Abstract
Nicotianamine (NA) is a non-protein amino acid derivative synthesized from S-adenosyl L-methionine able to bind several metal ions such as iron, copper, manganese, zinc, or nickel. In plants, NA appears to be involved in iron availability and is essential for the plant to complete its biological cycle. In graminaceous plants, NA is also the precursor in the biosynthesis of phytosiderophores. Arabidopsis lines accumulating 4- and 100-fold more NA than wild-type plants were used in order to evaluate the impact of such an NA overaccumulation on iron homeostasis. The expression of iron-regulated genes including the IRT1/FRO2 iron uptake system is highly induced at the transcript level under both iron-sufficient and iron-deficient conditions. Nevertheless, NA overaccumulation does not interfere with the iron uptake mechanisms since the iron levels are similar in the NA-overaccumulating line and wild-type plants in both roots and leaves under both sufficient and deficient conditions. This observation also suggests that the translocation of iron from the root to the shoot is not affected in the NA-overaccumulating line. However, NA overaccumulation triggers an enhanced sensitivity to iron starvation, associated with a decrease in iron availability. This study draws attention to a particular phenotype where NA in excess paradoxically leads to iron deficiency, probably because of an increase of the NA apoplastic pool sequestering iron. This finding strengthens the notion that extracellular NA in the apoplast could be a major checkpoint to control plant iron homeostasis.
Keywords: Apoplast, iron homeostasis, nicotianamine
Introduction
Iron is found associated with a huge range of metalloprotein active sites, in the chemical form of various prosthetic groups including haem, and in [Fe–S] clusters. Its activity is required in most of the basic redox reactions for both the production (photosynthesis) and the consumption (respiration) of oxygen. Iron is also involved in many enzymatic reactions required for nitrogen fixation, and DNA and plant hormones synthesis. At pH 7 in an oxygenated medium, the soluble iron concentration ranges between 10−11 M and 10–10 M (Lindsay, 1995), whereas the required iron concentration for a plant in order to complete its life cycle ranges from 10−9 M to 10−8 M (Guerinot and Yi, 1994). This is why iron deficiency occurs frequently. In contrast, iron reactivity with oxygen can promote iron-mediated oxidative stress through the Fenton reaction, leading to amino acid oxidation, lipid peroxidation, and DNA mutations (Briat et al., 2007). There is therefore a narrow optimal iron concentration that is required for plants to achieve their physiological functions enabling them to grow efficiently. In such a context, mechanisms to control iron uptake, transport in various organs, and storage have evolved to prevent both iron deficiency and toxicity, enabling optimal plant development and growth (Colangelo and Guerinot, 2006). Indeed, it is well documented that iron homeostasis in various plant tissues during growth and development is a dynamic process resulting from an integrated regulation of the expression of the various genes encoding proteins acting in the transport, storage, and utilization of iron (Curie and Briat, 2003; Briat et al., 2007).
However, not only proteins play key roles in iron homeostasis in plants. Small organic molecules, among which organic acids, and amino acids and their derivatives, often have a strong affinity constant for iron, with which they form chelates involved both in long distance trafficking and intracellular distribution of the metal (Briat et al., 2007). Nicotianamine (NA) is one of these molecules and has been described as a key compound of metal homeostasis in plants. NA is produced in most tissues, synthesized from S-adenosyl L-methionine by nicotianamine synthase (NAS). Insights into the function of NA in metal homeostasis come from the analysis of two types of plants in which NA production is impaired. First, the tomato chloronerva (chl) mutant contains a loss-of-function mutation in its unique NAS gene (Ling et al., 1996, 1999). Secondly, tobacco transgenic plants overexpressing a graminaceous gene encoding nicotianamine amino transferase (NAAT) consume NA (Takahashi et al., 2003). These NA-less plants harbour interveinal chlorosis in young leaves, a strongly reduced growth, no or abnormal inflorescences, and complete sterility. Studies of these plants showed that in the absence of NA, iron remains in vascular bundles and fails to reach the interveinal areas of leaves and more generally young growing tissues. Since young tissues are essentially fed by the phloem, phenotypic defects provoked by NA shortage strongly support a role for NA in loading/unloading iron to and from the phloem sap.
More recently, Arabidopsis transgenic lines, overexpressing an NAS from the metal overaccumulator Thlaspi caerulescens have been produced (Pianelli et al., 2005). In contrast to the NA-less plants described above, these transgenic plants expressing TcNAS1 overaccumulated NA, up to 100-fold more than wild-type plants. Furthermore, increased NA levels in different transgenic lines were quantitatively correlated with an increased nickel tolerance expressed at the cellular level, and are associated with an increased NA content (Pianelli et al., 2005).
In this study, the K1 and K8 Arabidopsis lines accumulating 100- and 4-fold more NA, respectively, than wild-type plants were used in order to evaluate the impact of such NA overaccumulation on iron homeostasis. In particular, the consequences of this engineering on the expression of iron-regulated genes including the IRT1/FRO2 iron uptake system and the basic helix–loop–helix (bHLH) transcription factors, and on the metal content of these plants were measured. Alteration of these parameters in response to iron deficiency was also evaluated.
Materials and methods
Plant growth conditions
Arabidopsis wild-type (ecotype Columbia-0) and transgenic seeds (K1 and K8 lines; Pianelli et al., 2005) were surface sterilized and sown on plates containing half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 1% sucrose, 0.5 g l−1 MES and 1% agar. The pH of the medium was adjusted to 5.7 with KOH. The plates were stored for 2 d at 4 °C in the dark and then placed in long-day conditions (16 h at 300 μE m−2 s−1, 21 °C). After 5 d, seedlings were transferred to either iron-sufficient [Fe(III)-EDTA, 50 μM] or iron-deficient (no iron added) medium for 12 d.
