Abstract
Nutrients destined for the developing cereal grain encounter several restricting barriers on their path towards their final storage sites in the grain. In order to identify transporters and chelating agents that may be involved in transport and deposition of zinc in the barley grain, expression profiles have been generated of four different tissue types: the transfer cells, the aleurone layer, the endosperm, and the embryo. Cells from these tissues were isolated with the ‘laser capture microdissection’ technology and the extracted RNA was subjected to three rounds of T7-based amplification. The amplified RNA was subsequently hybridized to Affymetrix 22K Barley GeneChips. Due to the short average length of the amplified transcripts and the positioning of numerous probe sets at locations more than 400 base pairs (bp) from the poly(A)-tail, a normalization approach was used where the probe positions were taken into account. On the basis of the expression levels of a number of metal homeostasis genes, a working model is proposed for the translocation of zinc from the phloem to the storage sites in the developing grain.
Keywords: Affymetrix, gene expression profiling, laser capture microdissection, microarray, RNA amplification, zinc homeostasis
Introduction
Zinc is a trace mineral essential to all life forms due to its key role in gene expression, cell development and replication. Zn2+ is redox-stable under physiological conditions and due to its small radius, Zn2+ is an efficient electron acceptor and therefore forms strong covalent bonds with S, N, and O donors. Zinc is consequently able to bridge amino acid residues like cysteine (Cys), histidine (His), aspartic acid (Asp), and glutamic acid (Glu) (Broadley et al., 2007). Because of this ability, zinc ions play an important role as cofactor by defining the three-dimensional structure and function of many proteins. In humans, zinc is an essential cofactor for more than 300 enzymes that are involved in the synthesis and degradation of carbohydrates, lipids, proteins, and nucleic acids as well as in the homeostasis of other micronutrients. Zinc stabilizes the molecular structure of cellular components and membranes and, in this way, contributes to the maintenance of cell and organ integrity (Broadley et al., 2007). Furthermore, more than 2000 transcription factors appear to require zinc as a co-factor (Prasad and Kucuk, 2002). A bioinformatics analysis of fully sequenced genomes and the predicted protein sequences indicated that 925 proteins in humans and 536 proteins in Arabidopsis thaliana bind zinc (Gladyshev et al., 2004).
Zn deficiency is increasingly recognized as being of the utmost importance for human health and quality of life. The Copenhagen Consensus Conference 2008 thus ranks the alleviation of zinc deficiency as a top priority (Copenhagen Consensus 2008, www.copenhagenconcensus.com). Based on analysis of diet compositions and nutritional needs, it has been estimated that 49% of the world's population is at risk of suffering from zinc deficiency. The various symptoms of severe zinc deficiency comprise growth retardation, delayed bone maturation, skin lesions, diarrhoea, impaired appetite, and increased susceptibility to infections caused by defects in the immune system. The groups most at risk for being affected by zinc deficiency are infants, young children, and pregnant women, especially in developing countries (Welch and Graham, 2004; Gibson, 2006).
The primary reason for zinc deficiency in humans is poverty, resulting in a narrow food base that is almost exclusively based on starch-rich staples like rice, wheat, maize, potatoes, and cassava that all have a suboptimal zinc content. In cereals, there are the additional problems of the presence of inhibitors like phytic acid that impedes bioavailability and the fact that the zinc deposited in the husk and embryo (typically 50% of the total zinc in the grain) is removed during milling and polishing leaving behind an endosperm fraction with a low zinc content (Brinch-Pedersen et al., 2007).
Over the last decade the international alliance HarvestPlus has been the major driver for the concept of biofortification, i.e. through modern plant breeding to increase the content of iron, zinc, and provitamin A in the edible parts of staples. For iron and zinc, this has stimulated a very substantial scientific interest in understanding the uptake, mobilization, transport, and deposition of the two micronutrients. Plant genomes encode large families of metal transporters that differ in their substrate specificities, expression patterns, and cellular localization to control metal distribution throughout the plant. These include transporters that are members of the ZIP (zinc/iron regulated protein) gene family, the cation diffusion facilitator (CDF) family, the P1B ATPase family, the natural resistance associated macrophage protein (NRAMP) family, and the Yellow Stripe1-Like (YSL) family (Colangelo and Guerinot, 2006). In addition, low molecular weight proteins and non-proteinogenic amino acids playing a role in metal chelation and detoxification have been identified, including metallothioneins, nicotianamine and derivatives of nicotianamine, (Cobbett and Goldsbrough, 2002; Haydon and Cobbett, 2007b). However, although many genes are involved in the transport pathway from the root to the grain, substantial progress has been achieved in understanding the specificities and temporal and spatial expression of the proteins involved (Ghandilyan et al., 2006; Kramer et al., 2007). It should, therefore, eventually be possible to identify the overall regulation of the zinc traffic from uptake to deposition and potential bottlenecks in the transport pathway that may be targeted by marker assisted breeding, TILLING, or transformation.
The present study is a first attempt to describe a road map for zinc trafficking in the developing cereal grain, using barley as a model and gene expression profiling as an indication of zinc transport and sequestration. In the developing barley grain, nutrients for grain-filling are provided through a single strand of phloem. Nutrients like zinc thereafter move symplastically from the phloem through plasmodesmata in the vascular parenchyma and the pigment strand until they reach the so-called nucellar projection, the unloading region for nutrients. The cell walls of the pigment strand are composed of lignin, phenolics, and suberin and constitute an apoplastic barrier. This prevents movement of solutes within the apoplast and restricts the transfer of the incoming nutrients to the symplast (Wang et al., 1994a). The cells located on the border of the maternal tissue have differentiated into transfer cells, which are characterized by the formation of wall ingrowths. The surface area of the plasma membrane is thereby increased up to 22-fold (Wang et al., 1994a). Passing the transfer cells, the nutrients are translocated across the plasma membrane and into the apoplastic space, the endosperm cavity. From there they are taken up by transporters of modified aleurone cells and distributed in the symplast to cells of the aleurone layer, the endosperm, and the embryo (Wang et al., 1994b). The intercellular transport chain of zinc from the phloem to the endosperm/embryo accordingly consists of relatively few elements that primarily involve the symplastic movement of sequestered zinc from the phloem, transport into the apoplast, uptake from the endosperm cavity and a second phase of symplastic transport in the filial tissues, the endosperm, and the aleurone. However, in addition to this transport chain between different tissues there must, as in other cells, be a range of intracellular zinc transport steps that involves temporal or permanent deposition in vacuoles, plastids, mitochondria, the endoplasmic reticulum, and the nuclei. At all stages, Zn2+ is probably sequestered by organic compounds like nicotianamine, 2′-deoxymugineic acid (DMA), and metallothionein.
Developing barley grains can, to some extent, be dissected into crude fractions for experimental analyses. However, in the current project, laser capture microdissection has been used (Emmert-Buck et al., 1996) in order selectively, to isolate cell sections of the transfer region the aleurone, the endosperm, and the embryo. Thereafter, the gene expression profile of the individual tissues has been assessed by microarray analyses using the Affymetrix platform. Laser capture microdissection of plant tissues for gene expression profiling is a technology still in its infancy and has so far only been performed on epidermal cells and vascular tissues of coleoptiles of maize for hybridization to a cDNA microarray (Nakazono et al., 2003), gene expression during embryogenesis of Arabidopsis using the ATH1 GeneChip (Casson et al., 2005), transcript analysis of the developing Arabidopsis seed on cDNA microarrays (Day et al., 2007), and, recently, in studies of phloem-derived RNAs in Arabidopsis using the ATH1 GeneChip (Deeken et al., 2008). In the present study, we therefore also report on technology optimization and the necessary measures to facilitate gene expression profiling using the Affymetrix barley microarray platform.
