Abstract
In the Scots pine (Pinus sylvestris L.) seed, embryos grow and develop within the corrosion cavity of the megagametophyte, a maternally derived haploid tissue, which houses the majority of the storage reserves of the seed. In the present study, histochemical methods and quantification of the expression levels of the programmed cell death (PCD) and DNA repair processes related genes (MCA, TAT-D, RAD51, KU80, and LIG) were used to investigate the physiological events occurring in the megagametophyte tissue during embryo development. It was found that the megagametophyte was viable from the early phases of embryo development until the early germination of mature seeds. However, the megagametophyte cells in the narrow embryo surrounding region (ESR) were destroyed by cell death with morphologically necrotic features. Their cell wall, plasma membrane, and nuclear envelope broke down with the release of cell debris and nucleic acids into the corrosion cavity. The occurrence of necrotic-like cell death in gymnosperm embryogenesis provides a favourable model for the study of developmental cell death with necrotic-like morphology and suggests that the mechanism underlying necrotic cell death is evolutionary conserved.
Keywords: Conifer, developmental cell death, embryogenesis, megagametophyte, necrotic cell death, seed development
Introduction
Generally, cell death can be divided into two classes, programmed cell death (PCD) and necrosis. PCD is a genetically encoded, innate physiological process that involves the selective death of individual cells, tissues or whole organs, whereas necrosis is generally caused in a passive manner by environmental perturbation (Pennell and Lamb, 1997). Both cell death events are well defined in animals, whereas in plants there seems to be more overlap between the phenotypic and molecular characteristics of PCD and necrosis, which makes the discrimination between the two forms more complicated (Van Breusegem and Dat, 2006). In plants, PCD occurs when cell suicide pathways are activated as a part of normal growth and development or in response to certain biotic and abiotic external factors (Beers, 1997). If cells are damaged by stresses at an overwhelming level, they undergo necrosis involving cell swelling, lysis, and inflammatory leakage of cell contents (Pennell and Lamb, 1997). Despite the initial idea that necrosis is an uncontrolled form of cell death, accumulating evidence suggests that necrotic cell death can also be a regulated event that contributes to development and to the maintenance of tissue and organismal integrity (Zong and Thompson, 2006).
During the PCD process, the main target of the cell degradation machinery is the nucleus. The degradation processes include events both in chromatin (i.e. chromatin condensation and DNA fragmentation) and in the nuclear envelope (i.e. the redistribution of the nuclear pore complex and the disassembling of nuclear lamina) (Earnshaw, 1995). The structural organization is conserved in plant and animal nuclei whereas the molecular composition of the nuclear envelope is not (Rose et al., 2004), which suggests that different molecular mechanisms may be responsible for the nuclear envelope events in plant and animal cells. Chromosome fragmentation and DNA degradation require the participation of endo- and exonucleases. The apoptotic nuclease Tat-D incises the double-stranded DNA without obvious specificity through its endonuclease activity and excises DNA from the 3′-end to the 5′-end via its exonuclease activity. Tat-D has been found to be evolutionarily conserved across the plant and animal kingdoms, potentially due to its indispensable biological function (Qiu et al., 2005). In animal cells, nuclear degradation during PCD is executed by a caspase family of cysteine proteases, whereas the canonical caspases are absent in plants. Plant genomes contain distant caspase-homologues, metacaspases, which have been classified as type I and type II based on their sequence and structural features (Sanmartín et al., 2005). During the somatic embryogenesis of Norway spruce [Picea abies (L.) Karst], a type II metacaspase, mcII-Pa, is required for the burst of the caspase activity that occurs in the cells undergoing PCD (Suarez et al., 2004).
The ability to self-destruct in response to environmental changes and cell signalling is shared by animal and plant cells, which indicates that PCD may have played an essential role in the development, survival and evolution of most, if not all, multicellular organisms (Ameisen, 2002). The development, reproduction, ecology, and genetics of the Pinus species are well documented, and they provide a model system for the gymnosperms, an evolutionary old group of vascular plants that last shared a common ancestor with angiosperms about 285 million years ago (Bowe et al., 2000; Lev-Yadun and Sederoff, 2000). The pine seed has been suggested as a model system for studying the origin and evolution of eukaryotic PCD machinery because natural cell death is essential for embryogenesis and seed development in gymnosperm plants (Zhivotovsky, 2002). The study of Filonova et al. (2002) suggested that PCD is the major mechanism responsible for the elimination of subordinate embryos in the Scots pine (Pinus sylvestris L.) seed and, once the dominant embryo is selected, the entire megagametophyte, a maternally derived haploid tissue, is also affected by PCD.
Living organisms are subject to a wide variety of genotoxic stresses arising from exposure to DNA damaging agents or from cell metabolism. The DNA lesions that result from these insults are rapidly detected in cells and responded to by the activation of an intricate web of signalling pathways that, depending on the type of the DNA damage, culminates in the activation of cell-cycle checkpoints and the appropriate DNA repair pathways or the initiation of PCD (Barzilai and Yamamoto, 2004). The DNA double helix can suffer damage to just one or to both of the strands. The lesions affecting only one strand (single-strand break, SSB) can be repaired using the intact complementary strand as a template, but in the case of DNA double-strand breaks (DSB), both strands are altered and the cell has no intact strand to direct repair. The mutation rate in long-lived plant species such as Pinus species is not, however, unexpectedly high, which indicates that the activities responsible for maintaining genome integrity must be efficient in plant somatic cells (Ann et al., 2007).
The repair of DSB is achieved either by homologous recombination (HR) involving extensive DNA sequence homology between the interacting molecules or by other straightforward pathways simply rejoining the two ends of the break, i.e. non-homologous recombination (NHR) (Bleuyard et al., 2006). Although the efficiencies of the two DSB repair pathways differ, the pathways themselves are well conserved among yeast, vertebrates, and Arabidopsis, and many of the DNA repair genes show a high level of sequence similarity (Bleuyard et al., 2006). In plants, HR is initiated by extensive resection to produce long 3′ tails and then followed by the formation of Rad51 filaments promoting the homology search and strand invasion. Non-homologous end joining (NHEJ) is catalysed by a complex of Ku70 and Ku80 that binds to double-stranded DNA ends to protect them from degradation, to align them before joining, and to promote the fastening of additional repair proteins before the ligation by a complex of DNA ligase IV and XRCC4 (Bray and West, 2005). The plant seed may provide a model system for studying the effects of a variety of endogenous DNA damaging agents and environmental stresses on genome integrity. From the early stages of development on the mother plant, the seed experiences developmentally programmed as well as environmental stresses, which are potentially damaging to the genome (Bray and West, 2005).