RNA preparation and Q-PCR analysis
For each experiment, after a 12 d treatment, roots of 10 plants were pooled and pulverized in liquid nitrogen. Total RNA was isolated from 50–75 mg of powder using TRI Reagent (Molecular Research Center, Inc.) following the manufacturer's instructions. Total RNA samples were DNase treated to eliminate genomic DNA (RQ1 DNase, Promega). The integrity of RNA was verified by agarose gel electrophoresis, and 10 μg of each RNA sample were reverse transcribed using M-MLV Reverse Transcriptase (Promega) and oligo(dT)15 primers (Promega).
Real-time PCR was performed with a LightCycler® FastStart DNA MasterPLUS SYBER GREEN I (Roche). A 2 μl aliquot of cDNA products was used as template with 2 μl of Q-PCR mix (Roche), 0.5 μl of primers (20 μM each), and 5.5 μl of distilled water. The gene-specific primers used were: IRT1for, 5′CGG TTG GAC TTC TAA ATG C3′; IRT1rev, 5′CGA TAA TCG ACA TTC CAC CG3′; FRO2for, 5′GCG ACT TGT AGT GCG GCT ATG3′; FRO2rev, 5′CGT TGC ACG AGC GAT TCT G3′; FIT1for, 5′ GGT TAG GCA AGT TTA AGC TCT G3′; and FIT1rev, 5′ GGA GAA GGT GTT TGT CCA TCT C3′. For AtbHLH38 and AtbHLH39, previously designed primers were used (Wang et al., 2007). The relative transcript level (RTL) values were calculated relative to the amount of transcript of the constitutively expressed clathrin gene (At4g24550) as follows: RTL=1000×2–DCt. The DCt values were calculated as follows: DCt(IRT1)=Ct(IRT1)–Ct(clat), where the Ct value represents the cycle number at which the PCR product reaches a set threshold.
Protein extraction and western blot analysis
Total protein extract was prepared from root tissues of 12 plantlets grown in the conditions described above and pulverized in liquid nitrogen. Extracts were obtained as already described (Seguela et al., 2008). Immunodetection of IRT1 protein was performed using an affinity-purified anti-peptide IRT1 antibody. The IRT1 polyclonal antibody was raised in rabbits against a synthetic peptide H2N-CPANDVTLPIKEDDSS-COOH (Eurogentec Liège, Belgium) as previously described (Connolly et al., 2002; Vert et al., 2002). After 1 h incubation with the first antibody diluted 1:5000 in blocking buffer, membranes were washed in blocking buffer three times for 30 min each and incubated for 1 h with anti-rabbit IgG conjugated to alkaline phosphatase (Promega) (diluted 1:20 000 in blocking buffer). Three different washes of 30 min each were performed: first in blocking buffer, secondly in 1× phosphate-buffered saline (PBS), and then in ‘Aurora Chemiluminescence Substrate solution’. Chemiluminescence was detected using KODAK BioMax XAR film. In parallel, 10 μg of total proteins were separated on a 13% polyacrylamide–0.1% SDS gel and stained with Coomassie blue to assess the loading control.
Ferric reductase activity determination
Fe(III) chelate reductase activity was determined for whole intact roots as described previously (Yi and Guerinot, 1996). In brief, the entire root system of five treated plants was submerged in a solution containing 0.1 mM Fe(III)-EDTA and 0.3 mM ferrozine. After 20 min in the dark at room temperature, the roots were removed and the absorbance of each solution sample was determined at 562 nm. The concentration of Fe(II)-ferrozine was calculated using a molar extinction coefficient of 28.6 mM−1 cm−1.
Measurement of metal content
The metal (Fe, Zn Mn, Cu, and K) concentrations of plantlets were determined by atomic absorbance spectrometry. Prior to mineralization, roots and shoots of ten 12-day-old treated plantlets were harvested separately. Roots were washed with 2 mM CaSO4, 10 mM EDTA for 10 min and with 0.3 mM BPDS (bathophenanthroline disulphonate), 5.7 mM dithionite sodium for 3 min, then rinsed twice with deionized water; shoots were just rinsed with deionized water. Samples were dried and their dry weight was determined. Dry plant material was digested in 67% HNO3 at 120 °C for 1 h, then diluted to 6.7% HNO3. The metal content was determined using a Varian SprectAA220-FS (Varian, Palo Alto, CA, USA).
For water-extractable iron, assays were performed on shoots of ten 17-day-old plantlets. After grinding in 3 vols of deionized water at room temperature, aliquots (300 μl) were centrifuged at low speed (2000 g). The supernatant was recovered and the pellet was resuspended in the same volume of water. The Fe content was measured in the supernatant and the pellet of each sample as already described (Lobréaux and Briat, 1991).
Determination of chlorophyll concentration
Plants were grown as described above in the ‘Plant growth conditions’ section, and leaves were sampled for chlorophyll concentration assay. The experiment was performed on a pool of 12 plantlets after acetone extraction (Lichtenthaler, 1987).