Materials and methods
Plant material
Plants of the barley (Hordeum vulgare) cv. Golden Promise were grown in pots in a controlled environment chamber with 16/8 h light/darkness at 23 °C and 18 °C, respectively. Seeds were harvested from two different plants 20 days after pollination (dap). The seeds were then either snap frozen in liquid nitrogen or sectioned into three pieces and immediately soaked in Farmers fixative (3:1 v/v ethanol:acetic acid).
Tissue preparation for paraffin embedding
After submerging the sectioned seeds in Farmers fixative, the fixation vials were placed in a vacuum to promote the infiltration of the fixative. The seeds were then left in Farmers fixative overnight at 4 °C. The following day the tissue was dehydrated in a graded series of ethanol [1 hour each (v/v) 70%, 90%, 100%, 100%]. The tissue was then shifted to a graduated series of ethanol:Histochoise Clearing Agent (Sigma) [1 h in each solution (v/v) (75:25%; 50:50%; 25:75%; 0:100%, and 0:100%)]. Flakes of Paraplast-X-Tra tissue embedding medium (Fluka) were added in the final step. The vials were immersed in water at 60 °C in order to melt the Paraplast. Once the flakes had dissolved, liquefied Paraplast-X-Tra was added to the vials, and kept in a 60 °C water bath for 30 min. The Paraplast-X-Tra was changed four times. Paraplast blocks were then prepared with one seed fragment in each block, and allowed to solidify overnight at 4 °C. 10 μm thick sections were cut from the blocks with a rotary microtome, and floated on diethyl pyrocarbonate (DEPC) (Sigma-Aldrich)-treated water on PALM® MembraneSlides at 42 °C for 2–4 h to stretch the ribbons. The slides were thereafter air-dried and stored in darkness at 4 °C under low humidity conditions.
Tissue preparation for cryosectioning
The frozen seeds were stored at –80 °C and embedded in Neg-50 Frozen Section Medium (Richard-Allan Scientific) just prior to sectioning. The seeds were sectioned at 10 μm in a cryostat at –20 °C and mounted on PALM® MembraneSlides (PALM Microlaser Technologies, Bernried, Germany). The slides were dipped in 100% ethanol to dehydrate the section completely just before the laser capture microdissection procedure.
Laser capture microdissection (LCM)
The PALM Microlaser System was used to isolate cells from the tissue sections. This system consists of a 337 nm pulsed nitrogen laser coupled to an inverted microscope. The energy and focus settings of the UV-laser were adjusted to cut through the tissue with as narrow a diameter as possible. The cells of interest were selected, dissected via UV-laser excision, and catapulted into the lid of an 0.5 ml Eppendorf tube filled with 30 μl of PicoPure RNA extraction buffer (Arcturus Engineering, CA). After collection, the tubes were centrifuged for 5 min. at maximum speed and placed in an –80 °C freezer until further processing. From each slide an additional sample was prepared in order to check the quality of the RNA of each tissue section. The integrity of the RNA from these samples was analysed on a 2100 Bioanalyser (Agilent Technologies) prior to commencing the amplification procedure. The cell samples isolated in the LCM procedure contained 300–1000 cells per sample. Five to twenty ng of RNA could be isolated from each sample.
RNA extraction
Prior to the extraction procedure, 20 μl of RNA extraction buffer was added to each sample giving a total of 50 μl in each tube. The tubes were then centrifuged for 5 min at maximum speed and subsequently incubated for 30 min at 42 °C. RNA extraction was carried out using the PicoPure RNA Isolation Kit according to the manufacturer's instructions, including treatment with the Rnase-Free DNase Set kit (Qiagen, USA) for 15 min at room temperature. The RNA was eluted from the purification column with 11 μl low-ionic-strength elution buffer.
RNA amplification
The RNA was amplified with the T7 polymerase-based linear amplification systems, RiboAmp (Arcturus Engineering) according to the manufacturer's advice. This system involves production of double-stranded cDNA with a T7 priming sequence at the 3′ end. This template is then used for in vitro transcription by T7 RNA polymerase to generate amplified RNA (aRNA) copies of the cDNA template. Three rounds of amplification were needed to generate sufficient aRNA for Affymetrix hybridizations, as a minimum of 15 μg was required. The yield of amplified RNA product was measured by RiboGreen fluorescence (Table 1).
Table 1.
Treatment of individual samples, yield after three rounds of amplification, present call values, and 3′/5′ ratios for tubulin α2-chain and β-actin. Samples were isolated from two different plants referred to as 1 and 2. Nos 17 and 18 are unamplified samples
| Sample | Tissue | Yield in μg after 3× amplification | %P | Tubulin 3’ UTR/3′ | Tubulin 3′/M | Actin 3′/5′ | Actin 3′/M-ratio |
| T_1 | Transfer cells | 17 | 27 | 159 | 2.2 | 2.6 | 1.3 |
| T_2 | 18 | 36 | 304 | 0.9 | 2.6 | 1.2 | |
| A_1 | Aleurone | 32 | 35 | 375 | 0.6 | 3.2 | 2.3 |
| A_2 | 25 | 31 | 391 | 0.5 | 3.3 | 1.7 | |
| E_1 | Endosperm | 31 | 26 | 303 | 1.3 | 2.3 | 0.8 |
| E_2 | 24 | 38 | 385 | 1.0 | 3.6 | 2.4 | |
| Em_1 | Embryo | 27 | 35 | 429 | 0.7 | 7.0 | 3.0 |
| Em_2 | 44 | 34 | 557 | 0.6 | 4.9 | 2.8 | |
| No. 17 | Endosperm | – | 52 | 1.2 | 0.3 | 14.7 | 1.3 |
| No. 18 | Embryo | – | 64 | 0.4 | 0.6 | 8.8 | 1.5 |
RNA quantification
The concentration of amplified RNA was measured fluorometrically using a microtitre plate reader (Synergy 2, BioTek). Samples were measured after 5 min of incubation with the Ribogreen reagent (Molecular Probes, Eugene, OR) in black microtitre plates, with 485 nm excitation and 535 nm emission wavelengths. This enables accurate measurements of RNA concentrations as low as 1.0 ng ml−1.
Generation of biotinylated aRNA
20 μg of each amplified RNA sample was labelled with biotin using the TURBO Labeling Biotin kit (Molecular Devices, Sunnyvale, CA, USA). The labelling procedure was carried out according to the manufacturer's instructions. Further probe preparation and hybridizations were performed according to Affymetrix's guidelines, undertaken at the RH Microarray Centre, Copenhagen, Denmark.
Real-time PCR
50–100 ng of aRNA from the second and third rounds of amplification was used for the synthesis of cDNA using the oligo-(dT) primer provided in the RiboAmp amplification kit and the reverse transcriptase Superscript II (Invitrogen). For quantification of gene expression, the Power SYBR Green reagent (Applied Biosystems, Warrington, UK) was used with the ABI Prism 7900 Sequence Detection System (Applied BioSystems). The PCR primers for expression analysis were designed to fit the 3′ end of the EST, preferentially within the target sequence used for generation of the probe sets on the 22K Barley GeneChip. The primer software AlleleID 2 was used for the primer design.