The fertilization process and the structure of the female gametophyte are the distinctive features of the two phylogenetic groups of seed plants. Unlike angiosperms, gymnosperms display single fertilization. Thus, their megagametophyte undergoes development before fertilization and represents a significant investment of maternal resources, which are wasted if fertilization does not occur (Steeves, 1983). Another hallmark feature of the gymnosperms’ seed development is polyembryony, which can be either monozygotic or caused by the fertilization of two or more egg cells within the same ovule. Polyembryony is, however, a transient stage during seed development and the opportunity for embryo replacement is brief. During the progress of embryogenesis, the growth of subordinate embryos is retarded. Finally, only one embryo survives, while the others are eliminated (Filonova et al., 2002). In gymnosperms, the overall embryo development pathway can be divided into three distinct phases, called proembryogeny, early embryogeny, and late embryogeny. Proembryogeny includes the stages before the elongation of the suspensor system. Early embryogeny initiates with the elongation of the suspensor system and terminates with the appearance of the root meristem. Late embryogeny culminates with the maturation of the embryo (Singh, 1978). Embryos grow and develop within the corrosion cavity of the megagametophyte, which houses the majority of the storage reserves of the seed (King and Gifford, 1997).
Developmental PCD of cereal endosperm, the functional homologue of the megagametophyte, is a commonly acknowledged phenomenon (Young et al., 1997; Young and Gallie, 1999), and the Scots pine megagametophyte has also been suggested to be affected by PCD during embryo development (Filonova et al., 2002). In the present study, it is shown that a major part of the Scots pine megagametophyte cells stay alive and the megagametophyte is viable and metabolically active tissue from the early phases of embryo development until the imbibition phases of mature seeds. However, specific megagametophyte cells in the embryo surrounding region (ESR) face sudden destruction with apparent necrotic features during embryogenesis. Our results suggest that developmentally programmed necrotic cell death or PCD with necrotic morphology plays an important role in Scots pine embryogenesis.
Materials and methods
Collection of immature and mature Scots pine seeds
One-year-old immature seed cones were collected from two open-pollinated elite Scots pine (Pinus sylvestris L.) clones during one growing season. The elite clones, K818 and K884, were grafted in the Scots pine clone collection in Punkaharju, Finland (61°48′ N, 29°17′ E), and one representative graft was selected for cone collections for both clones. The collection was repeated four times in July throughout the period of embryo development. The cones were collected on 5 July (sampling date I), 12 July (sampling date II), 19 July (sampling date III), and 26 July (sampling date IV). In gymnosperms including Scots pine, zygotic embryo development depends on the effective temperature sum (d.d.) (i.e. the heat sum unit based on the daily mean temperatures minus the adapted +5 °C base temperature) (Sirois et al., 1999; Vuosku et al., 2006). On the sampling dates I, II, III, and IV, the effective temperature sums were 436.3, 509.1, 587.4, and 678.9 d.d., respectively. Immature seeds were dissected from the developing cones and the seed coat was removed. For the microscopic examination, the seeds were fixed immediately and embedded in paraffin, as described in a previous paper (Vuosku et al., 2006). For the gene expression studies, the seeds were frozen in liquid nitrogen and stored at -80 °C until use.
Mature cones were collected from the same grafts as the immature cones and represented the Scots pine clones K884 and K818. The cones were collected and seeds extracted in late autumn of the same growing season. Mature seeds were used for the DNA extraction, seed viability test, and for in vitro culturing of the megagametophyte tissue.
Nucleic acid detection
The paraffin sections were dewaxed in Histochoice (Sigma) and rehydrated through a graded series of ethanol. For the observation of nucleic acids, the sections were stained by a dual fluorescence dye, acridine orange (1.6 mM), according to Bouranis et al. (2003) and examined with a microscope (Laser Scanning Microscope LSM 5 Pascal, Carl Zeiss) using an HBO 103 mercury lamp. Adobe Photoshop CS was used to adjust contrast, brightness, and colour uniformly to entire images.
TUNEL assay
TUNEL (terminal deoxyribonucleotidyl transferase (TdT)-mediated deoxyuridine triphosphate (dUTP) nick end labelling) assay was used for the in situ detection of nuclear DNA fragmentation. The dewaxed and rehydrated sections were digested with 10 μg ml−1 proteinase-K (Roche) for 30 min and washed twice with PBS buffer. The sections were labelled with the TMR red (red fluorescence) in situ cell death detection kit (Roche) according to the manufacturer's protocol. Prior to the labelling procedure, the positive control sections were incubated with recombinant DNase I (Roche) to induce DNA strand breaks. The label solution without terminal transferase, instead of the TUNEL reaction mixture, was used for the negative controls. The labelled sections were examined in a confocal laser scanning microscope (LSM 5 Pascal, Carl Zeiss) with an HBO 103 mercury lamp and using the HeNe laser 543 nm line, dichroic beam splitter (HFT 488/543/633; Carl Zeiss), and LP 560 emission filter (Carl Zeiss).
Seed viability test
The tetrazolium test that is based on the reduction of tetrazolium salts to red coloured endproducts in viable tissues was performed by using ISTA (International Seed Testing Association) prescriptions (1993) modified by Savonen (1999). Dry, mature seeds were transversally cut at both ends before immersing them into a tetrazolium staining solution (1% 2,3,5-triphenyl tetrazolium chloride in 67 mM phosphate buffer) in a vacuum desiccator. Vacuum was induced three times by a vacuum pump to accelerate the staining of the seeds, which were then incubated for 4 h at 30 °C in the staining solution. Seeds, boiled for 30 min before the tetrazolium test, were used as a negative control.
Megagametophyte in vitro culture
Mature seeds were surface-sterilized for 12 h in 3% (v/v) PPM™ solution (Preservative for Plant Tissue Culture, Plant Cell Technology) and imbibed for 2 d on filter paper moistened with distilled water. The seeds were opened with a scalpel, and the embryos and megagametophytes were separated from each other. 112 megagametophytes in total were cultured on basal DCR medium (Gupta and Durzan, 1985) modified by Becwar et al. (1990) and gelled by 1.25 g l−1 Phytagel™ (Sigma) at room temperature in the dark for 10 d. Thereafter the number of proliferating megagametophytes was documented.
Nucleic acid extraction
Genomic DNA was extracted to test DNA integrity by the NucleoSpin® Plant kit (Macherey-Nagel) according to the manufacturer's protocol for gymnosperms. DNA was extracted from the immature seeds on the sampling dates I, II, III, and IV as well as from the embryos and megagametophytes of imbibed mature seeds. DNA (2 μg from each sample) was electrophoretically separated on a 2% agarose gel, stained with ethidium bromide and visualized and photographed under UV light.