Results
NA overaccumulation up-regulates iron uptake mechanisms
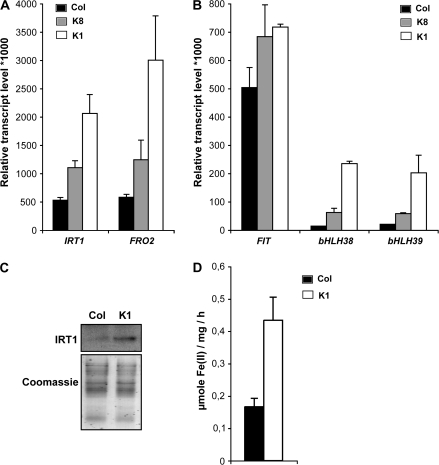
In order to evaluate the role of NA in metal homeostasis, a T. caerulescens cDNA encoding NAS had been expressed in Arabidopsis. The resulting NA-overaccumulating lines were only characterized for heavy metal resistance, mainly Ni, and the phytoremediation process (Pianelli et al., 2005). However, the clear role of NA in iron homeostasis (see Introduction) prompted the analysis of the effect of a deregulation of the plant NA content on the expression of several well characterized target genes such as the root iron transporter AtIRT1 (Eide et al., 1996; Vert et al., 2002) and the root iron chelate reductase AtFRO2 (Robinson et al., 1999). Wild-type Col-0 and transgenic plants with a 4- or a 100-fold increased NA content (Pianelli et al., 2005), K8 and K1 lines, respectively, were grown in vitro under control conditions where iron is not limiting [50 μM Fe(III)-EDTA]. In wild-type Col-0 plants, the genes encoding the iron transporter AtIRT1 and the ferric reductase AtFRO2 were both expressed at a low level (Fig. 1A). In the NA-overaccumulating lines, however, expression of both genes was observed at higher levels (Fig. 1A). A 2-fold increase in AtIRT1 and AtFRO2 was observed in the K8 line, whereas their expression was 4- and 5-fold higher, respectively, in the K1 NA-overaccumulating line compared with the wild-type plants (Fig. 1A).
Fig. 1.
Induction of iron uptake mechanisms in Arabidopsis lines overaccumulating NA. Wild-type plants (Columbia ecotype Col-0, black bars) and plants with a 4-fold (K8, grey bars) or a 100-fold (K1, white bars) increase in their NA content were sown on half-strength MS medium and after 5 d transferred to an iron-sufficient medium [50 μM Fe(III)-EDTA] for 12 d. Real-time RT-PCR determination of the relative transcript levels corresponding to the genes involved in the iron uptake, AtIRT1 and AtFRO2 (A), or in the transcriptional regulation of the iron starvation response, AtFIT1 (bHLH29), bHLH38, and bHLH39 (B). Error bars represent the SE of four repetitions. IRT1 protein accumulation in roots (C) was detected using an IRT1 affinity-purified peptide antibody (upper panel). The Coomassie staining shows equal loading (lower panel). Root Fe(III) chelate reductase activity (D) was performed on a mix of five plantlets. Error bars represent the SE of four repetitions.
The expression of AtIRT1 and AtFRO2 is controlled by the transcription factor AtFIT1 (AtbHLH29; Colangelo and Guerinot, 2004) through interaction with two other bHLH proteins (AtbHLH38 and AtbHLH39; Yuan et al., 2008). In order to determine if the variation of the NA levels also has an impact on these regulatory genes, the accumulation of the corresponding transcript was measured by real-time RT-PCR. As observed above for AtIRT1 and AtFRO2, the two transcription factors AtbHLH38 and AtbHLH39 were expressed to much higher levels in the K8 and K1 line (Fig. 1B). Interestingly, the AtFIT1 expression does not appear to be affected to the same extent as that of the other two bHLH genes. As AtFIT1 protein appears to be involved in the post-transcriptional and transcriptional regulation of the AtIRT1 and the ATFRO2 proteins, respectively (Connolly et al., 2002, 2003), the presence of the AtIRT1 protein was verified by western blot and the root ferric reductase activity was quantified in the most affected line (K1). As shown in Fig. 1, the abundance of AtIRT1 protein was slightly increased in the K1 genetic background overaccumulating NA, compared with what is observed with wild-type Col-0 Arabidopsis plants (Fig. 1C), and the ferric reductase activity was 2.6-fold higher in the NA-overaccumulating Arabidopsis line compared with control plants (Fig. 1D).
Increased sensitivity to iron deficiency in Arabidopsis overaccumulating NA
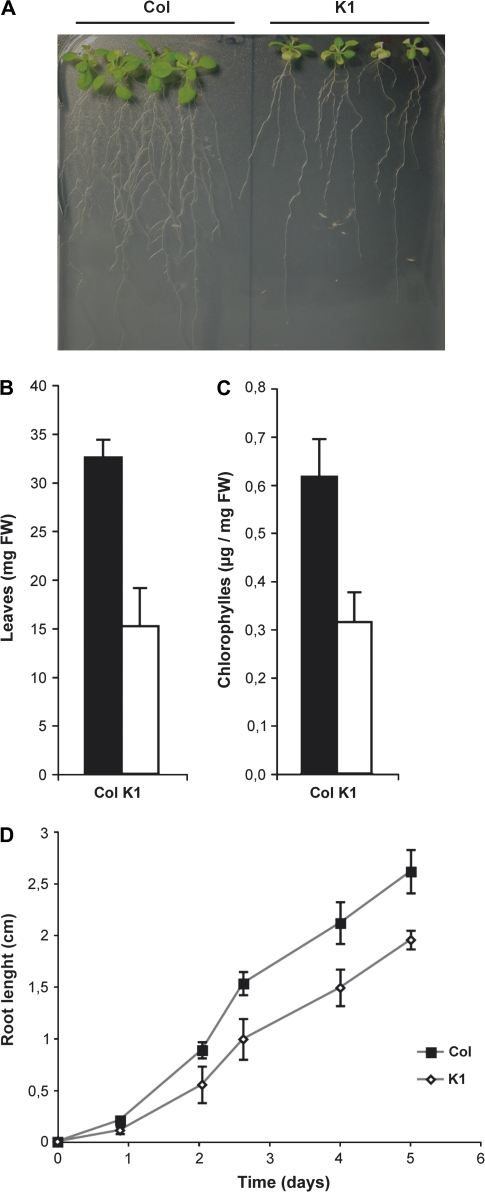
Given the deregulation of the iron acquisition systems in the K1 NA-overaccumulating line under iron-sufficient conditions, it was decided to focus on this line and to investigate its behaviour under iron starvation further. After transfer to a medium without added iron for 14 d, the wild-type plants displayed delayed growth but the K1 line appeared highly sensitive to iron starvation (Fig. 2A). To evaluate the effect of the deficiency, the leaf biomass and the chlorophyll content of both wild-type and K1 plants grown under iron-deficient conditions were quantified. As shown in Fig. 2B and C, both the leaf biomass and the chlorophyll content were decreased by 50% in the K1 line compared with wild-type plants under iron starvation. This lower tolerance to iron deficiency was also observed at the root level since the root growth of the K1 line was clearly reduced compared with wild-type plants (Fig. 2D).