The primers sequences were: Jekyll (forward 5′-AAGGTCTCTGGTTGTGGTTTGC-3′ and reverse 5′-CAGTGCGTACCATGTGCTTGT-3′); Ltp2 (forward 5′-GCATGTTCCTGTTATGTGGATGTT-3′ and reverse 5′-TGCTGGTGATCTATGGCGG-3′); HMA1 (forward 5′- GCAGTGCCTAGCATCCTATAATCC-3′ and reverse 5′-CTGTTGGCTGAGATTTGTTTGGTC-3′); HMA2 (forward 5′-GCTGCTCTTGCTGTCTTCTCC-3′ and reverse 5′-GAGAAGCCGCAAAGGGAGATAC-3′); Nramp3 (forward 5′-CTGCCTATTTGTATTAGCTCCGG-3′ and reverse 5′-AGGTCTATATTGTATATCAGTTGCTGCTG-3′); NAAT B (forward 5′-CCCCATCTATATCTTTCAATAAAATGG-3’ and reverse 5′-TATGAATAGAAGTCAACATCTCCTTGAATA-3′); GADPH (forward 5′-GCTCAAGGGTATCATGGGTTACG-3′ and reverse 5′-GCAATTCCAGCCTTAGCATCAAAG-3′); Tubulin alpha2 chain (forward 5′-AGGATGGTGACGAGGGTGATG-3′ and reverse 5′-CAGTAGTACGCCTTGGGAAAGC-3′). The housekeeping genes tubulin and GADPH were chosen as references to normalize the expression ratios of each gene. Each reaction was performed in a total volume of 10 μl containing 1 μl diluted cDNA, 0.6 μl of each 5 mM primer, 2.8 μl H2O, and 5 μl Power SYBR Green reagent. All reactions were performed in triplicate. To be able to compare the expression ratios for all the genes analysed, the same qualibrator sample was included in every run. The relative expression levels were calculated using the Pfaffl equation (Pfaffl, 2001) where the efficiency of each primer pair was taken into account.
Microarray data analysis
The 22K Barley GeneChip contains 22 840 probe sets and was used for generating expression profiles. In order to assess the technical performance of the RNA amplification and labelling procedures a single chip analysis, the MAS5 algorithm, was used to calculate background, noise, scaling factors, and present/absent calls. The default parameters for the 22K Barley GeneChip were used and a global scaling normalization to a target intensity value of 100 was performed for all probe sets. The expression levels of the probe sets on each chip were calculated using a modified version of the Robust Multi-chip Average (RMA) methodology where only signal intensities from the last five probes of each probe set were used for normalization of the data. The RMA method is implemented in the Affymetrix Bioconductor package (Irizarry et al., 2003).
Results
The purpose of the present study was to decide whether it is possible to establish a road map for zinc transport and deposition from the phloem into the barley grain based on laser capture microdissection and expression profiling. Barley (Hordeum vulgare) is a diploid cereal crop that, besides its importance as animal feed, human food, and as raw material for beer production, also serves as a model for the small-grained cereals. A substantial amount of genetic information is available for this crop, including more than 470 000 public ESTs. Seeds from the barley cultivar Golden Promise were harvested 20 dap. Cells from the unloading region (T_1 and T_2), the aleurone layer (A_1 and A_2), the endosperm (E_1 and E_2), and the embryo (Em_1 and Em_2) were isolated with the LCM technology (Fig. 1) and amplified RNA from each sample was hybridized to Affymetrix 22K Barley GeneChips.
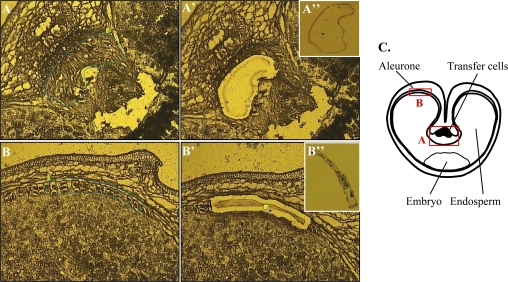
Fig. 1.
Laser capture of transfer cells (A) and aleurone cells (B). The cells of interest are marked with a line (A, B); the laser cuts along the line and catapults the isolated tissue into the lid of a tube (A′, B′); the captured tissue can be visualized in the cap (A′′, B′′). (C) Schematic drawing showing a cross-section of a cereal grain and the localization of the isolated tissues. (This figure is available in colour at JXB online.)
Bioinformatics
A literature survey and database search was performed in order to identify genes with a documented or putative function in metal homeostasis in plants. Relevant proteins were first identified in Arabidopsis and rice (Oryza sativa) and the corresponding barley orthologues were subsequently obtained by BLASTing the sequences against TIGRs (The Institute for Genomic Research) Barley Gene Indices. The database searches identified 68 different ESTs grouped into 11 different metal homeostasis families. The different families and the numbers identified from each gene family are given in Table 2. On the 22K Barley GeneChip all the gene families are represented, but out of the 68 genes identified in the database search only 48 are present on the array.
Table 2.
Overview of metal homeostasis genes identified in barley
| Gene superfamily | Subfamily | Common name | No. identified in barley databases | No. present on 22K Barley GeneChip |
| P-type ATPase | P1B-type ATPase | HMA | 10 | 4 |
| Zrt-, Irt-like Protein | ZIP | 8 | 8 | |
| Cation Diffusion Facilitator (CDF) | MTP | 12 | 4 | |
| Natural resistance associated macrophage proteins | Nramp | 4 | 3 | |
| Ca2+-sensitive cross-complementer 1 (CCC1) | Vacuolar iron transporter 1 | VIT1 | 2 | 2 |
| Major facilitator superfamily of membrane proteins | Zinc-Induced Facilitator1 | ZIF1 | 1 | 1 |
| Ca2+/cation antiporter (CaCA) | Cation exchanger | CAX | 3 | 3 |
| Oligopeptide transporter family | Yellow Stripe Like | YSL | 6 | 5 |
| Metallothionein | MT | 10 | 9 | |
| Nicotianamine synthase | NAS | 8 | 7 | |
| Nicotianamine aminotransferase | NAAT | 4 | 2 | |
| Total | 68 | 48 |
Tissue isolation and integrity of extracted RNA from paraffin and frozen tissue sections
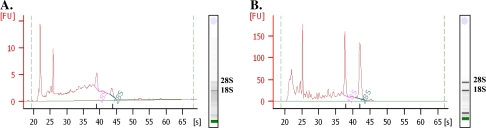
Two tissue preparation procedures (paraffin embedding and snap freezing) were implemented to assess the effects of different fixation and sectioning regimes on RNA quality in the microdissected tissue sections. Agilent BioAnalyser capillary electropherograms showed that the RNA isolated from the paraffin sectioned material had a low quality resulting in high baselines and rather diffuse bands corresponding to 28S rRNA and 18S rRNA on the virtual gel image. By contrast, RNA extracted from the cryosections revealed a high quality with low baselines and distinct bands representing the 28S rRNA and 18S rRNA molecules (Fig. 2).
Fig. 2.
Quality control of the integrity of LCM-derived total RNA prior to amplification. (A) Electropherogram of RNA extracted from tissue sections of paraffin embedded barley grains. (B) Electropherogram of RNA extracted from cryosectioned grains. (This figure is available in colour at JXB online.)