Total RNA was extracted for the gene expression studies from immature seeds using the automated magnetic-based KingFisher™ ml method (Thermo Electron Corporation) with MagAttract® RNA Tissue Mini M48 kit (Qiagen) according to the manufacturer's instructions. In addition to RNA binding and washing steps, a DNase treatment for the elimination of contaminating genomic DNA was included in the extraction procedure. The RNA samples were precipitated with ethanol, and the RNA yields were measured three times with OD260 analysis using GeneQuant pro (Amersham). RNA integrity was checked by agarose gel electrophoresis. Five independent RNA extractions were done per clone and per sampling date, except for the seeds of the clone K884 on sampling date III and IV when only two and four intact RNA samples, respectively, could be extracted.
PCR primer designing
The PCR primers for the amplification of the cDNA fragments of the putative Scots pine RAD51, KU80, LIG, MCA, and TAT-D were designed based on the Pinus EST sequences. EST-sequences were selected on the basis of the similarity at the nucleotide or amino acid level with the genes isolated earlier from other plant species. The query sequences used for the BLAST searches in the EST database of the NCBI GeneBank were Arabidopsis Rad51 protein (NP_568402), Arabidopsis Ku80 homologue protein (NP_564520), Arabidopsis DNA ligase IV protein (AAF91284), Picea abies mRNA for metacaspase type II (AJ534970), and Arabidopsis Tat-D-related deoxyribonuclease family protein (NP_974418). The Pinus EST sequences found were aligned together to yield either an entire protein coding sequence of the corresponding gene or a cDNA fragment that was as long as possible. The PCR primers used for the amplification of the cDNA fragments are presented in Supplementary Table S1 at JXB online.
The PCR primers for the expression studies of the putative Scots pine RAD51, KU80, LIG, MCA, and TAT-D were designed on the basis of the sequence data discovered in this study. Sequence data can be found in the GenBank data libraries under accession numbers EU513162, EU513164, EU513165, EU513166, and EU513167. The primers for the expression studies of the housekeeping genes ACT, UBI, and GAPDH were based on the lodgepole pine (Pinus contorta) ACT sequence (M36171), maritime pine (Pinus pinaster) putative UBI sequence (AF461687), and Scots pine GAPDH sequence (L07501), respectively. The PCR primers for the gene expression studies are presented in Supplementary Table S2 at JXB online.
RT-PCR and cDNA cloning
The reverse transcription step was primed using anchored oligo-dT primers. cDNA was reverse-transcribed from 1 μg of total RNA by SuperScript II reverse transcriptase (Invitrogen) using standard methods in a reaction volume of 20 μl. Fragments of the putative Scots pine RAD51, KU80, LIG, MCA, and TAT-D were amplified by standard PCR using zygotic embryo cDNA as a template and DyNAzyme™EXT polymerase (Finnzymes). The fragments with appropriate length were gel-purified using a Montage DNA Gel Extraction Kit (Millipore Corporation) and cloned using a TOPO TA Cloning Kit (Invitrogen). Five clones from each gene fragment were sequenced using an Applied Biosystems 3730 DNA analyser. The sequences were aligned and the predicted protein sequences were submitted to search for conserved domains (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Real-time RT-PCR analyses of mRNA transcripts
A first-strand cDNA template was synthesized from 1 μg of total RNA as described above. The PCR amplification conditions of the gene fragments were optimized for the LightCycler® 2.0 instrument (Roche Diagnostics), and the subsequent PCR runs showed a single PCR product during melting curve and electrophoretic analysis. The real-time PCR amplifications were performed using the FastStart DNA Master SYBR Green I real-time PCR kit (Roche Molecular Biochemicals), 50 nM gene-specific primers and 2 μl cDNA (1:10 dilution) in the reaction volume of 20 μl. The real-time PCR amplification was initiated by incubation at 95 °C for 10 min followed by 40 cycles of 10 s at 95 °C, 10 s at 58 °C, and 5 s at 72 °C. For removing possible differences in the amount of starting material and enzymatic efficiencies, the geometric mean of three selected reference genes (ACT, UBI, and GAPDH) belonging to different functional classes was used for the normalization of the gene expression levels (Vandesompele et al., 2002).
Statistical analyses
The development of RAD51, KU80, LIG, MCA, and TAT-D expressions, each as a function of an effective temperature sum, were analysed by fitting an ordinary regression model for the logarithm of relative expression (LRE) against the linear term of the effective temperature sum (T), scaled per 100 d.d. Separate slopes for the regression lines were estimated for the two clones (C), and the assumption of a common slope was evaluated by adding an appropriate interaction term C×T in the model and investigating its error margin. In each case, the goodness-of-fit of the simple linear model was assessed against a model allowing curvature in the functional relationship between T and LRE by including both a 2nd order and a 3rd order polynomial term of T in the model formula.
The models were fitted using function lm() in the R statistical environment version 2.6.2 (R Development Core Team 2008). Assessment of the common model assumptions was based on an informal inspection of the four diagnostic graphs provided by the default plot methods for the fitted model objects. The results were summarized by giving the point estimates and the 95% confidence intervals (CI) of the exponentiated regression coefficients associated with T for both clones from the linear model in each case. These quantities represent the estimated relative increase or average rate of change (and the associated error margin) in the pertinent gene expression per an increase of 100 d.d. in the effective temperature sum. When presenting these summaries in Table 1, possible deviations from linearity are ignored but commented in the text if needed.
Table 1.
Estimated average relative changes in expressions (with 95% confidence intervals, CI) per 100 d.d. increase in the effective temperature sum for the DNA repair and PCD related genes (RAD51, KU80, LIG, MCA, and TAT-D) during embryo development in the Scots pine clones K818 and K884
| K818 |
K884 |
|||
| Estimate | 95% CI | Estimate | 95% CI | |
| RAD51 | 0.67 | 0.50–0.90 | 0.49 | 0.36–0.68 |
| KU80 | 0.91 | 0.71–1.15 | 0.85 | 0.66–1.10 |
| LIG | 0.96 | 0.81–1.15 | 0.94 | 0.77–1.14 |
| MCA | 0.97 | 0.77–1.22 | 0.66 | 0.51–0.85 |
| TAT-D | 0.80 | 0.67–0.97 | 0.62 | 0.51–0.76 |
Results
Occurrence of PCD with necrotic morphology during Scots pine zygotic embryogenesis
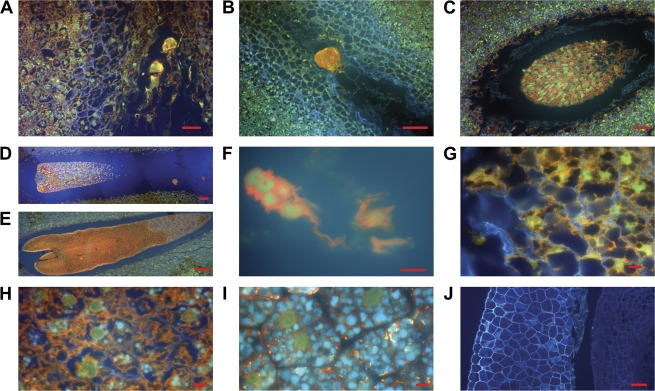
In the present study, acridine orange, a dual fluorescence dye, was applied in order to detect nucleic acids in the embryo and megagametophyte cells during Scots pine embryogenesis. The embryos were at the early embryogeny stage (Fig. 1A, B) before the appearance of the root meristem. During late embryogeny, the dominant embryo was maturating (Fig. 1C), whereas the growth of the subordinate embryos was retarded, and they fell more and more behind the leading embryo (Fig. 1D). Finally, the subordinate embryos disappeared and the leading embryo occupied the corrosion cavity (Fig. 1E). In the dying subordinate embryos, the cells broke down, resulting in a leakage of nucleic acids into the surrounding extracellular space (Fig. 1F).