Fig. 2.
The NA-overaccumulating line is highly sensitive to iron deficiency. (A) The phenotype of Col-0 plants and K1 plants overaccumulating NA sown and transferred after 5 d to an iron-deficient medium (no iron added) for 12 d. (B) Fresh weight and total chlorophyll content (C) of the entire rosette part of 10 wild-type plants (black bars) or 10 NA-overaccumulating plants (white bars) grown under iron-deficient conditions as described above. (D) Kinetics of the root growth of wild-type plants (filled squares) or the NA-overaccumulating line (open squares) transferred 5 d after germination on half-strength MS medium without iron added. Error bars represent the SE of 10 repetitions.
Overexpression of iron-regulated genes upon iron deficiency in the NA-overaccumulating line
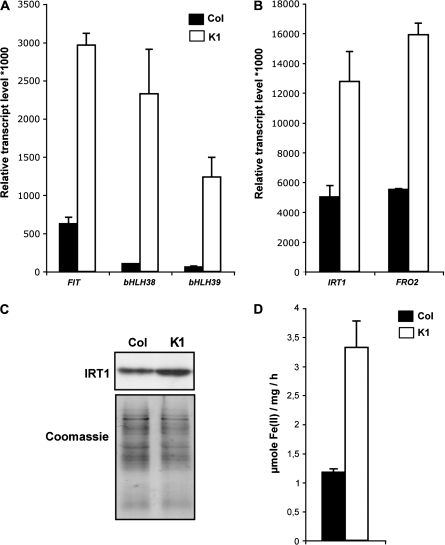
The K1 NA-overaccumulating line is affected in the perception of its iron status under non-limiting iron conditions (Fig. 1), and is more sensitive to iron deficiency than wild-type plants (Fig. 2). It was therefore decided to check if the K1 line was able to sense and to respond properly to the iron starvation conditions. For this purpose, the expression of AtIRT1, AtFRO2, and the three bHLH transcription factor genes (AtFIT1, AtbHLH38, and AtbHLH39) was monitored by real-time RT-PCR. Expression of the three transcription factors appear to be overinduced upon iron starvation in the K1 line compared with the wild-type background (Fig. 3A), even though these genes are still induced in wild-type plants (compare Figs 1B and 3A), as already observed (Colangelo and Guerinot, 2004; Yuan et al., 2008). The same behaviour was observed for the target genes AtIRT1 and AtFRO2, both genes being highly induced in response to iron deficiency in Col-0 plants (compare Figs 1A and 3B), as previously described (Eide et al., 1996; Vert et al., 2002; Connolly et al., 2003). In the K1 line, the increase in abundance of AtIRT1 and AtFRO2 transcripts was also observed (compare Figs 1A and 3B). However, both genes are expressed 2.5-fold more than in wild-type plants. The corresponding AtIRT1 protein level (Fig. 3C) and the ferric reductase activity (Fig. 3D) were also determined and found to be higher in the K1 line than in wild-type plants.
Fig. 3.
Overinduction of iron uptake mechanisms upon iron starvation in plants overaccumulating NA. Col-0 plants (black bars) and K1 plants overaccumulating NA (white bars) were sown for germination on half-strength MS medium and were transferred after 5 d to an iron-deficient medium (no iron added) for 12 d. Real-time RT-PCR determination of FIT1, bHLH38, and bHLH39 (A) or IRT1 and FRO2 (B) transcript levels in roots of iron-starved plants. Error bars represent the SE of four repetitions. IRT1 protein accumulation (C) was detected in a crude protein extract from roots by using an IRT1 affinity-purified peptide antibody. The Coomassie staining shows equal loading. Root Fe(III) chelate reductase activity (D) was determined on a mix of five plantlets. Error bars represent the the SE of four repetitions.