RNA integrity after three rounds of amplification
A single microarray hybridization using the 22K Barley GeneChip from Affymetrix requires 15 μg of RNA. As each sample of isolated cell sections only generated 5–20 ng total RNA three rounds of RNA amplification were performed to obtain sufficient quantities for the hybridizations. However, each amplification round inherently truncates the transcripts due to the use of random primers to initiate the synthesis of the first strand of cDNA in the second and third amplification rounds. The probe sets on the 22K Barley GeneChip consist of 10 perfect match 25mer oligonucleotids distributed across the 3′-most 600 nucleotides of an EST contig, singleton, or cloned gene (Close et al., 2004) Accordingly, very short transcripts will only hybridize to a few of the probes in the probe set. To account for this problem the expression levels were calculated using a modified version of the Robust Multi-chip Average (RMA) methodology. To compensate for the 3′ end bias of the amplified aRNA, only signal intensities from the last five probes closest to the 3′ end of each probe set were used for normalization of the data. RMA is a quantile normalization method that is better suited for comparison analysis of amplified RNA samples than the linear normalization, which is implemented in the MAS5 method. RMA consists of three steps: (i) convolution background correction, (ii) quantile normalization, and (iii) summarization based upon a multi-array model of the normalized and log (base 2) transformed probe set data. The MAS5 software is based on the concept of averaging signals from the different probes on each chip separately. The Robust Multi-chip Analysis (RMA) fits a linear model for both transcript abundance and probe affinity to the log intensities of probes across the whole set of chips and the RMA algorithm only uses the values from the perfect match (PM) probes and ignores the mismatch (MM) probes (Irizarry et al., 2003).
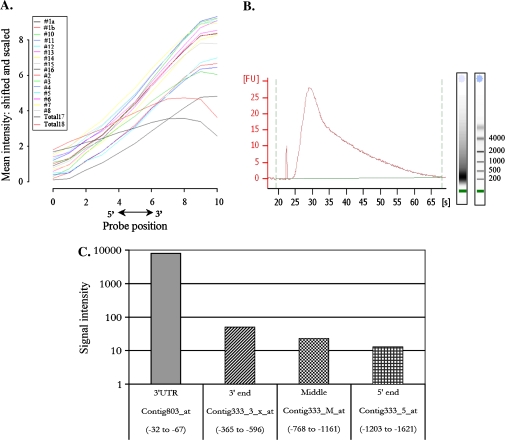
The level of degradation in individual samples was visualized from the results of the microarray hybridizations by examining all the probe sets on a chip. By computing the average signal intensity at each probe position from the 3′ end to the 5′ end of the probe sets, a degradation curve was generated where the steepness of the slopes reflects the level of degradation (Fig. 3A). For all of the amplified samples the signal intensity declined from the 3′ end towards the 5′ end, indicating low representation of the 5′ end of the genes. As a control, two samples were included of unamplified RNA extracted from crude fractions of the endosperm and the embryo. For these samples, the mean signal intensity levels were fairly balanced for all probes within the probe sets. The 3′/5′-ratios of signals from probe sets corresponding to the 3′ end of genes compared to signals from the 5′ or middle section of the same transcript also provide an estimation of the level of 5′ end representation in the amplified samples. The Barley 22K GeneChip from Affymetrix contains probe sets designed to the 5′ end, the middle region, and the 3′ end of the housekeeping genes actin (contig1390) and α-tubulin (contig333), which can be used for the 3′/5′-ratio calculations. The tubulin gene has an additional probe set in the 3′ untranslated region (UTR) (contig803) located within the last 67 nucleotides of the transcript, whereas the 3′ probe set Contig333_3_x is positioned 365–464 nt from the end of the coding region of the gene. Comparison of the signal intensities from the tubulin 3′ UTR probe set to the 3′ probe set, which is located less than 300 nt further upstream, show for all hybridizations that there is a dramatic decrease in the intensities exemplified by sample T_1 in Fig. 3C. As opposed to that, the 3′/5′-ratios of both actin and tubulin in the two samples prepared from non-amplified RNA fell within the manufacturer's recommendations for the 3′/5′ ratio (Table 1) (Affymetrix Gene Expression Service Laboratory Report, 2008, see http://ipmb.sinica.edu.tw/affy).
Fig. 3.
(A) RNA degradation plot showing the average signal intensity at each position from the 5′ end to the 3′ end of the probe set. The general pattern shows lower intensities towards the 5′ end. (B) Profile of biotin-labelled aRNA prior to fragmentation. The average sizes of the transcripts are 200-400 nucleotides. (C) Histogram showing signal intensities from four different probe sets located in the 5′ end (Contig333_5_at), in the middle region (Contig333_M_at), in the 3′ end (Contig333_3_x_at), and in the 3′ untranslated region (Contig803_at) of the gene encoding the tubulin alpha2-chain (TC156922) from the T_1 sample. The positions of the target sequences that comprise each probe set relative to the termination of the coding region are shown. (This figure is available in colour at JXB online.)
Reliability of amplified samples
The integrity of the amplified samples was determined by the amount of present calls (%P) in the MAS5 data analysis, which is a measure of the percentage of genes on the array that are marked as being expressed. A significant decline was found in the present call values when comparing the values for the amplified samples that ranged from 25% to 42% P with values of the unamplified RNA samples extracted from crude fractions of endosperm or embryo tissue that were rated to have 52% and 64% expressed genes, respectively (Table 1).
Gene expression and validation
Due to the short length of the amplified transcripts, the exact positions of the probe sets of all the 48 identified metal homeostasis genes that were represented on the 22K Barley GeneChip had to be determined. Considering that most of the genes were represented by at least five probes within the last 400 nucleotides counted from the poly(A)-tail, a modified RMA algorithm was applied for normalization of the expression data where only signals from the last five probes of each probe set were taken into account. Due to this modified normalization approach, the expression data of 44 (92%) of the metal homeostasis genes identified were considered to be reliable since they all have at least five individual 25mer probes located within the 400 most nucleotides of the 3′ end.
A road map for zinc trafficking
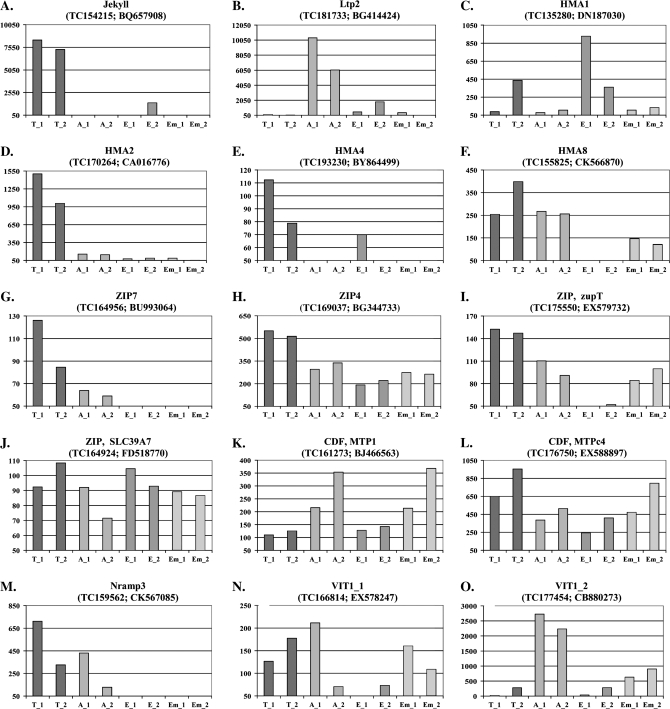
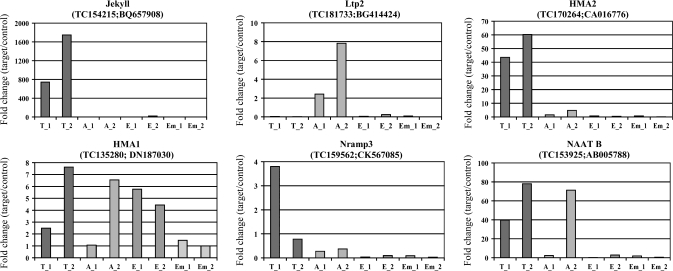
As an initial approach for validating the laser capture microdissection gene expression profiling results, the expression levels of two tissue specific genes were analysed. Jekyll is a transfer cell specific gene (Radchuk et al., 2006) and Ltp2 has been shown to be highly expressed in cells of the aleurone layer (Kalla et al., 1994). Our expression data for both genes confirmed that both of them were expressed in a highly tissue-specific manner and, further, that the expression levels were roughly the same in the two samples of materials derived from two different plants. (Fig. 4A, B)
Fig. 4.