Fig. 1.
Acridine orange stained Scots pine embryos and megagametophyte tissue during embryogenesis. The double-stranded nucleic acids (i.e. DNA) fluoresce green and the single-stranded (i.e. RNA) fluoresce red. (A) Dominant and subordinate early embryos in the corrosion cavity on sampling date I. (B) Dominant early embryo in the corrosion cavity on sampling date II. (C) Dominant late embryo in the corrosion cavity on sampling date III. (D) Dominant late embryo and the dying subordinate embryos in the corrosion cavity on sampling date III. (E) Late embryo filling the corrosion cavity on sampling date IV. (F) Dying cells of subordinate embryos broke down, resulting in a leakage of nucleic acids into the surrounding extracellular space. (G) During embryo development, the cell wall, plasma membrane, and nuclear envelope of the megagametophyte cells lining the corrosion cavity broke down with the release of cell debris and nucleic acids into the corrosion cavity (early embryogeny, sampling date II). (H) During early embryogeny, nuclei in megagametophyte cells stained green, except for rRNA containing nucleoli, which were red. In the cytoplasmic region, the red colour indicated the presence of mRNA and active gene expression. (I) During late embryogeny, nuclei in megagametophyte cells appeared normal with the presence of nucleoli and with no sign of DNA fragmentation. (J) Control sample with no acridine orange staining. Bars: (G, H, I) 10 μm, (F) 20 μm, (A, C, J) 50 μm, and (B, D, E) 100 μm.
In the megagametophyte, the ESR lining of the corrosion cavity could easily be distinguished from the rest of the megagametophyte tissue. During all the developmental stages, the megagametophyte cells in ESR were destroyed by cell death that showed morphologically necrotic features. Their cell wall, plasma membrane and nuclear envelope broke down with the release of cell debris and nucleic acids into the corrosion cavity (Fig. 1G; see Supplementary Fig. S1A, B, and C at JXB online). Except for the megagametophyte cells in ESR, the rest of the megagametophyte tissue stayed alive with no symptoms of PCD or necrosis throughout embryo development. There were signs of mRNA and active gene expression in the cytoplasm of the megagametophyte cells (Fig. 1H, I; see Supplementary Fig. S1D and E at JXB online).
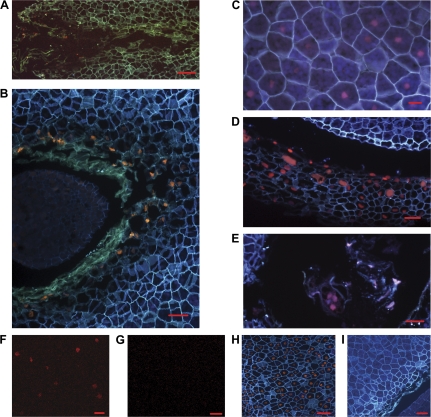
Nuclear DNA fragmentation, i.e. DNA strand breaks leaving free 3′-OH termini, was shown by the TUNEL assay. TUNEL-positive nuclei were detected in the cells that displayed morphological features of necrotic cell death in ESR and in the arrow-shaped megagametophyte region in front of the dominant embryo throughout the development of the embryo (Fig. 2A, B). By contrast, no TUNEL-positive cells were found in the inner parts of the megagametophyte before the developmental stage of late embryogeny, when a weak signal could be detected (Fig. 2C). Furthermore, TUNEL-positive nuclei were also found in the cells of the nucellar layer covering the megagametophyte (Fig. 2D) as well as in the degrading suspensor cells (Fig. 2E). The TUNEL signals were located identically when examined by fluorescence microscopy as well as excitation at 543 nm (Fig. 2F, G).
Fig. 2.
Nuclear DNA fragmentation detected by TUNEL assay in Scots pine seed sections. (A) TUNEL-positive signals in the megagametophyte in the vicinity of the corrosion cavity at the early embryogeny stage. (B) TUNEL-positive signals in the megagametophyte in the arrow-shaped region in front of the embryo at the late embryogeny stage. (C) Slightly TUNEL-positive nuclei appeared in the inner part of the megagametophyte during the late embryogeny stage (sampling date IV). (D) TUNEL-positive nuclei in the nucellar layer at the late embryogeny stage. (E) TUNEL-positive nuclei in the suspensor cells at the early embryogeny stage. (F) TUNEL-positive nuclei detected by excitation at 543 nm in the inner part of the megagametophyte at the late embryogeny stage. (G) Lack of DNA fragmentation detected by excitation at 543 nm in the inner part of the megagametophyte at the early embryogeny stage. (H) Positive control (DNase treatment). (I) Negative control (omission of TdT). Bars: (C, D, F) 20 μm, (B, E, G, H, I) 50 μm, and (A) 100 μm.
Intranuclear DNA fragmentation in megagametophyte tissue is not a hallmark of PCD
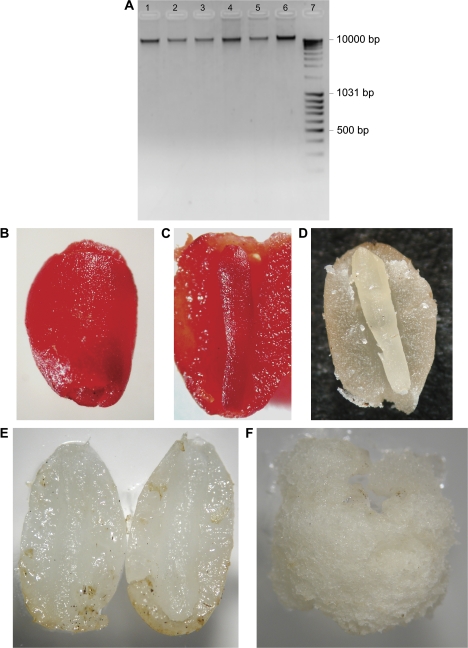
Although slightly TUNEL-positive nuclei accumulated in the megagametophyte tissue at late embryogeny, no fragmentation was observed in the DNA which was extracted from the developing Scots pine seeds. Moreover, both the megagametophyte and embryo DNA also looked intact in mature seeds after 2 d of imbibition (Fig. 3A).
Fig. 3.