Impact of NA overaccumulation on metal content
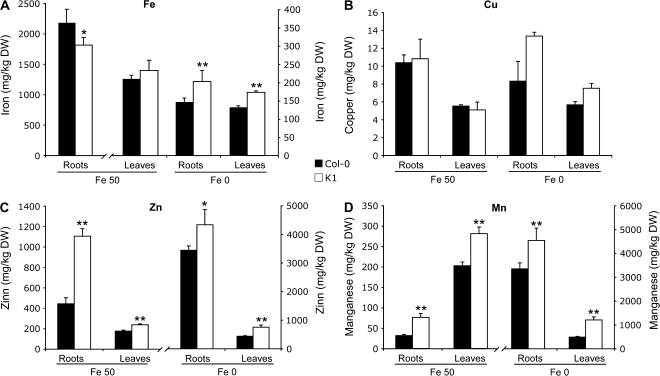
AtIRT1 and AtFRO2 are the major players involved in iron uptake in Arabidopsis. AtIRT1 is also able to transport other metals such as Zn, Mn, or Cd (Korshunova et al., 1999). On the other hand, NA is able to bind the same metals that can be transported by AtIRT1 (Stephan and Scholz, 1993). It was therefore of interest to determine whether or not the deregulation of AtFIT1, AtbHLH38, AtbHLH39, AtIRT1, and AtFRO2 gene expression, together with the NA overaccumulation, in the K1 line could have any impact on the plant metal content. The root and shoot iron, zinc, manganese, and copper contents were determined in the wild-type and the K1 line grown on iron-sufficient or iron-deficient medium.
Concerning iron (Fig. 4A), similar levels were measured in roots and shoots of both wild-type and K1 lines. However, the lower levels of iron in the roots of the K1 line were statistically significant, and reproducibly observed. The iron starvation treatment indeed reduces the iron content of both lines. In spite of this, the Fe levels were elevated by 30% in the K1 line compared with the wild-type plants in both the roots and the leaves (Fig. 4A).
Fig. 4.
Metal contents in an Arabidopsis line overaccumulating NA. Col-0 plants (black bars) and plants overaccumulating NA (white bars) were sown on half-strength MS medium and after 5 d transferred to an iron-sufficient [50 μM Fe(III)-EDTA] or iron-depleted (no iron added) medium for 12 d. Iron (A), copper (B), zinc (C), and manganese (D) concentrations were determined by atomic absorption spectrometry in roots and rosettes. Error bars represent the SE of four repetitions. Asterisks above the bars indicate significant differences (P <0.01; Student's test) between genotypes within the same organs under the same treatment.
The overinduction of the iron uptake system observed in the K1 line also impacted the content of the other metals. The copper root and shoot content was not different between the wild-type and the K1 line under iron-sufficient conditions (Fig. 4B). Determination of the Zn and Mn contents revealed a different behaviour since both metals accumulated in the K1 line (Fig. 4C, D). The Zn and Mn levels were increased 2.4- and 1.4-fold in the roots and the shoots, respectively, under control condition. These latter results were in good agreement with the already observed ability of the K1 line to accumulate other metals such as Ni (Pianelli et al., 2005). Upon iron starvation, it was observed that the iron-starved wild-type plants contained more Cu, Zn, and Mn (Fig. 4B–D) compared with the situation in iron-sufficient conditions, in agreement with previous results (Vert et al., 2002). However, in the NA-overaccumulating line, a supplemental increase of the Cu, Zn, and Mn levels was observed compared with the wild-type plants (Fig. 4B–D). As a control, the concentration of K+, an ion not chelated by NA, was also determined. No variations were observed between the K1 line and wild-type plant except a minor decrease in the K1 leaves under iron starvation (Supplementary Fig. S1 available at JXB online) that could be due to the growth delay of this line after 12 d in vitro (see Fig. 2A).
Overaccumulation of NA results in a decreased iron availability
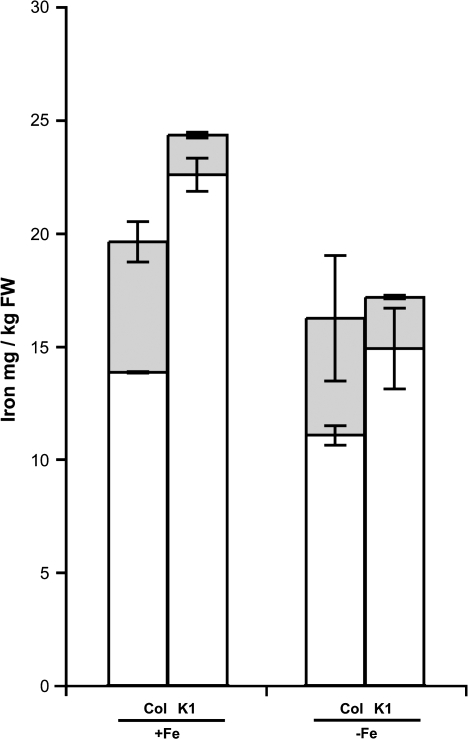
To gain a better insight into the iron phenotype of the K1 NA-overaccumulating line (constitutive induction of the iron uptake mechanisms, enhanced sensitivity to iron starvation despite an increase iron content), the iron availability in this Arabidopsis line was investigated. Fresh leaves of plants grown under control or iron-starved conditions were ground in water. After centrifugation, the pellet was assumed to represent the insoluble fraction whereas the supernatant represented the soluble, available iron. The total iron content related to the fresh weight appeared comparable between the two genetic backgrounds (wild-type versus the K1 line), being slightly higher in the K1 line (Fig. 5), which was consistent with the data presented in Fig. 4A. For the wild-type leaves, however, 30% of the total iron was found in the water-extractable fraction, whereas in the K1 line the soluble iron fraction represented only 6% of the total iron (Fig. 5). Upon iron starvation, although the total iron content decreased in wild-type leaves (as observed in Fig. 4A), the water-extractable iron fraction remained at 30% (Fig. 5), which corresponded to 5 μg of iron (g FW)−1. In the K1 line, the total iron content was still slightly higher than in the wild type (as in Fig. 4A) and the water-extractable iron fraction increased to 12% of this amount (Fig. 5). However, this remained below the value of the wild type and represented 2 μg of iron (g FW)−1, which corresponded to a 2.5-fold decrease of the soluble iron fraction compared with Col-0 plants.