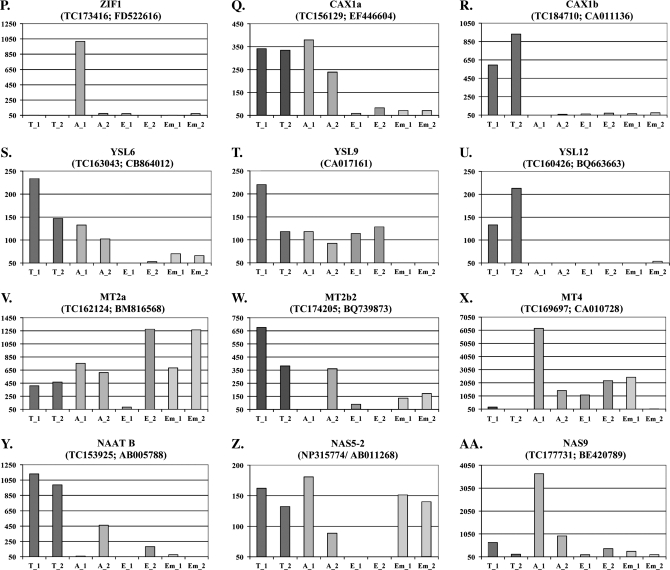
Histograms showing gene expression levels of 25 different metal homeostasis genes and of the transfer cell-specific gene Jekyll and the aleurone-specific gene Ltp2. The tissue samples were isolated from grains collected from two different plants. Each gene is identifyed by its assigned name, the TIGR accession number and the GenBank accession number, respectively. The background intensity was arbitrarily set to 50 and only values above this threshold are shown on the y-axis. T_1 and T_2: transfer cell samples from plants 1 and 2; A_1 and A_2: aleurone samples from plants 1 and 2; E_1 and E_2: endosperm samples from plants 1 and 2; Em_1 and Em_2: embryo samples from plants 1 and 2.
In Fig. 4, the expression data have been compiled for 25 genes involved in metal homeostasis, divided into the different gene families.
HMA
The HMAs are members of the family of P1B-type Heavy Metal ATPases. They are divided into two groups based on their substrate specificities: Zn/Cd/Co/Pb and Cu/Ag transporters. The superfamily of P-type ATPases uses ATP to pump a wide range of cations across membranes against their electrochemical gradient. In barley, 10 HMAs have been identified of which four (HMA 1–3 + 10) belong to the Zn/Cd/Co/Pb group and the rest (HMA 4–9) to the Cu/Ag group (Williams and Mills, 2005). The expression data show that HMA1 particularly is expressed in the endosperm while HMA2 and HMA4 appear to be largely expressed in the transfer cells. The highest expression levels for HMA8 are found in transfer cells and aleurone (Fig. 4C–F).
ZIP
Most members of the ZIP (Zrt-, Irt-like Protein) family are predicted to have eight transmembrane domains with their N- and C-termini located on the cytoplasmic side of the membrane. Without any known exceptions these proteins transport zinc and/or other metal ions from the extracellular space or organellar lumen into the cytoplasm (Eide, 2006). It is apparent that ZIP7 is only expressed in the transfer cells and in the aleurone cells while ZIP4 and ZIP SLC39A7 are transcribed in all tissues. ZupT appears not to be expressed in the endosperm but transcripts are detected in all other tissues of the grain (Fig. 4G–J).
CDF
Members of the CDF (Cation Diffusion Facilitator) family are involved in efflux of transition metals from the cytoplasm into cellular compartments or to the outside of cells. Most members of this family have six transmembrane domains with a topology where both the amino and carboxy termini are located on the cytoplasmic side of the plasma membrane. They serve as secondary active transporters using the gradient of other ions to drive the transport. The substrate for these transporters are particularly Zn, Cd, and Co (Eide, 2006). Four out of the 12 known barley members of this gene family are present on the array. According to our data MTPc4 is expressed throughout the grain with the greatest expression found in the transfer cells. MTP1 is also expressed in all tissues of the grain but with the lowest expression in the transfer cells (Fig. 4K, I). The other two CDF members are expressed at very low levels in the seed (data not shown).
Nramp
Nramp (Natural resistance associated macrophage proteins) are implicated in the transport of divalent metal ions. Plant Nramp transporters have 12 predicted transmembrane domains and a long C-terminal tail which is unique to these proteins (Williams et al., 2000). Three out of the four different barley Nramp genes are present on the 22K Barley GeneChip. Nramp3 is expressed in the transfer cells and to a lesser extent in the aleurone layer (Fig. 4M). Data for Nramp6-1 and Nramp6-2 are not shown since fewer than five of the probes were positioned within the last 400 nucleotides of their coding regions.
VIT1
The yeast CCC1 (Ca2+-sensitive Cross-Complementer1) gene encodes an iron/Mn2+ transporter responsible for storing these two metals in the vacuole. The Arabidopsis orthologue of the CCC1 gene is the VIT1 (vacuolar iron transporter 1) gene (Li et al., 2001). Two VIT1 transporters were identified in barley. VIT1_1 is expressed in all tissues but the endosperm. VIT1_2 is highly expressed in the aleurone cells and to a minor extent in the embryo (Fig. 4N, O).
CAX
The CAX (Cation Exchanger) family is one of the five subfamilies that make up the CaCA superfamily. CAXs are a group of integral membrane proteins with 10–11 transmembrane domains that export cations out of the cytosol to maintain an optimal ionic concentration in the cell. This translocation process is energized by the pH gradient established by proton pumps (Shigaki et al., 2006). In our analysis, CAX1a is mainly expressed in the transfer cells and the aleurone layer and CAX1b is specifically expressed in the transfer cells (Fig. 4P, Q). CAX2 does not seem to be expressed in the grain (data not shown).
ZIF1
ZIF1 (Zinc-Induced Facilitator1) belong to the Major Facilitator Superfamily of membrane proteins, which are implicated in the transport of a wide range of small organic molecules. It is located in the tonoplast and is believed to mediate zinc transport from the cytosol into the vacuole (Haydon and Cobbett, 2007a). ZIF1 was only expressed in the aleurone sample from one of the plants (Fig. 4R). Interestingly, preliminary experiments indicate that ZIF1 is inducible in the embryo following foliar sprays with 5 mM ZnSO4 (data not shown).
YSL
YSL (Yellow Stripe Like) transporters are predicted to have 12 transmembrane domains and are involved in the translocation of metals that are complexed with phytosiderophores (PS) or nicotianamine (NA), a non-proteinogenic amino acid that serves as a precursor for PS synthesis in grasses (Curie et al., 2001). YSL12 appears to be expressed exclusively in the transfer cells whereas YSL6 and YSL9 are expressed throughout the grain with the exception of the endosperm (YSL6) and the embryo (YSL9) (Fig. 4S–U). YSL11 and YSL15 are not expressed in the grain.
Metallothionein
Metallothioneins (MTs) are low-molecular weight proteins with a high cysteine content and a high affinity for binding metal cations like zinc, cadmium, and copper. With a metal:MT stoichiometry of around 7:1 MTs play a role in plant metal homeostasis (Kramer and Chardonnens, 2001). In this study, the expression of eight out of the 10 MTs that are present in barley has been examined. The MT1a was not represented on the array and the expression data for MT3 were not reliable due to the position of the corresponding probe set. MT1b1, MT1b3, MT2b1, and MT2b3 were apparently not expressed in the grain. MT1b2 was present in all tissues at a low level. MT2a was expressed throughout the grain. In all tissues but the transfer cells MT4 also displayed high expression levels. MT2b2 was expressed in all tissues (Fig. 4V–X). Initial experiments with foliar zinc applications revealed that MT2b2 was expressed at extremely high levels in the aleurone and in the embryo 6 h after the treatment (data not shown). This indicates that MT2b2 may be involved in zinc homeostasis as a possible chelator of this mineral in the aleurone layer and in the embryo when zinc is present in excess.