Intact DNA of immature as well as mature Scots pine seeds and viable, proliferating megagametophytes of mature seeds. (A) The agarose gel shows intact DNA extracted from immature Scots pine seeds on sampling date I (lane 1), sampling date II (lane 2), sampling date III (lane 3), and sampling date IV (lane 4) as well as from the embryos (lane 5) and megagametophytes (lane 6) of mature seeds after 2 d imbibition. (B) Megagametophyte tissue of mature Scots pine seed was tested for viability using tetrazolium. The megagametophyte showed deep red when the seed coat was removed after the tetrazolium test. (C) Both the embryo and the megagametophyte were deep red in the mature seed, split in half after the tetrazolium test. (D) Tetrazolium test performed on mature seed boiled for 30 min as a negative control. (E) At the beginning of the proliferation test, embryos were excised from mature seeds and the halves of the megagametophytes were set on DCR medium. (F) After 10-d culture megagametophytes showed proliferation.
According to the tetrazolium test, the megagametophytes of the imbibed mature seeds were alive. In the viable seeds, both the megagametophyte and the embryo stained strongly red, indicating metabolic activity (Fig. 3B, C), whereas the artificially destroyed, boiled control seeds remained white (Fig. 3D). About 60% of the mature megagametophytes were able to proliferate on the tissue culture medium when embryos were excised (Fig. 3E, F), which confirmed that mature megagametophytes are viable tissues in which cells are able to proliferate when propagated in vitro.
DNA repair and PCD related genes in Scots pine
Due to the fact that the survival of organisms depends on the accurate transmission of genetic information from one cell to its daughters, the mechanisms responsible for DNA integrity show remarkable evolutionary conservation across kingdom borders (Zhou and Elledge, 2000). In this study, five Scots pine genes related to DNA repair or PCD were identified.
Among the best known HR genes are the Escherichia coli recA gene and its eukaryotic homologues Rad51s (Lin et al., 2006). A 528 bp long cDNA fragment of the putative Scots pine RAD51 was sequenced. The predicted 176 amino acid protein sequence contained a Rad51 domain structure and showed 91–93% identity to the Rad51 proteins of other plant species (see Supplementary Fig. S2 at JXB online).
NHEJ is primarily responsible for the repair of DSBs in higher organisms, and the mechanism has been highly conserved throughout eukaryotic evolution. The process is mediated by the DNA-dependent protein kinase (DNA–PK) complex that contains a Ku70-Ku80 heterodimer and a catalytic subunit and the DNA ligase IV–XRCC4 complex (Critchlow and Jackson, 1998). A 672 bp fragment of the putative Scots pine KU80 and a 505 bp fragment of the putative Scots pine LIG were sequenced. The predicted 224 amino acids long Ku80 sequence contained a Ku70/Ku80 N-terminal domain and was 50% identical to the Arabidopsis Ku80 homologue and 56% identical to the Vigna radiata Ku80-like protein (see Supplementary Fig. S3 at JXB online). On the basis of the sequence similarity as well as the conserved functional domain, it is likely that the cDNA fragment obtained was part of the Scots pine KU80. However, the isolated fragment also showed high similarity to plant H-box binding proteins, i.e. KAP2 proteins (see Supplementary Fig. S3 at JXB online). The predicted 168 amino acids sequence of the Scots pine DNA ligase was 83% identical to the putative Oryza sativa DNA ligase (see Supplementary Fig. S4 at JXB online) and included the ATP dependent DNA ligase C terminal region, which suggests that the isolated cDNA fragment belonged to a member of the DNA ligase family.
Previously, metacaspase (MCA) has been shown to have a role in the activation and/or execution of PCD in plants (Suarez et al., 2004). In this study a 1257 bp long cDNA fragment covering the coding sequence of the putative Scots pine MCA was sequenced. The predicted metacaspase protein of 418 amino acids contained caspase domain structures. The alignment of the Scots pine metacaspase with plant metacaspase protein sequences revealed that the predicted protein was 89% identical to the Norway spruce type-II metacaspase, which suggests that the Scots pine protein was a type-II metacaspase (see Supplementary Fig. S5 at JXB online).
Tat-D is an evolutionarily conserved apoptotic nuclease that exists in organisms across all kingdoms (Qiu et al., 2005). An 813 bp fragment of the putative Scots pine TAT-D was sequenced. The predicted 270 amino acid long protein sequence contained Tat-D nuclease domain and showed 69% identity to the Oryza sativa Tat-D nuclease and 52% identity to Homo sapiens a Tat-D nuclease, which implies that the isolated fragment was a part of the Scots pine Tat-D nuclease (see Supplementary Fig. S6 at JXB online).
DNA repair and PCD related gene expression during embryo development
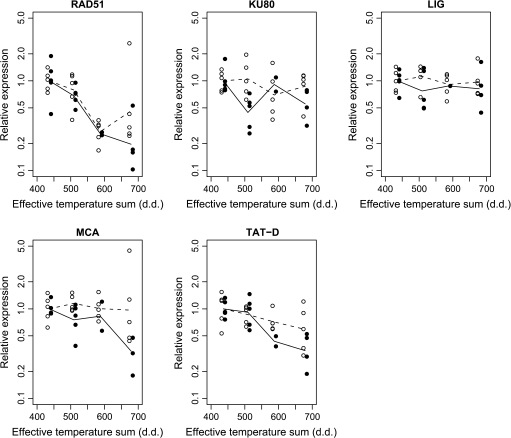
The cell death process and DNA fragmentation were subsequently studied by investigating the expression levels of the putative Scots pine RAD51, KU80, LIG, MCA, and TAT-D genes. The relative gene expressions by the effective temperature sum (d.d.) are presented in Fig. 4, and the estimated average relative changes in gene expressions by each increase of 100 d.d. are reported in Table 1. During Scots pine embryo development RAD51 expression was suppressed in both clones (33% and 50% decrease per 100 d.d. in K818 and K884, respectively) with no evidence of difference in the average rates of changes between the clones. However, there was some indication of deviation from linearity mainly due to one outlying observation. For the KU80 and LIG genes, no evidence of any trend in the expression profiles was found. MCA gene expression showed a downward trend (34% decrease per 100 d.d) in pine clone K884, whereas in clone K818 the trend was not as clear, maybe due to one outlier observation. The omission of the outlier observation produced an estimate for the average rate of change (15% decrease per 100 d.d.), which was statistically consistent with assuming a common slope for both clones. Likewise, TAT-D gene expression had a downward trend in both clones (20% and 38% decrease per d.d. in K818 and K884, respectively), which was also consistent with a common slope assumption.
Fig. 4.