Fig. 5.
Decreased water-extractable iron in the NA-overaccumulating line. Col-0 plants and plants overaccumulating NA were sown on half-strength MS medium and were transferred after 5 d to an iron-sufficient [50 μM Fe(III)-EDTA] or iron-deficient medium (no iron added) for an additional 12 d (as indicated below the graph). Soluble iron was extracted in water, then soluble iron (supernatant, grey bars) and the remaining iron (pellet, white bars) were measured by absorbance of Fe2+-o-phenanthroline using thioglycolic acid as the reducing agent. Error bars represent the SE of four repetitions. Differences were found to be highly significant (P <0.01; Student's test) between the soluble pools of Col-0 and the K1 line under iron-sufficient condition as well as between the K1 line soluble pools. Differences between the soluble pools of Col-0 and the K1 line under iron starvation are significant (P <0.05; Student's test).
Discussion
The amino acid derivative NA is known to play an important role in metal, and in particular iron, homeostasis in plants (Briat et al., 2007). Most of the data illustrating this function were obtained from plants depleted in NA, either by a mutation in the single NAS gene in the tomato chl mutant (Stephan and Grün, 1989; Higuchi et al., 1996; Ling et al., 1999) or by overexpression of the NAAT of a graminous plant (barley) in tobacco, leading to NA consumption in these transgenic plants (Takahashi et al., 2003). To document the function of NA in plant iron homeostasis further, transgenic Arabidopsis lines were produced which overexpressed TcNAS and consequently accumulated more NA than wild-type plants (Pianelli et al., 2005). In this study, two of these lines were used, named K8 and K1, which overaccumulate 4- and 100-fold more NA, respectively, than wild-type plants. In the K1 line, the NA content is 10-fold higher than the highest value measured in plants: 690 nmol (g FW)−1 in Lycium chinense leaves (Stephan and Scholz, 1993). No obvious macroscopic phenotype could be associated with this large increase in NA content when plants were grown under iron-sufficient conditions, and the iron concentrations remain almost similar between the K1 line and the wild-type plants (Fig. 4A). At the molecular level, an iron-starved phenotype was revealed. Indeed, the two genes involved in the iron uptake mechanisms, AtIRT1 and AtFRO2, were induced at the transcript level, as were the bHLH transcription factors involved in the transcriptional regulation of these uptake genes. This up-regulation of the root iron uptake system and of its regulators was observed for both the K1 and K8 lines (Fig. 1A, B). Although the transcript levels for each of these genes increased in agreement with the amount of NA in the various lines analysed, this relationship is not strictly linear. Indeed, there is only a 2- to 3-fold increase in abundance of these transcripts between the K8 and K1 lines, whereas there is a 25-fold difference in NA content between these two lines. A plausible explanation for this observation could be that a saturating NA concentration to trigger iron deficiency responses does exist, and that it is already exceeded in the K1 plants. This observation was intriguing because it mimicked the ‘apparent iron deficiency’ phenotype observed with the NA-free tomato mutant chl (Pich et al., 1991). However, in the case of chl, which has a higher iron content than the wild-type plant, the deficiency was due to the lack of iron bioavailability because of the absence of NA. The strong expression of AtIRT1 and AtFRO2 at the transcript level in the K1 and K8 lines (Fig. 1A) does not correlate with an obvious increased iron content (Fig. 4A), and this is consistent with the fact that the IRT1 protein abundance and the ferric reductase activity are only ∼2-fold higher in the K1 line compared with wild-type plants, when grown under iron-sufficient condition (Fig. 1C, D). The fact that the IRT1 protein and FRO2 activity were no more than 2-fold increased in the K1 line which contains 25-fold more NA than the K8 lines, led to a complete analysis to be performed only with the K1 line. Given the post-transcriptional control of IRT1 and FRO2 proteins (Connolly et al., 2002, 2003) it is likely that the presence of iron at the root level is perceived by the plant, leading to a low level of the corresponding IRT1 and FRO2 activities, in order to prevent excess iron uptake which would lead to iron overload and toxicity (Vert et al., 2003). However, under the same conditions, it was observed that Mn and Zn accumulated, but not Cu (Fig. 4). This suggests, therefore, a possible involvement of IRT1 whose substrate specificity matches this pattern of metal accumulation (Korshunova et al., 1999). A possible explanation could be that the slightly increased activity of the IRT1/FRO2 system in the K1 line contributes to increase the availability of Zn and Mn (among other ions). It has been proposed that Mn(III) can be reduced by the plasma membrane ferric chelate reductase activity under iron deficiency, leading to its higher availability (Norvell et al., 1993; Delhaize, 1996). Iron starvation leads to accumulation of heavy metals (Vert et al., 2002). It is also possible that other uptake systems with lower specificity for iron are induced to overcome its depletion. To verify this point, accumulation of the transcripts of the ZIP 1, 2, 3, and 4 transporter genes (Grotz et al., 1998; Guerinot, 2000) was measured (Supplementary Fig. S2A at JXB online). The ZIP 3 and 4 genes were found to be induced in the K1 genetic background, suggesting that they are in the same regulatory network as genes involved in root iron uptake which are activated upon iron starvation. The expression of the ZIP 1 and 2 genes was not affected, most probably because of the involvement of other regulatory mechanisms. Because of their higher Zn and Mn content, the behaviour of the NA-overaccumulating K1 line in response to a deficiency in these two metals was analyzed. As expected, the K1 line was more resistant to Zn and Mn deficiency than wild-type plants (Supplementary Fig. 2B at JXB online). However, this phenotype could be considered as a consequence of the ‘iron-starved’ behaviour of the K1 line, associated with the induction of several uptake systems to overcome iron deficiency, rather than a specific response. A transcriptomic analysis of the K1 line would be required to identify such regulatory networks.
