PKA affect these repressors in different ways, which likely accounts for the different kinetics of induction we observed for these genes upon Sch9 versus PKA activation (Lippman and Broach, unpublished observations).
Our results are consistent with the model that core growth-related genes—those encoding ribosomal proteins, ribosomal biogenesis proteins and core metabolic enzymes—and stress-related genes are regulated independently in response to the two primary nutritional inputs. As shown here, the quality and availability of carbon source impinge on the expression of these genes through the PKA pathway. As shown previously, Sch9 activity is directly dependent on phosphorylation by TORC1, which in turn responds to nitrogen availability by an undefined mechanism (Urban et al, 2007). Thus, the quality and amount of nitrogen source impinge on the expression of growth- and stress-related genes through Sch9. The site at which and the means by which these two pathways are integrated remain to be determined.
Our results establish unexpected connections between Sch9 and Gpr1/Gpa2. First, diminished Sch9 activity or deletion of GPR1 induces a common set of sterol and cell wall biosynthesis genes during growth on a non-fermentable carbon source. We do not fully understand the mechanism of this regulation. We note, though, that rapamycin treatment induces the expression of at least a subset of these genes, indicating that TORC1 plays a role in maintaining repression of these genes. The second connection between Sch9 and Gpr1/Gpa2 is our observation that attenuation of Sch9 activity enhanced the transcriptional repression response attendant on Gpa2 activation. As attenuation of Sch9 activity had no effect on the basal activity of those genes subject to enhanced repression, we conclude that Sch9 normally mitigates the signaling activity of Gpa2. Thus, these results suggest potential cross-talk between the TOR and Gpa2/PKA pathways by which diminished TOR signaling enhances the signaling response through the PKA pathway (Figure 8). We have not pinpointed the precise mechanistic basis of this cross-talk. However, this cross-talk may account for the enhanced transcriptional response of gpa2 cells during nitrogen source downshift conditions that elicit filamentous growth (Medintz et al, 2007). Thus, this cross-talk may play an important role in the developmental program yeast pursues under limiting nutrient conditions.
Transcription, nutrition and growth
Cells adapt to reduced nutritional availability, in part by altering their transcriptional profile. A subset of the nutrient-regulated genes changes expression in response to nutritional status strictly dependent on growth rate but independent of the particular nutrient that limits the growth of the cells. The expression levels of some of the genes—such as ribosome biogenesis genes—are directly proportional to the growth rate and those of others are inversely proportional. The levels of expression of members of this collection of genes provide a ‘growth rate signature' that is highly predictive of the growth rate of the cells from which the sample was taken (Brauer et al, 2008). Thus, nutrient availability establishes the growth rate of the cell as well as a corresponding highly stereotypic transcriptional pattern. However, causality in this correlation is unclear: does metabolism of the available nutrients determine growth rate, which in turn establishes the transcriptional pattern, or does the cell's perception of the available nutrients set the transcriptional pattern, which in turn allows metabolism of those nutrients to achieve a certain growth rate?
Our results provide an unequivocal answer to that question: the growth rate-specific transcriptional pattern is determined by what the cell perceives as its nutritional environment rather than the actual availability of nutrients or their metabolic products. Slattery et al (2008) recently reached a similar conclusion using a different genetic approach. From this observation, we can conclude that the cell sets its cellular growth and metabolism machinery to handle, and depend on, the nutrients it recognizes as being present. Normally, this dependence causes no problems, as what the cell perceives to be present is usually present. However, as a result of drug treatment or genetic manipulation, discordance in the perception and the reality has lethal consequences, either when the cell perceives a rich nutritional environment that is not present or perceives a poor environment when in fact a rich one exists. Consistent with this interpretation, strains lacking PKA activity fail to grow but can be rescued by eliminating the Msn2/Msn4 transcription factors (Smith et al, 1998), which normally elicit a major portion of the ‘poor growth'-specific gene transcriptional pattern. Thus, cells rely on their perception of the nutritional state to set the appropriate growth rate transcriptional pattern and doing so when perception does not match reality is lethal, unless the cell can ignore the signal. This mechanism may extend to other cells, perhaps accounting for the sensitivity to growth-inhibiting chemotherapeutic agents of mammalian cells containing activated oncogenes, such as K-ras mutation.