The expression of the putative RAD51, KU80, LIG, MCA, and TAT-D genes in developing Scots pine seeds. The expressions were quantified by real-time PCR and normalized by the expression derived from the housekeeping genes ACT, UBI, and GAPDH. The observed relative gene expressions are presented in relation to the effective temperature sum (d.d.) in the clones K818 (open circles) and K884 (closed circles), and the geometric mean values of the replicates pertaining to the same clone and sampling date are connected by dashed lines for K881 and solid lines for K884. There are five replicates per clone and per sampling date, except for the seeds of clone K884 on sampling dates III and IV, from which there are two and four replicates, respectively.
Discussion
In gymnosperms the megagametophyte accumulates nutrients before fertilization and nourishes the developing embryo after fertilization. The megagametophyte of gymnosperms and the endosperm of angiosperms, characterized by haploid and triploid genomes and produced before and after fertilization, respectively, can be considered as functional homologues due to their role as a nutrient source of the developing embryo (Costa et al., 2004). PCD in the endosperm is well described especially in cereals, maize (Young et al., 1997) and wheat (Young and Gallie, 1999). The PCD programmes exhibit both common and unique elements, but in both species PCD is accompanied by an increase in nuclease activity and the internucleosomal degradation of nuclear DNA. In gymnosperms, there are only a few reports on PCD occurring in the megagametophyte tissues. In white spruce [Picea glauca (Moench) Voss] seeds, the megagametophyte undergoes PCD during post-germinative seedling growth and is also characterized by nuclear fragmentation, internucleosomal DNA cleavage and sequential activation of several nucleases (He and Kermode, 2003). In the present study, it is shown that Scots pine megagametophyte cell destruction via PCD or necrotic cell death occurs only in the ESR during embryo development and the imbibition phase of the mature seed germination.
The DNA ladder is a very specific hallmark of PCD in both animal and plant cells, and it occurs by an activation of the endogenous endonucleases that cleave the DNA in the linker region between histones on the chromosomes. On the other hand, in necrosis, DNA degrades randomly and results in a smear (Bortner et al., 1995; Danon et al., 2000). The TUNEL assay is a method for detecting PCD via DNA fragmentation in various tissues. The method takes advantage of the multiple free 3′ hydroxyl termini that are generated by activated endonucleases to insert labelled dUTP, which can be detected by microscopy. In our study, nuclear DNA fragmentation was found in the Scots pine seed in the cells of the nucellar layer and in the degrading suspensor tissue, the results of which are in agreement with previous reports revealing that both tissues are dying via PCD during pine seed development (Filonova et al., 2002; Hiratsuka et al., 2002). Moreover, a TUNEL-positive signal was located in the megagametophyte at the ESR layer that contained broken cells. At the developmental stage of late embryogeny, it was also possible to find some slightly TUNEL-positive nuclei in the inner part of the megagametophyte. Thus, our results do not support the results reported by Filonova et al. (2002), in which the entire Scots pine megagametophyte was affected by DNA fragmentation shortly before the first signs of PCD in the subordinate embryos. Filonova et al. (2002) suggested that once the dominant embryo was selected, the entire megagametophyte was affected by PCD, and that it required approximately 2 weeks to programme all the megagametophyte cells to die. Despite slight DNA fragmentation at late embryogeny, neither other morphological signs of PCD in the cells of the inner part of the megagametophyte nor a DNA ladder in the DNA samples extracted from the immature and mature seeds were found. Conversely, in the inner part of the megagametophyte most of the cells appeared viable, with the presence of nucleoli also during late embryogeny. Previously, the presence of a visible nucleolus has been connected with the transcriptional activity in a cell (Medina et al., 2000; Pasternak et al., 2001). In the present study, the mRNA levels of the putative MCA or TAT-D genes decreased during late embryogeny. This suggested that no large-scale PCD or nucleic acid fragmentation occurred in megagametophyte tissue, but might also reflect the increasing contribution of the more developed embryo to the total amount of mRNA. Moreover, the megagametophyte tissue of mature seeds was both metabolically active and capable of in vitro proliferation, which confirms that the free 3′ hydroxyl termini in nuclear DNA during late embryogeny were not a hallmark of PCD. In the loblolly pine (Pinus taeda L.) seed, the megagametophyte has also been found to be a living metabolically active tissue that interacts with the germinating embryo and young seedling (Brownfield et al., 2007).
Currently, the TUNEL assay is one of the most frequently used methods to detect PCD and it has provided valuable information about PCD in various tissues, even though the sensitivity and specificity of the technique have been criticized. False-positive TUNEL staining has been detected in mouse kidney and liver tissues (Pulkkanen et al., 2000), and in plant tissues, fixation and embedding have been reported to cause sufficient nicking of nuclear DNA to produce false TUNEL positive nuclei (Wang et al., 1996). The TUNEL method is presumed to be unable to discriminate apoptotic from necrotic cells, given that the latter also have free DNA ends (Kelly et al., 2003). In our study, the possibility that the positive TUNEL labelling would have been caused by sample preparation was ruled out by positive and negative controls as well as by associating the TUNEL assay with the time-course data. Thus, it is proposed that the positive labelling in the TUNEL assay in the inner parts of the megagametophyte at the developmental stage of late embryogeny may be a consequence of DNA strand breaks caused by maturation drying or by the DNA breaks with free 3′-OH ends that appear during DNA repair. The decrease in RAD51 expression and the constant expression of KU80 and LIG suggest that, during late embryogeny, the proportion of mitotic cells decreased and the DNA breaks were mainly repaired by the NHEJ pathway. The maintenance of genetic information during the seed dehydration and rehydration cycle is essential for cell survival (Osborne et al., 2002). Both DNA stability on dehydration and the ability for DNA repair on rehydration are well-known features of desiccation-tolerant seeds (Boubriak et al., 1997).
The exact function of the ESR is still unknown, but several roles have been suggested for the endosperm ESR cells. They may function in embryo nutrition, establish a physical barrier between the embryo and the endosperm during seed development, or provide a zone for communication between the embryo and the endosperm (Olsen, 2004). In gymnosperms, the corrosion cavity is filled with fluid, serving as a nutritional and hormonal interphase between the developing embryos and megagametophyte (Carman et al., 2005). Dying cells were found in the Scots pine seed in the ESR of the megagametophyte as well as in the subordinate embryos, i.e in the tissues that are closely apposed to the leading embryo and in contact with the corrosion cavity fluid. In both cases, the cells were not able to maintain their discrete identity from the environment but broke down with the release of cell debris and fragmented nucleic acids into the corrosion cavity. The result implies that the cell death signal transmitted via the corrosion cavity fluid may originate from the leading embryo making space for growth and utilizing the megagametophyte and subordinate embryos for its own nutrition. In the Scots pine seed, there is a physiological interaction between the embryo and the megagametophyte, since the existence of an embryo appeared to be the prerequisite for the megagametophyte to stay alive (Plym Forshell, 1974). After fertilization, the maternal-to-zygotic transition entails an extensive reprogramming of gene expression and an establishment of an embryo-specific developmental programme. The timing and mechanisms of this major developmental switch in higher plants are still unclear and controversial, and there are no reports concerning gymnosperm seeds. In maize, the maternal-to-zygotic transition is suggested to occur several days after fertilization (Grimanelli et al., 2005).