The most unexpected feature of the NA-overaccumulating K1 line is its ‘iron-starved’ molecular phenotype associated with the induction of the iron uptake system, AtIRT1 and AtFRO2, among others. However, if IRT1 were indeed involved in the metal accumulation observed in the K1 line, an increase in the iron content at least to the same extent as Mn and Zn would have been found, which was not the case, as mentioned above. It therefore suggests that the K1 line is most probably affected in the mobilization of its endogenous iron pool rather than in its uptake abilities. It is therefore not surprising to observe that the NA-overaccumulating line is highly sensitive to iron starvation (Fig. 2). When grown in iron-depleted medium, the K1 line iron uptake system (IRT1 and FRO2), as well as the corresponding regulatory genes (AtFIT1, AtbHLH38, and AtbHLH39) are heavily induced, revealing an even higher transcript level than that observed with wild-type plants (Fig. 3). In spite of the overinduction of its iron uptake system, the K1 line appeared highly sensitive to iron starvation (Fig. 2), even though its iron content is higher than that of the wild-type plants (Fig. 4A). The main difference between the two genotypes concerns the localization of the iron. Indeed, a decrease of the water-extractable iron fraction, assumed to be the available iron, is correlated with an increase of the non-extractable (apoplastic) iron fraction in the K1 line (Fig. 5). Given the behaviour of the K1 plants already observed under iron-sufficient conditions, the simplest explanation of the K1 phenotype is that the NA overaccumulation observed in this line interferes with the iron localization and availability. Indeed, it has previously been reported that leaf K1 protoplasts contained only a 10-fold excess of NA compared with wild-type protoplasts, whereas total NA is present in a 100-fold excess at the foliar level (Pianelli et al., 2005). Most of the accumulated NA in the K1 line appeared therefore to be apoplastic. It has been shown that NA is able to bind iron efficiently over a wide range of pH values from 4 to 9 (Von Wirén et al., 1999; Rellan-Alvarez et al., 2008). The large amount of apoplastic NA observed in the K1 line could be responsible for the chelation of iron in the weak acidic compartment, leading to its sequestration outside of the cell. In support of this hypothesis, in the chl tomato NA-free mutant it has been reported that the lack of NA is associated with a drop of the apoplastic iron pool compared with the wild-type plants (Becker et al., 1995). The application of exogenous NA to chl plants enabled the reconstitution of this iron apoplastic fraction (Becker et al., 1995), suggesting a role for NA in the establishment of this extracellular iron pool. As an alternative to this hypothesis, the large excess of NA molecules in both intra- and extracellular compartments could lead to a continuous shuttle of NA and NA-Fe across the plasma membrane in both directions. Such a shuttle would restrict the delivery of NA-linked iron atoms to terminal biological targets.
Further experiments will be required to decipher the mechanisms involved in the NA overaccumulation in the apoplast of the K1 plants, responsible for an increased iron sequestration within this compartment. In particular, the mechanism by which NA molecules are localized outside of the cell appears to be a crucial question that remains to be addressed. Only a few hypotheses can be envisaged for the excretion of NA. According to the calculations of dissociation curves and the complex formation constant, it has been proposed that at the neutral cytoplasmic pH, free NA is a lipophilic molecule that could permeate biological membranes (Stephan et al., 1996). However, this seems unlikely to happen and members of the oligopeptide transporter family, that encompass the YSL (yellow-stripe like) transporter family, have to be considered as candidates potentially involved in the transport of free NA across membranes, in order to furnish the apoplast compartment (Briat et al., 2007). A second hypothesis comes from the study of graminaceous plants, and would involve polar traffic of vesicles and exocytosis. In graminaceous plants, NA is used as a precursor for the synthesis of phytosiderophores whose secretion into the rhizosphere allows iron chelation and uptake through YS transporters (Curie and Briat, 2003). It has been proposed that the synthesis of phytosiderophores and of their NA precursor could occur in intracellular vesicles (Nishizawa and Mori, 1987). Furthermore, microarray analyses of genes induced during iron deficiency in barley have identified candidate genes potentially involved in the polar vesicle transport of siderophores (Negishi et al., 2002). In support of this hypothesis, the NAS protein was shown to be hydrophobic and probably membrane bound (Higuchi et al., 1999).
In conclusion, the use of an NA-overaccumulating Arabidopsis line enabled some insights to be gained into the potential role of this metal-chelating agent in iron homeostasis. NA overaccumulation does not interfere with the iron uptake mechanisms since the K1 line contains iron levels comparable with that of the wild-type at both the root and the leaf levels under both sufficient and deprivation conditions. This observation also suggests that the translocation of the iron from the root to the shoot is also unaffected in the NA-overaccumulating line. However, NA overaccumulation triggers a decrease of the iron availability most probably because of a sequestration in the apoplast at the leaf level. This iron deficiency is perceived by the plant to trigger the induction of the iron uptake genes in the roots, as has been recently demonstrated for tobacco (Enomoto et al., 2007). The present findings strengthen the possibility that extracellular NA could be a major checkpoint to control plant iron homeostasis. They indicate the necessity in future experiments to investigate, at a cellular and molecular level, the mechanisms involved in the excretion of NA.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Material
Acknowledgments
The work of GC was supported by a thesis fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche. This work was supported by Centre National de la Recherche Scientifique (CNRS), by the Institut National de la Recherche Agronomique (INRA), by the Toxicologie Nucléaire et Environnementale Programme (ToxNucE) of the Réseau Inter-Organismes (RIO), and by ANR CIDS (ANR-06-BLAN-0).