Materials and methods
Strains
The strains used in this study were derived from W303-1B and are listed in Supplementary Table S6. We created an analog-sensitive allele of SNF1 by mutating the wild-type gene in plasmid B2716 to generate the snf1I132G allele (plasmid B2717) using the QuikChange® XL Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). We digested plasmid B2717 with BamHI and XhoI and integrated into the SNF1 locus of strain Y2896 to obtain strain Y3505. We crossed Y3505 with Y2865, sporulated the diploid and obtained Y3504. We PCR amplified the SNF1 locus from strain Y3504 and sequenced it to confirm the presence of the I132G allele. pMT3366, a URA3 integrating plasmid carrying sch9as, and yeast strain yMT2422 (sch9∷KAN-PGAL1-SCH9) (Jorgensen et al, 2004) were kindly provided by Mike Tyers. We PCR amplified sch9∷KAN- PGAL1-SCH9 from yMT2422 and integrated it into Y3554. We sporulated the resulting strain to obtain strain Y3506. We sequenced the PGAL1-SCH9 region from strain Y3506 and confirmed it contained no mutations in the SCH9 ORF but found a single-point mutation (A to G) at position −313 in the GAL1 promoter region. To introduce sch9as into W303-1B strain background, we digested pMT3366 with AflII, transformed it into Y3554 to generate a circular integration of the plasmid at the SCH9 locus, and then transformed the resulting strain with the SCH9 LEU2 CEN ARS plasmid B2718. The resulting strain was sporulated to obtain a Ura+ Leu+ segregant, from which URA3 and the wild-type SCH9 locus were excised by selecting on FOA. After segregating off plasmid B2718 to obtain strain Y3562, we PCR amplified the SCH9 locus and sequenced it to confirm the presence of the sch9as allele. We crossed Y3562 to Y3565 (MATα sch9∷KAN-PGAL1-SCH9) and sporulated the diploid to obtain strain Y3507. Construction of PGAL10-RAS2G19V (Y2866) has been described by Fedor-Chaiken et al (1990). We constructed strain Y3168 (PGAL10-RAS2S24N) by digesting plasmid B2365 carrying PGAL10-RAS2S24N with SnaBI and integrating into Y2864. We generated a tpk1M164G allele, a tpk2M147G allele and a tpk3M165G allele by mutating the corresponding cloned wild-type genes in an integrating URA3 plasmid using the QuikChange XL Site-Directed Mutagenesis Kit from Stratagene. These were used to replace separately the corresponding wild-type allele in W303 by a two-step replacement protocol. The remaining two TPK genes were deleted from each strain to generate Y3383 (tpk1M164G tpk2∷kanMX tpk3∷TRP1), Y3384 (tpk1∷HIS3 tpk2M147G tpk3∷TRP1) and Y3386 (tpk1∷HIS3 tpk2∷kanMX tpk3M165G [TPK2-LEU2]). Strains Y3383 and Y3384 grew normally in synthetic complete (SC) medium but were completely inhibited by the addition of 0.15 μM 1NM-PP1. Leu− segregants of strain Y3386 grew very slowly, consistent with the relatively low level of Tpk3 in the cell. Strain Y3561 was obtained as a segregant from crosses among the above three strains. Gene deletions were created by amplification of approximately 500 bp upstream and downstream of the respective deletion mutant from Yeast Consortium Deletion Strain collection (Invitrogen, Carlsbad, CA) and integrating into the Y2864 background. All other strains were created using standard yeast genetic techniques.
For functional analysis of 3′UTR motifs in genes UTP5, ICL1, CAT8 and PAU13, GFP-TADH1-TRP1 was PCR amplified from MR5091 and fused to UTP5 (Y3591), ICL1 (Y3592), CAT8 (Y3593) and PAU13 (Y3588) at their genomic loci with replacement of approximately 200–300 bp of the 3′UTR region.