PCD is a generic term for different programmes of active cell destruction, and so far at least four morphologically distinct groups of genetically encoded cell death, i.e. apoptosis, autophagy, oncosis, and pyroptosis, have been identified in dying eukaryotic cells (Fink and Cookson, 2005). There is also growing evidence of physiological cell death with necrotic-like morphology (Kitanaka and Kuchino, 1999). Recently, developmental stimuli-induced necrotic cell death was found in Dictyostelium discoideum, a protist that emerged in evolution after plants and from an ancestor common to fungi and animals (Laporte et al., 2007). Our results suggested that the developmental and physiological cell death with necrotic-like morphology exists also in plants and that the mechanism underlying necrotic-like cell death is evolutionarily old. Gymnosperms belong to the most ancient seed plants, dating back to the Devonian period (400–360 million years ago) (González-Martínez et al., 2006). Necrotic cell death has, in effect, been revealed to be a heterogeneous entity containing both active and passive cell death and, almost certainly, such developmentally programmed cell deaths should, in a broad sense, be under genetic control (Kitanaka and Kuchino, 1999). In Scots pine seed the unique occurrence of developmentally regulated necrotic-like cell death in a maternally derived haploid tissue provides a useful model for the further study of the genetic control of necrotic-like cell death. An interesting question is whether the genetic control of developmentally programmed necrotic cell death resides in the dying cell itself or, as we propose in the case of the Scots pine megagametophyte, outside the cell, i.e. in the developing embryo, which utilizes the megagametophyte for its own nutrition.
Supplementary data
See Supplementary data at JXB online.
Supplementary Table S1. PCR primers for the amplification of the cDNA fragments of the putative Scots pine RAD51, KU80, LIG, MCA, and TAT-D.
Supplementary Table S2. PCR primers for the real-time PCR.
Supplementary Fig. S1. Acridine orange-stained Scots pine megagametophyte tissue during embryogenesis.
Supplementary Fig. S2. Alignment of the partial amino acid sequence of the putative Scots pine Rad51 with the Rad51 sequences of other plant species.
Supplementary Fig. S3. Alignment of the partial amino acid sequence of the putative Scots pine Ku80 with the Ku80 sequences of other plant species.
Supplementary Fig. S4. Alignment of the partial amino acid sequence of the putative Scots pine LIG with the LIG sequences of other plant species.
Supplementary Fig. S5. Alignment of the amino acid sequence of the putative Scots pine MCA with the MCA sequences of other plant species.
Supplementary Fig. S6. Alignment of the partial amino acid sequence of the putative Scots pine Tat-D nuclease with the Tat-D sequences of other species.
Supplementary Material
Acknowledgments
We thank our colleagues Dr Anneli Kauppi from the University of Oulu and Eira-Maija Savonen and Dr Eila Tillman-Sutela from the Finnish Forest Research Institute for their help, enthusiasm, and interest during the work. We are also grateful to Ms Eeva Pihlajaviita, Finnish Forest Research Institute, for technical help, and to the personnel of the Finnish Forest Research Institute at the Punkaharju Research Unit for conducting the collections of the research material. The research was funded by the Academy of Finland (Project 121994 to TS and Senior Scientist grant 120146 to EL), by the Biological Interactions Graduate School (to JV and SS), and by grants from the Oulu University Scholarship Foundation, the Taina Kuusi Foundation, the Finnish Cultural Foundation, and the Niemi Foundation (to JV).
References
- Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death and Differentiation. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- Ann W, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Molecular Biology and Evolution. 2007;24:90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- Barzilai A, Yamamoto KI. DNA damage responses to oxidative stress. DNA Repair. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Becwar MR, Nagmani R, Wann SR. Initiation of embryogenic cultures and somatic embryo development in loblolly pine (Pinus taeda) Canadian Journal of Forest Research. 1990;20:810–817. [Google Scholar]
- Beers EP. Programmed cell death during plant growth and development. Cell Death and Differentiation. 1997;4:649–661. doi: 10.1038/sj.cdd.4400297. [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI. Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair. 2006;5:1–12. doi: 10.1016/j.dnarep.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Bortner CD, Oldenburg NBE, Cidlowski JA. The role of DNA fragmentation in apoptosis. Trends in Cell Biology. 1995;5:21–26. doi: 10.1016/s0962-8924(00)88932-1. [DOI] [PubMed] [Google Scholar]
- Boubriak I, Kargiolaki H, Lyne L, Osborne DJ. The requirement for DNA repair in desiccation tolerance of germinating embryos. Seed Science Research. 1997;7:97–105. [Google Scholar]
- Bouranis DIL, Chorianolopoulou SN, Siyiannis VF, Protonotarios VE, Hawkesford MJ. Aerenchyma formation in roots of maize during sulphate starvation. Planta. 2003;217:382–391. doi: 10.1007/s00425-003-1007-6. [DOI] [PubMed] [Google Scholar]
- Bowe LM, Coat G, dePamphilis CW. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales’ closest relatives are conifers. Proceedings of the National Academy of Sciences, USA. 2000;97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray CM, West CE. DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytologist. 2005;168:511–528. doi: 10.1111/j.1469-8137.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- Brownfield DL, Todd CD, Stone SL, Deyholos MK, Gifford DJ. Patterns of storage protein and triacylglycerol accumulation during loblolly pine somatic embryo maturation. Plant Cell, Tissue and Organ Culture. 2007;88:217–223. [Google Scholar]
- Carman JG, Reese G, Fuller RJ, Ghermay T, Timmis R. Nutrient and hormone levels in Douglas-fir corrosion cavities, megagametophytes, and embryos during embryony. Canadian Journal of Forest Research. 2005;35:2447–2456. [Google Scholar]
- Costa LM, Gutièrrez-Marcos JF, Dickinson HG. More than a yolk: the short life and complex times of the plant endosperm. Trends in Plant Science. 2004;9:507–514. doi: 10.1016/j.tplants.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends in Biochemical Sciences. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Danon A, Delorme V, Mailhac N, Gallois P. Plant programmed cell death: a common way to die. Plant Physiology and Biochemistry. 2000;38:647–655. [Google Scholar]
- Earnshaw WC. Nuclear changes in apoptosis. Current Opinion in Cell Biology. 1995;7:337–343. doi: 10.1016/0955-0674(95)80088-3. [DOI] [PubMed] [Google Scholar]