NAAT
Nicotianamine aminotransferase (NAAT) catalyses the transfer of an amino residue to nicotianamine (NA) resulting in the production of 2′-deoxymugineic acid (DMA), the precursor of all mugineic acids (Curie and Briat, 2003). NAAT B was expressed in all four tissues with the highest expression observed in the transfer cells (Fig. 4Y). NAAT A was expressed at a very low level in the aleurone and endosperm samples (data not shown).
NAS
Nicotianamine synthase catalyses a reaction where three molecules of S-adenosyl methionine are joined to produce one molecule of nicotianamine (Curie and Briat, 2003). In barley, eight nicotianamine synthase (NAS) genes have been identified of which seven are present on the 22K Barley GeneChip. NAS2, NAS4, NAS7, and NAS8 are not expressed in the grain. NAS1 and NAS5-2 are expressed at very low or moderate levels, respectively. NAS9 is highly expressed in the aleurone and at low levels in the other tissues of the grain (Fig. 4Z, AA).
To validate the microarray results real-time PCR analysis was performed for a subset of the genes investigated. The primers were designed to fit the very 3′ end of the EST to account for the problems with truncated transcripts. However, the sequence amplified was not in all cases equivalent to the target sequence used for probe design for the 22K Barley GeneChip. As illustrated by a comparison of the results obtained from the microarray (Fig. 4) with those from the real-time PCR (Fig. 5) there is a good agreements between the results obtained by the two different approaches, the only major exception being HMA1 where the real-time data indicate that the gene is expressed in all tissue while the microarray analysis suggest a preferential expression in the endosperm.
Fig. 5.
Histograms showing relative gene expression levels calculated from real-time PCR data. The experiments were performed with the same amplified materials that were used for the hybridizations. T_1 and T_2: transfer cell samples from plants 1 and 2; A_1 and A_2: aleurone samples from plants 1 and 2; E_1 and E_2: endosperm samples from plants 1 and 2; Em_1 and Em_2: embryo samples from plants 1 and 2.
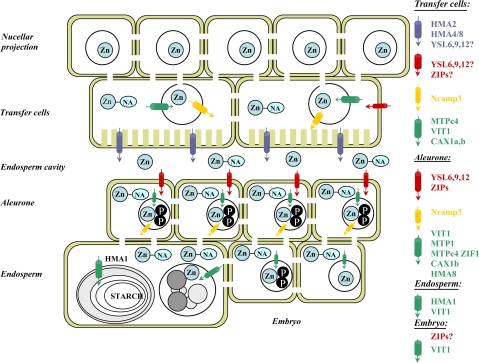
Fig. 6.
Proposed road map for zinc trafficiking from the phloem to the final sequestring sites in the developing grain. Blue cylinders indicate cellular efflux transporters, red cylinders indicate cellular importing transporters, green cylinders indicate vacuolar uptake transporters, and yellow cylinders indicate vacuolar efflux transporters: Zn, zinc; NA, nicotianamine; P, phytate.
Discussion
The purpose of the present study was (i) to assess the possibilities for using laser capture microdissection and gene expression profiling to get an overview of the expression patterns of genes encoding primarily zinc transporters and zinc sequestration compounds and (ii) on the basis of the expression data to generate a road map for zinc trafficking in the barley grain. As gene expression profiling of laser capture microdissected tissue still is at its infancy, a detailed analysis of the RNA quality was undertaken using different types of fixation and sectioning regimes and further implemented alternative statistical approaches to the Affymetrix data.
Clear evidence was obtained that cryosectioning is superior to chemical fixation, embedding, and sectioning. Plant material fixed in a fixative such as Farmers fixative (ethanol:acetic acid (3:1 v/v)) and embedding in paraffin is usually considered to provide tissue sections of high morphological integrity and with good preservation of biological molecules. This method is accordingly widely used (Kerk et al., 2003; Ramsay et al., 2004; Inada and Wildermuth, 2005). The alternative cryosectioning method involves freezing of the fresh tissue, which can cause formation of ice crystals and eventually the destruction of cellular structures upon thawing. Since the large central vacuole contains lysosomal enzymes and maintains turgor pressure, disruption of this organelle during the freeze–thaw process will result in a severe loss of tissue integrity (Inada and Wildermuth, 2005). The optimized embedding protocol reported by Ramsay et al. (2004) was followed and it was found that the RNA extracted from our paraffin sections was partly degraded. On the electrophoretic fluorescence profile and the virtual gel image obtained with the Agilent 2100 Bioanalyser the high baseline and diffuse 28S rRNA and 18S rRNA bands demonstrate that the quality of the RNA is poor and therefore not suitable for RNA expression analysis. The difficulty in obtaining high quality RNA from paraffin sections has also been reported by other groups (Perlmutter et al., 2004; Ishimaru et al., 2007) and a study performed by Cai and Lashbrook (2006) suggest that the mounting of the paraffin sections on a drop of water on the membrane slides and the ensuing drying period is responsible for the observed degradation of RNA. By comparison, the RNA extracted from the frozen cryosections was of higher quality with sharp ribosomal RNA bands and could therefore be subjected to the amplification procedure (Fig. 2A, B).
The cell samples isolated in the LCM procedure contained 300–1000 cells per sample from which 5–20 ng total RNA could be isolated. After two rounds of amplification the yield of aRNA was between 2 μg and 7 μg per sample. As 15 μg aRNA is required for hybridization to the 22K Barley GeneChip, it was necessary to perform a third round of amplification. The third amplification round has previously been shown to decrease the average distribution size of aRNA but without seriously affecting the reproducibility (Scherer et al., 2003).
The degradation plot revealed that the average signal intensities were dramatically lower for probes located in the 5′-end of the probe sets as opposed to probes in the 3′-end (Fig. 3A). This reflects the gradual 5′ shortening of amplified RNA that occurs during the amplification process caused by the use of random primers to initiate the synthesis of the first cDNA strand in the second and third amplification rounds (Nygaard and Hovig, 2006). The significant decrease in signal intensity from probe sets located more than 400 bp from the poly(A)-tail (Fig. 3C) demonstrates the truncation of the amplified transcripts and reveals that not all probes representing a transcript will provide valid information of the actual presence or absence of a given transcript in a sample. The probe sets on the 22K Barley GeneChip are based on the 3′-most 600 nucleotides of an EST contig, singleton or cloned gene (Close et al., 2004). Due to that, a sample of amplified RNA that contains transcripts varying in size from 100–500 nucleotides, with an average length of approximately 300 nt (Fig. 3B), will fail to hybridize to single probes or entire probe sets positioned outside this narrow region. When normalizing the data using either the MAS5 or the RMA normalization algorithm, genes without probe representation within the last 300 bases of the coding region will incorrectly be called absent or will generate signal intensities that do not correspond to the actual expression level of the gene. The normalized results obtained from hybridization of samples that are amplified three and possibly also two times will therefore not provide an accurate gene expression profile, as a large number of probe sets will generate false negative data. A similar observation has been made by Boelens et al. (2007), who reported a significant linear decrease in signal intensity from probes located at increased distances from the poly(A)-tail resulting from truncation of the aRNA after two rounds of amplification. The issue of the very short transcripts was taken into account by applying a modified RMA algorithm to our data where only signal intensities from the last five probes of each probe set were used for the normalization. Out of the 48 metal homeostasis genes on the array, 44 genes (92%) were represented by at least five probes within the last 400 nucleotides of the coding region. Despite the pronounced 3′ bias of the amplified transcripts, the modified normalization approach enabled us to obtain reliable expression data for most metal homeostasis genes in the different tissue of the developing barley grain. Furthermore, it was possible to validate the results by real-time PCR on a subset of genes where primers could be designed to amplify a sequence equivalent to the target sequence used for the Affymetrix probe design.