Glossary
Abbreviations
- bHLH
basic helix–loop–helix
- BPDS
bathophenantroline
- chl
chloronerva
- NA
nicotianamine
- NAS
nicotianamine synthase
- NAAT
nicotianamine amino transferase
- YSL
yellow-stripe like
References
- Becker R, Fritz E, Manteuffel R. Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva. Plant Physiology. 1995;108:269–275. doi: 10.1104/pp.108.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Curie C, Gaymard F. Iron utilization and metabolism in plants. Current Opinion in Plant Biology. 2007;10:276–282. doi: 10.1016/j.pbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. The Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. Put the metal to the petal: metal uptake and transport throughout plants. Current Opinion in Plant Biology. 2006;9:322–330. doi: 10.1016/j.pbi.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiology. 2003;133:1102–1110. doi: 10.1104/pp.103.025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF. Iron transport and signaling in plants. Annual Review of Plant Biology. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- Delhaize E. A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiology. 1996;111:849–855. doi: 10.1104/pp.111.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y, Hodoshima H, Shimada H, Shoji K, Yoshihara T, Goto F. Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta. 2007;227:81–89. doi: 10.1007/s00425-007-0596-x. [DOI] [PubMed] [Google Scholar]
- Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proceedings of the National Academy of Sciences, USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML. The ZIP family of metal transporters. Biochimica et Biophysica Acta. 2000;1465:190–198. doi: 10.1016/s0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiology. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Nishizawa NK, Römheld V, Marschner H, Mori S. Absence of nicotianamine synthase activity in the tomato mutant chloronerva. Journal of Plant Nutrition. 1996;19:1235–1239. [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiology. 1999;119:471–480. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology. 1987;148:350–382. [Google Scholar]
- Lindsay WL. Chemical reactions in soil that affect iron availability to plants. A quantitative approach. In: Abadia J, editor. Iron nutrition in soils and plants. Dordrecht: Kluwer Academic Publishers; 1995. pp. 7–14. [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proceedings of the National Academy of Sciences, USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Pich A, Scholz G, Ganal MW. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Molecular and General Genetics. 1996;252:87–92. doi: 10.1007/BF02173208. [DOI] [PubMed] [Google Scholar]
- Lobréaux S, Briat JF. Ferritin accumulation and degradation in different organs of pea (Pisum sativum) during development. Biochemical Journal. 1991;274:601–606. doi: 10.1042/bj2740601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Negishi T, Nakanishi H, Yazaki J, et al. cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. The Plant Journal. 2002;30:83–94. doi: 10.1046/j.1365-313x.2002.01270.x. [DOI] [PubMed] [Google Scholar]
- Nishizawa N, Mori S. The particular vesicle appearing in barley root cells and its relation to mugineic acid secretion. Journal of Plant Nutrition. 1987;10:1013–1020. [Google Scholar]
- Norvell WA, Welch RM, Adams ML, Kochian LV. Reduction of Fe(III), Mn(III), and Cu(II) chelates by roots of pea (Pisum sativum L.) or soybean (Glycine max) Plant and Soil. 1993;156:123–126. [Google Scholar]
- Pianelli K, Mari S, Marquès L, Lebrun M, Czernic P. Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Research. 2005;14:739–748. doi: 10.1007/s11248-005-7159-3. [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G, Seifert K. Effect of nicotianamine on iron uptake and citrate accumulation in two genotypes of tomato, Lycopersicon esculentum Mill. Journal of Plant Physiology. 1991;137:323–326. [Google Scholar]
- Rellan-Alvarez R, Abadia J, Alvarez-Frenandez A. Formation of metal–nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange. A study by electrospray ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22:1553–1562. doi: 10.1002/rcm.3523. [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Seguela M, Briat JF, Vert G, Curie C. Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. The Plant Journal. 2008;55:289–300. doi: 10.1111/j.1365-313X.2008.03502.x. [DOI] [PubMed] [Google Scholar]
- Stephan UW, Grün M. Physiological disorders of the nicotianamine-auxotroph tomato mutant chloronerva at different levels of iron nutrition. II. Iron deficiency responses and heavy metal metabolism. Biochemie und Physiologie der Pflanzen. 1989;185:189–200. [Google Scholar]
- Stephan UW, Schmidke I, Stephan VW, Scholz G. The nicotianamine molecule is made-to-measure for complexation of metal micronutrients in plants. Biometals. 1996;9:84–90. [Google Scholar]
- Stephan UW, Scholz G. Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiologia Plantarum. 1993;88:522–529. [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. The Plant Cell. 2003;15:1263–1280. doi: 10.1105/tpc.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C. Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiology. 2003;132:796–804. doi: 10.1104/pp.102.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wirén N, Klair S, Bansal S, Briat J, Khodr H, Shiori T, Leigh R, Hider R. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiology. 1999;119:1107–1114. doi: 10.1104/pp.119.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P. Iron deficiency-mediated stress regulation of four subgroup Ib bHLH genes in Arabidopsis thaliana. Planta. 2007;226:897–908. doi: 10.1007/s00425-007-0535-x. [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. The Plant Journal. 1996;10:835–844. doi: 10.1046/j.1365-313x.1996.10050835.x. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.