- Filonova LH, von Arnold S, Daniel G, Bozhkov PV. Programmed cell death eliminates all but one embryo in a polyembryonic plant seed. Cell Death and Differentiation. 2002;9:1057–1062. doi: 10.1038/sj.cdd.4401068. [DOI] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infection and Immunity. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Martínez SC, Krutovsky KV, Neale DB. Forest-tree population genomics and adaptive evolution. New Phytologist. 2006;170:227–238. doi: 10.1111/j.1469-8137.2006.01686.x. [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Perotti E, Ramirez J, Leblanc O. Timing of the maternal-to-zygotic transition during early seed development in maize. The Plant Cell. 2005;17:1061–1072. doi: 10.1105/tpc.104.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Durzan DJ. Shoot multiplication from mature trees of Douglas-fir (Pseudotsuga menziesii) and sugar pine (Pinus lambertiana) Plant Cell Reports. 1985;4:643–645. doi: 10.1007/BF00269282. [DOI] [PubMed] [Google Scholar]
- He X, Kermode AR. Nuclease activities and DNA fragmentation during programmed cell death of megagametophyte cells of white spruce (Picea glauca) seeds. Plant Molecular Biology. 2003;51:509–521. doi: 10.1023/a:1022319821591. [DOI] [PubMed] [Google Scholar]
- Hiratsuka R, Yamada Y, Terasaka O. Programmed cell death of Pinus nucellus in response to pollen tube penetration. Journal of Plant Research. 2002;115:141–148. doi: 10.1007/s102650200019. [DOI] [PubMed] [Google Scholar]
- International Seed Testing Association. International rules for seed testing. Seed Science and Technology. 1993;21:1–288. [Google Scholar]
- Kelly KJ, Sandoval RM, Dunn KW, Molitoris BA, Dagher PC. A novel method to determine specificity and sensitivity of the TUNEL reaction in the quantitation of apoptosis. American Journal of Physiology: Cell Physiology. 2003;2842:C1309–C1318. doi: 10.1152/ajpcell.00353.2002. [DOI] [PubMed] [Google Scholar]
- King JE, Gifford DJ. Amino acid utilization in seeds of loblolly pine during germination and early seedling growth. Plant Physiology. 1997;113:1125–1135. doi: 10.1104/pp.113.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanaka C, Kuchino Y. Caspase-independent programmed cell death with necrotic morphology. Cell Death and Differentiation. 1999;6:508–515. doi: 10.1038/sj.cdd.4400526. [DOI] [PubMed] [Google Scholar]
- Laporte C, Kosta A, Klein G, Aubry L, Lam D, Tresse E, Luciani MF, Golstein P. A necrotic cell death model in a protist. Cell Death and Differentiation. 2007;14:266–274. doi: 10.1038/sj.cdd.4401994. [DOI] [PubMed] [Google Scholar]
- Lev-Yadun S, Sederoff R. Pines as model gymnosperms to study evolution, wood formation, and perennial growth. Journal of Plant Growth Regulation. 2000;19:290–305. [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proceedings of the National Academy of Sciences, USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina FJ, Cerdido A, de Carcer G. The functional organization of the nucleolus in proliferating plant cells. European Journal of Histochemistry. 2000;44:117–131. [PubMed] [Google Scholar]
- Olsen OA. Nuclear endosperm development in cereals and Arabidopsis thaliana. The Plant Cell. 2004;16:214–227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne DJ, Boubriak I, Leprince O. Rehydration of dried systems: membranes and the nuclear genome. In: Black M, Pritchard HW, editors. Desiccation and survival in plants: drying without dying. Wallingford, Oxon, UK: CABI Publishing; 2002. pp. 343–364. [Google Scholar]
- Pasternak T, Potters G, Caubergs R, Jansen MAK. Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level. Journal of Experimental Botany. 2001;56:1991–2001. doi: 10.1093/jxb/eri196. [DOI] [PubMed] [Google Scholar]
- Pennell RI, Lamb C. Programmed cell death in plants. The Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plym Forshell C. Seed development after self-pollination and cross-pollination of Scots pine, Pinus sylvestris L. Studia Forestalia Suecica. 1974;118:1–37. [Google Scholar]
- Pulkkanen KJ, Laukkanen MO, Naarala J, Yla-Herttuala S. False-positive apoptosis signal in mouse kidney and liver detected with TUNEL assay. Apoptosis. 2000;5:329–333. doi: 10.1023/a:1009631424351. [DOI] [PubMed] [Google Scholar]
- Qiu J, Yoon JH, Shen B. Search for apoptotic nucleases in yeast: role of Tat-D nuclease in apoptotic DNA degradation. Journal of Biological Chemistry. 2005;280:15370–15379. doi: 10.1074/jbc.M413547200. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Rose A, Patel S, Meier I. The plant nuclear envelope. Planta. 2004;218:327–336. doi: 10.1007/s00425-003-1132-2. [DOI] [PubMed] [Google Scholar]
- Sanmartín M, Jaroszewski L, Raikhel NV, Rojo E. Caspases. Regulating death since the origin of life. Plant Physiology. 2005;137:841–847. doi: 10.1104/pp.104.058552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savonen EM. An improvement to the topographic tetrazolium testing of Scots pine (Pinus sylvestris L.) seeds. Seed Science and Technology. 1999;27:49–57. [Google Scholar]
- Singh H. Embryology of gymnosperms. Berlin: Borntrager; 1978. [Google Scholar]
- Sirois L, Bégin Y, Parent J. Female gametophyte and embryo development of black spruce along a shore-hinterland climatic gradient of a recently created reservoir, northern Quebec. Canadian Journal of Botany. 1999;77:61–69. [Google Scholar]
- Steeves TA. The evolution and biological significance of seeds. Canadian Journal of Botany. 1983;61:3550–3560. [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Borzhkov PV. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Current Biology. 2004;14:339–340. doi: 10.1016/j.cub.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. Reactive oxygen species in plant cell death. Plant Physiology. 2006;141:384–390. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuosku J, Jokela A, Läärä E, Sääskilahti M, Muilu R, Sutela S, Altabella T, Sarjala T, Häggman H. Consistency of polyamine profiles and expression of arginine decarboxylase in mitosis during zygotic embryogenesis of Scots pine. Plant Physiology. 2006;142:1027–1038. doi: 10.1104/pp.106.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li J, Bostock RM, Gilchrist DG. Apoptosis: a functional paradigm for programmed plant cell death induced by host-selective phytotoxin and invoked during development. The Plant Cell. 1996;8:375–391. doi: 10.1105/tpc.8.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Molecular Biology. 1999;39:915–926. doi: 10.1023/a:1006134027834. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiology. 1997;115:737–751. doi: 10.1104/pp.115.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky B. From the nematode and mammals back to the pine tree: on the diversity and evolution of programmed cell death. Cell Death and Differentiation. 2002;9:867–869. doi: 10.1038/sj.cdd.4401084. [DOI] [PubMed] [Google Scholar]
- Zhou BBS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zong WX, Thompson CB. Necrotic death as a cell fate. Genes and Development. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.