As an additional validation approach, the gene expression profile of two genes known to have a tissue-specific expression in the grain was analysed, namely the aleurone-specific gene Ltp2 and the transfer cell-specific gene Jekyll. Ltp2 is a non-specific putative lipid transfer protein, which is expressed in aleurone cells from shortly after the onset of aleurone cell differentiation to mid-maturity of the grain (Kalla et al., 1994). The Jekyll gene encodes a 140-residue protein that does not resemble any other known protein but is proposed to be necessary for the establishment of the nucellus structure in the barley caryopsis and for the subsequent cell autolysis (Radchuk et al., 2006). Very high levels of the Jekyll protein were detected in the transfer cells. For both genes specific expression was found in the aleurone and the transfer cells, respectively.
The transfer cells constitute the first bottleneck for nutrient, for example, zinc, translocation, as they are the interface between the symplast of the nucellar projection—the phloem strand of the maternal tissue and the apoplast of the endosperm cavity. Interestingly, it was found that members of the HMA, ZIP, CDF, Nramp, VIT1, CAX, YSL, MT, and NAAT are all expressed at high levels in the transfer cells. Among the genes that were highly expressed in one of the other examined tissues, HMA1, MTP1, VIT1_2, ZIF1, NAS9, and MT4 were expressed at a low level in the transfer cells. Tentatively we would like to propose that zinc is unloaded from the phloem complexed with nicotianamine or another derivative such as mugineic acid. Furthermore, we would like to propose that the transfer cells accumulate the zinc in vacuoles for temporary storage from where it is mobilized, transferred to the cytosol, and pumped into the endosperm cavity. In agreement with this suggestion, X-ray energy spectra show a high density of zinc as well as other minerals in the unloading region (Mazzolini et al., 1985). Putative candidates for transporters moving zinc into vacuoles include the expressed members of the CDF, VIT1, ZIF1, and CAX families while HMA2 is a putative candidate for transport from the transfer cells into the apoplastic endosperm cavity. AtHMA2 and AtHMA4 in Arabidopsis belong to the group of Zn/Cd/Pb/Co transporters and have been shown to be responsible for translocation of zinc from the pericycle cells and into the xylem vessels for long-distance transport to the shoots (Hussain et al., 2004). Nramp3 is a possible candidate for a vacuolar efflux transporter as Nramp3 and 4 have been shown to be essential for the mobilization of minerals from the protein storage vacuoles of the Arabidopsis embryo (Lanquar et al., 2005). The observed expression of the NAS5-2 and NAS9 genes in the transfer cells might relate to zinc-nicotianamine trafficking in the cytosol for temporary storage in the vacuoles and subsequent transfer to the plamalemma facing the endosperm cavity. The high activity of the NAAT B gene indicates that a part of the synthesized NA molecules are converted to other precursors of mugineic acids (MAs) such as 2′-deoxymugineic acid. However, other sequestering agents like metallothioneins may also be involved in metal chelation in this tissue. Interestingly, three of the five YSL genes on the array are expressed in the transfer cells. YSL transporters are usually considered to translocate metal–phytosiderophore complexes into cells. A possible explanation for the presence of these YSL transporters in the transfer cells could be that they can capture zinc–nicotianamine complexes diffusing from the endosperm cavity into the apoplast of the transfer cells. The ZIP transporters found to be active in the transfer cells may have a similar function of capturing zinc flowing back into the apoplast.
The embryo and the aleurone are major depositories for zinc as revealed by chemical analyses, histochemical staining and X-Ray microanalysis (Mazzolini et al., 1985; Ozturk et al., 2006; Choi et al., 2007). It has been generally believed that zinc, together with iron and manganese, was complexed with phytic acid in the protein storage vacuoles of the aleurone and the embryo. In agreement with this, zinc is known to bind strongly to phytic acid. However, it is also considered likely that zinc may be bound to proteins in the two tissues. In agreement with this, it is apparent from histological staining experiments that zinc is also present in the outer part of the endosperm, which contain high amounts of protein (Ozturk et al., 2006). Aleurone and embryo share many other characteristics as they contain a lot of additional nutrients to be utilized during germination. Furthermore, the scutellum of the embryo as well as the aleurone serve as secretory tissues for a variety of hydrolytic enzymes for endosperm degradation during germination.
The aleurone and embryo expression profile for genes encoding zinc transporters and sequestering compounds have a number of similarities. The only HMA gene expressed in the two tissues is the Cu/Ag transporting HMA8. All four ZIP transporters and the two CDF transporter genes are transcribed at the same level. On the other hand the Nramp3, ZIF1, CAX1a, VIT1_2, NAS9, and possibly NAAT B, genes show a higher transcription rate in the aleurone than in the embryo. We tentatively propose that these gene expression profiles indicate that the ZIP transporters ensure an efficient uptake of zinc in aleurone and embryo and that the CDF transporters move zinc across the tonoplast into the vacuole. Very little zinc efflux across the plasmalemma is predicted, as judged by the low expression of the HMA2 gene. Additional transport into the storage protein vacuole of the aleurone may be ensured by ZIF1, CAX1a, and VIT1_2 while Nramp3 mediates flux out of the vacuole. Interestingly, initial data indicate that ZIF1 expression is induced by foliar zinc applications in the embryo (data not shown). As described above, NAS9 and NAS5-2 are active in the grain with very high expression of NAS9 in the aleurone tissue. Both YSL6 and YSL9 are transcribed in the aleurone. This may indicate that zinc–nicotianamide complexes are taken up from the apoplast between the testa layer and the aleurone. All three MT genes appear to be expressed in the aleurone and embryo at high levels. Initial experiments indicate that MT2b2 is strongly induced in the embryo after foliar application of zinc (data not shown).
The expression of transporter and sequestration genes is generally lower in the endosperm than in other tissues. This is very well in line with the endosperm having a low concentration of zinc except in the outer layers. Notable exceptions are HMA1, ZIP (SLC39A7), MT2A, and YSL9. The very high activity of MT2a in one of the endosperm samples may reflect that zinc in the endosperm is being sequestered primarily by this particular metallothionein protein.
In summary, a roadmap has been defined for zinc trafficking in the barley grain from phloem unloading until deposition based on gene expression profiles of laser microdissected tissues. We would like to emphasize that the roadmap should only be taken as a working hypothesis where the exact function and contribution needs to be validated for each of the genes putatively identified to have a role in zinc transport and sequestration. Transgenic approaches involving up-regulation and RNAi-mediated down-regulation of several of the genes are currently in progress. Surprisingly, in initial investigations it has been found that most of the genes are not transcriptionally enhanced following foliar application of 5 mM ZnSO4. However, only one time point was analysed (6 h after application). Furthermore, it is conceivable that many of the proteins are post-translationally regulated in which case the Zn status of the cells will not have a direct influence of the level of transcription. This is, for example, the case for the P-type ATPases whose activity status is regulated by phosphorylation (Bækgaard et al., 2005).
Acknowledgments
The scientific work was financed by a project grant from The Danish Agency for Science, Technology and Innovation. Ole Braad Hansen is thanked for taking care of the barley plants.