Cell growth
Cells pregrown in SC medium supplemented with 3% glycerol as the only carbon source were diluted into fresh medium and grown at 30°C to an OD600 of 0.25, at which time 20 ml of cells was collected representing the 0 min time point. The remaining culture was then induced with the treatment specific to the experiment and 20-ml cells were collected at various times after initiation of the experiment, generally at 20, 40, 60 and 80 min. snf1as cells were treated with 0.4 or 1 μM of 4-methylnapthyl-1-tert-butyl-3-phenylpyrazolo[3,4-d] pyrimidine (mMe). sch9as, tpk1astpk2astpk3as and sch9astpk1astpk2astpk3as cells were treated with 100 nM of C3-1′-naphthyl-methyl PP1 (1NM-PP1). PGAL10-RAS2G19V, PGAL10-RAS2S24N, sch9∷KAN-PGAL1-SCH9 and PGAL10-GPA2Q300L strains were induced with 2% galactose. Glucose was added to a final concentration of 2% to the control experiments. Cells were collected by filtration and then frozen in liquid nitrogen.
mRNA analysis
Total RNA was extracted using the Qiagen RNeasy® Mini Kit (Valencia, CA). For northern blot analysis, samples were fractionated on a 0.8% agarose gel (0.1 M borate buffer (pH 8.3), 7.5% formaldehyde) and transferred to Nytran SuperCharge membrane (Schleicher and Schuell BioScience Inc., Keene, NH). The membrane was crosslinked at 80°C for 2 h and probed with PCR generated, agarose gel purified, 32P-labeled DNA (Random Primers DNA Labeling System; Invitrogen). The intensities of the signal were visualized using a STORM 860 Imaging System (Molecular Dynamics, Sunnyvale, CA).
For microarray hybridization, RNA samples were treated with an additional DNase I purification step. cRNA was synthesized using the standard protocol of the Agilent Low RNA Input Linear Amplification Kit (Agilent Technologies, Palo Alto, CA). Briefly, 100 ng of total RNA was used as a template for first and second strand cDNA synthesis with reverse transcriptase using a primer containing poly dT and T7 polymerase promoter. Labeled cRNA was synthesized from cDNA using T7 RNA polymerase and cyanine3- (Cy3-) or Cy5-labeled CTP (PerkinElmer Life and Analytical Sciences, Boston, MA). The amount of cRNA synthesized and incorporation of Cy3- and Cy5-CTP into cRNA were measured using a NanoDrop (NanoDrop Technologies, Wilmington, DE). Equal amounts of Cy3- and Cy5-labeled cRNA were combined, mixed with the control target and fragmented for 30 min. Each sample was then hybridized to an Agilent yeast oligo microarray for 17 h at 60°C. The arrays were washed and scanned using Agilent Microarray Scanner (Agilent Technologies) at 100% PMT for red and green channels and at 10 μm resolution. The feature information was extracted from the microarrays using Agilent Feature Extraction Software with the default settings. The data were stored and analyzed on the Princeton University Microarray database. Dye normalization for each array was determined by the rank consistency method and then spot intensities were calculated by the LOWESS method. Spots were retained for further analysis only if both the Cy3 and Cy5 channels were greater than 2.6σ of mean background intensity and were uniform in intensity. Only those genes for which 80% of the arrays yielded good data were retained for analysis. R2 values between experimental duplicate were greater than 0.99. All data described in this study can be viewed and downloaded from the PUMAdb website (http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=524).
Computational methods
Expression data were clustered using the Iclust algorithm with default parameters (Slonim et al, 2005). The obtained gene clusters were used as input to the FIRE software with its default configuration to predict putative regulatory elements, or motifs, with a presence/absence pattern across 5′ and 3′ promoter sequences that is relatively informative about the associated gene cluster indices (Elemento et al, 2007).