References
- Bækgaard L, Fuglsang A, Palmgren MG. Regulation of plant plasma membrane H+- and Ca2+-ATPases by terminal domains. Journal of Bioenergetics and Biomembranes. 2005;37:369–374. doi: 10.1007/s10863-005-9473-0. [DOI] [PubMed] [Google Scholar]
- Boelens MC, Meerman GJT, Gibcus JH, Blokzijl T, Boezen HM, Timens W, Postma DS, Groen HJ, van den Berg A. Microarray amplification bias: loss of 30% differentially expressed genes due to long probe-poly(A)-tail distances. BMC Genomics 8. 2007 doi: 10.1186/1471-2164-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Borg S, Tauris B, Holm PB. Molecular genetic approaches to increasing mineral availability and vitamin content of cereals. Journal of Cereal Science. 2007;46:308–326. [Google Scholar]
- Broadley MR, White PJ, Hammond JP, Zelko I, Lux A. Zinc in plants. New Phytologist. 2007;173:677–702. doi: 10.1111/j.1469-8137.2007.01996.x. [DOI] [PubMed] [Google Scholar]
- Cai SQ, Lashbrook CC. Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. The Plant Journal. 2006;48:628–637. doi: 10.1111/j.1365-313X.2006.02886.x. [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. The Plant Journal. 2005;42:111–123. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- Choi EY, Graham R, Stangoulis J. Semi-quantitative analysis for selecting Fe- and Zn-dense genotypes of staple food crops. Journal of Food Composition and Analysis. 2007;20:496–505. [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP. A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiology. 2004;134:960–968. doi: 10.1104/pp.103.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. Put the metal to the petal: metal uptake and transport throughout plants. Current Opinion in Plant Biology. 2006;9:322–330. doi: 10.1016/j.pbi.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Curie C, Briat JF. Iron transport and signaling in plants. Annual Review of Plant Biology. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Day R, McNoe L, Macknight RC. Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm. International Journal of Plant Genomics. Article. 2007;61028:1–17. doi: 10.1155/2007/61028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. The Plant Journal. 2008;55:746–759. doi: 10.1111/j.1365-313X.2008.03555.x. [DOI] [PubMed] [Google Scholar]
- Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochimica et Biophysica Acta. 2006;1763:711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang ZP, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Ghandilyan A, Vreugdenhil D, Aarts MGM. Progress in the genetic understanding of plant iron and zinc nutrition. Physiologia Plantarum. 2006;126:407–417. [Google Scholar]
- Gibson RS. Zinc: the missing link in combating micronutrient malnutrition in developing countries. Proceedings of the Nutrition Society. 2006;65:51–60. doi: 10.1079/pns2005474. [DOI] [PubMed] [Google Scholar]
- Gladyshev VN, Kryukov GV, Fomenko DE, Hatfield DL. Identification of trace element containing proteins in genomic databases. Annual Review of Nutrition. 2004;24:579–596. doi: 10.1146/annurev.nutr.24.012003.132241. [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiology. 2007a;143:1705–1719. doi: 10.1104/pp.106.092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. Transporters of ligands for essential metal ions in plants. New Phytologist. 2007b;174:499–506. doi: 10.1111/j.1469-8137.2007.02051.x. [DOI] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. The Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N, Wildermuth MC. Novel tissue preparation method and cell-specific marker for laser microdissection of Arabidopsis mature leaf. Planta. 2005;221:9–16. doi: 10.1007/s00425-004-1427-y. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru T, Nakazono M, Masumura T, Abiko M, San-oh Y, Nishizawa NK, Kondo M. A method for obtaining high integrity RNA from developing aleurone cells and starchy endosperm in rice (Oryza sativa L.) by laser microdissection. Plant Science. 2007;173:321–326. [Google Scholar]
- Kalla R, Shimamoto K, Potter R, Nielsen PS, Linnestad C, Olsen O-A. The promotor of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell-specific expression in transgenic rice. The Plant Journal. 1994;6:849–860. doi: 10.1046/j.1365-313x.1994.6060849.x. [DOI] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiology. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer U, Chardonnens AN. The use of transgenic plants in the bioremediation of soils contaminated with trace elements. Applied Microbiology and Biotechnology. 2001;55:661–672. doi: 10.1007/s002530100631. [DOI] [PubMed] [Google Scholar]
- Kramer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Letters. 2007;581:2263–2272. doi: 10.1016/j.febslet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LT, Chen OS, Ward DM, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. Journal of Biological Chemistry. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- Mazzolini AP, Pallaghy CK, Legge GJF. Quantitative microanalysis of Mn, Zn and other elements in mature wheat seed. New Phytologist. 1985;100:483–509. [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. The Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Research. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk L, Yazici MA, Yucel C, Torun A, Cekic C, Bagci A, Ozkan H, Braun HJ, Sayers Z, Cakmak I. Concentration and localization of zinc during seed development and germination in wheat. Physiologia Plantarum. 2006;128:144–152. [Google Scholar]
- Perlmutter MA, Best CJM, Gillespie JW, Gathright Y, Gonzalez S, Velasco A, Linehan WM, Emmert-Buck MR, Chuaqui RF. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. Journal of Molecular Diagnostics. 2004;6:371–377. doi: 10.1016/S1525-1578(10)60534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AS, Kucuk O. Zinc in cancer prevention. Cancer and Metastasis Reviews. 2002;21:291–295. doi: 10.1023/a:1021215111729. [DOI] [PubMed] [Google Scholar]
- Radchuk V, Borisjuk L, Radchuk R, Steinbiss HH, Rolletschek H, Broeders S, Wobus U. Jekyll encodes a novel protein involved in the sexual reproduction of barley. The Plant Cell. 2006;18:1652–1666. doi: 10.1105/tpc.106.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay K, Wang ZH, Jones MGK. Using laser capture microdissection to study gene expression in early stages of giant cells induced by root-knot nematodes. Molecular Plant Pathology. 2004;5:587–592. doi: 10.1111/j.1364-3703.2004.00255.x. [DOI] [PubMed] [Google Scholar]
- Scherer A, Krause A, Walker JR, Sutton SE, Seron D, Raulf F, Cooke MP. Optimized protocol for linear RNA amplification and application to gene expression profiling of human renal biopsies. Biotechniques. 2003;34:546–556. doi: 10.2144/03343rr01. [DOI] [PubMed] [Google Scholar]
- Shigaki T, Rees I, Nakhleh L, Hirschi KD. Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. Journal of Molecular Evolution. 2006;63:815–825. doi: 10.1007/s00239-006-0048-4. [DOI] [PubMed] [Google Scholar]
- Wang HL, Offler CE, Patrick JW. The cellular pathway of photosynthate transfer in the developing wheat grain. II. A structural analysis and histochemical studies of the pathway from the crease phloem to the endosperm cavity. Plant, Cell and Environment. 1994a;18:373–388. [Google Scholar]
- Wang HL, Offler CE, Patrick JW, Ugalde TD. The cellular pathway of photosynthate transfer in the developing wheat-grain. I. Delineation of a potential transfer pathway using fluorescent dyes. Plant, Cell and Environment. 1994b;17:257–266. [Google Scholar]
- Welch RM, Graham RD. Breeding for micronutrients in staple food crops from a human nutrition perspective. Journal of Experimental Botany. 2004;55:353–364. doi: 10.1093/jxb/erh064. [DOI] [PubMed] [Google Scholar]
- Williams LE, Mills RF. P-1B-ATPases: an ancient family of transition metal pumps with diverse functions in plants. Trends in Plant Science. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochimica et Biophysica Acta. 2000;1465:104–126. doi: 10.1016/s0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]