Supplementary Material
Supplementary Tables and Figures
Data Set 1 - Supplementary Table 1b
Data Set 2 - Supplementary Table S4
Acknowledgments
We thank Mike Tyers and Paul Jorgensen for providing plasmids and strains, Kevan Shokat for donating a generous supply of a number of PP1 derivatives, Curtis Huttenhower for growth rate gene signature analysis, Xiuying Zhang for excellent technical assistance, Olivier Elemento for expert assistance with FIRE and Iclust, and Donna Storton for assistance with microarray technology. This research was supported by an NIH grant GM076562 to JRB and a Center for Quantitative Biology/NIH grant P50 GM071508.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM (2003) Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem 278: 34998–35015 [DOI] [PubMed] [Google Scholar]
- Bishop AC, Buzko O, Shokat KM (2001) Magic bullets for protein kinases. Trends Cell Biol 11: 167–172 [DOI] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP (1997) The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 21: 85–111 [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D (2008) Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR, Deschenes RJ (1990) The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res 54: 79–139 [DOI] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DC, Cornell MJ, Petty J, Hakes L, Wardleworth L, Rash B, Brown M, Dunn WB, Broadhurst D, O'Donoghue K, Hester SS, Dunkley TP, Hart SR, Swainston N, Li P (2007) Growth control of the eukaryote cell: a systems biology study in yeast. J Biol 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BS, Rine J (2006) A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S (2007) A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor-Chaiken M, Deschenes RJ, Broach JR (1990) SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61: 329–340 [DOI] [PubMed] [Google Scholar]
- Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ (2005) Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci USA 102: 17675–17680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR (2004) Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha N, Hellauer K, Turcotte B (1996) Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding. Nucleic Acids Res 24: 1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M (1992) Carbon regulation in Saccharomyces. In Molecular and Cellular Biology of the Yeast Saccharomyces, Broach JR, Pringle JR, Jones EW (eds) Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400 [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniak A, Xue Z, Macool D, Kim JH, Johnston M (2004) Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell 3: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Brachet V, Moriya H, Johnston M (2006) Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell 5: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E, Mosch HU, Rupp S, Lisanti MP (1997) Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem 272: 20321–20323 [DOI] [PubMed] [Google Scholar]
- Lai LC, Kosorukoff AL, Burke PV, Kwast KE (2005) Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol Cell Biol 25: 4075–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris R, Bussemaker HJ, Boorsma A, Piper M, van der Spek H, Grivell L, Blom J (2003) Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol 4: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko D, Slattery MG, Heideman W (2007) Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J Biol Chem 282: 26623–26628 [DOI] [PubMed] [Google Scholar]
- Medintz IL, Vora GJ, Rahbar AM, Thach DC (2007) Transcript and proteomic analyses of wild-type and gpa2 mutant Saccharomyces cerevisiae strains suggest a role for glycolytic carbon source sensing in pseudohyphal differentiation. Mol Biosyst 3: 623–634 [DOI] [PubMed] [Google Scholar]
- Nath N, McCartney RR, Schmidt MC (2003) Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol 23: 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 19: 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M (1998) Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J 17: 2566–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M (1995) Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol 15: 1564–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino A, Herrero P, Moreno F (2006) Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res 34: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi N, Martinez MJ, Barre P, Blondin B (2000) Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol Microbiol 35: 1421–1430 [DOI] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther O, Fausboll A, Akesson M, Bro C, Hansen LK, Brunak S, Nielsen J (2006) Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol 7: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger E, Boles E, Ciriacy M (1997) Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem 245: 324–333 [DOI] [PubMed] [Google Scholar]
- Santangelo GM (2006) Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M (2000) Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol 20: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery MG, Liko D, Heideman W (2008) Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae. Eukaryot Cell 7: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim N, Atwal GS, Tkacik G, Bialek W (2005) Information-based clustering. Proc Natl Acad Sci USA 102: 18297–18302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S (1998) Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J 17: 3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J (2006) Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET (2005) Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol 25: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H, Miwa T, Shinozaki M, Saito M, Yun CW, Yamamoto K, Kumagai H (2000) GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 267: 164–168 [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Wigler M (1988) SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev 2: 517–527 [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Guldal CG, Zhang X, Tavazoie S, Broach JR (2004) Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol 2: E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG (1996) Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
- Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D (1994) Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem 269: 19509–19515 [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP (1998) GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J 17: 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Figures
Data Set 1 - Supplementary Table 1b
Data Set 2 - Supplementary Table S4