Abstract
Aim
To identify genetic variants underlying six anthropometric traits: body height, body weight, body mass index, brachial circumference, waist circumference, and hip circumference, using a genome-wide association study.
Methods
The study was carried out in the isolated population of the island of Korčula, Croatia, with 898 adult examinees who participated in the larger DNA-based genetic epidemiological study in 2007. Anthropometric measurements followed standard internationally accepted procedures. Examinees were genotyped using HumanHap 370CNV chip by Illumina, with a genome-wide scan containing 316 730 single nucleotide polymorphisms (SNP).
Results
A total of 11 SNPs were associated with the investigated traits at the level of P < 10−5, with one SNP (rs7792939 in gene zinc finger protein 498, ZNF498) associated with body weight, hip circumference, and brachial circumference (P = 3.59-5.73 × 10−6), and another one (rs157350 in gene delta-sarcoglycan, SGCD) with both brachial and hip circumference (P = 3.70-6.08 × 10−6). Variants in CRIM1, a gene regulating delivery of bone morphogenetic proteins to the cell surface, and ITGA1, involved in the regulation of mesenchymal stem cell proliferation and cartilage production, were also associated with brachial circumference (P = 7.82 and 9.68 × 10−6, respectively) and represent interesting functional candidates. Other associations involved those between genes SEZ6L2 and MAX and waist circumference, XTP6 and brachial circumference, and AMPA1/GRIA1 and height.
Conclusion
Although the study was underpowered for the reported associations to reach formal threshold of genome-wide significance under the assumption of independent multiple testing, the consistency of association between the 2 variants and a set of anthropometric traits makes CRIM1 and ITGA1 highly interesting for further replication and functional follow-up. Increased linkage disequilibrium between the used markers in an isolated population makes the formal significance threshold overly stringent, and changed allele frequencies in isolate population may contribute to identifying variants that would not be easily identified in large outbred populations.
Interest in anthropometric traits has a long history, ever since the ancient philosophical debates and pioneering attempts to understand human anatomy (1). Modern research on these traits has largely shifted from fundamental morphological anatomy toward the understanding of environmental and genetic factors which determine, affect, and modify these traits. While environmental factors are known to have an important effect on some of these traits, primarily weight and other weight-related traits (2), genetic factors remained elusive to researchers for a long time. Population genetics theory hypothesized, based on the pre-genomic knowledge and comparative studies in plants and animals (3), that majority of these traits will be complex and have highly polygenic background (2,4,5). Additionally, some of these traits have been reported to have very high heritability, with that of height being used as the example of an extremely heritable trait (generally in the range of 0.90 to 0.95) (2). However, the search for the genetic variants underlying height has long been futile, casting doubt on the fundamental assumption that highly heritable traits will be good candidates for gene mapping studies. It is only lately that very large consortia were formed and achieved the needed statistical power to identify the first candidate loci using genome-wide association studies and expanded our understanding of the determinants of human stature (6-11).
The growing world-wide epidemic of obesity is one of the major issues in modern public health. Search for genes underlying obesity has also been rather unsuccessful, due to an even greater degree of complexity than for height, with large environmental effects and variation over time (12). The search for obesity genes is further complicated by the differences in the central and peripheral type of obesity (13,14). The genetic background of skinfolds has largely been under-investigated (15), with only a few genes implied in their regulation (15,16), some of which do seem to show strong interaction with the environment (17,18). Skinfold measurements may also show variation in time (19), making this research area highly complex and usually considered secondary to the more classical estimates of the amount of fat tissue, including body mass index and waist circumference or waist-to-hip ratio.
In this article, we report on a comprehensive genome-wide association study of height, weight, body mass index, and three circumference measurements – brachial, waist, and hip – in the isolated population of the island of Korčula in Croatia. The study is a part of a larger genetic epidemiology research program in Croatian island isolates, “10 001 Dalmatians.” The genetic epidemiology research program in Croatian island isolates began in 1999 (20,21), then expanded to study human genetic variation and effects of isolation and inbreeding (22-29), and finally entered the phase of focusing on diseases and gene mapping studies (8,30-35). By now, the research project has included more than 3000 examinees from isolated populations, and eventually it aims to reach 10 001 examinees.
Subjects and methods
This study was carried out in the adult population of the island of Korčula, Croatia. The field work was performed in 2007 in the eastern parts of the island, targeting healthy volunteers from the town of Korčula and villages Lumbarda, Žrnovo, and Račišće (Figure 1). Participants were invited by mail, posters, radio, and personal contacts. The sampling scheme for this study was convenient sampling, as the study aim was to include approximately 1000 of adult island inhabitants for the purpose of the genome-wide association analysis, regardless on the sample representativeness and demographic structure.
Figure 1.
Settlements on the island of Korčula, Croatia, included in the study
All examinees were aged 18 and over and had signed informed consent before entering the study, which was approved by the Ethical Committee of the Medical School, University of Zagreb. A total of 944 examinees were included in the study between March and December 2007. In all examinees, a large number of quantitative phenotypic traits were measured, including height, weight (and derived body mass index), and brachial, waist, and hip circumference. For these anthropometric measurements, standard methods were used (36).
DNA extraction was performed using Nucleon kits (Tepnel, Manchester, UK) and a total of 944 samples were genotyped in Institute of Human Genetics, Helmholtz Zentrum München, Germany. Genotyping was performed using Illumina HumanHap 370CNV (Illumina, San Diego, CA, USA), with a total of 346 027 single nucleotide polymorphism (SNP) markers. Quality control of the genotype data, excluding markers with a call rate <98%, with minor allele frequency <2%, or out of Hardy-Weinberg equilibrium (P < 10−10), left 316 730 SNPs in the analysis. Exclusion of individuals carrying markers with a call rate of <98% and those sampled twice in the study, left 898 people for inclusion in the analysis.
Relatedness between examinees was estimated from their whole genome data using the sharing of genome identical by descent (IBD) in PLINK (37). This method is robust to pedigree information errors, undeclared relationships, and samples swap, and gives realized sharing rather than expected one given by the pedigree information (for the same pedigree-based relationship, sharing varies within a range due to segregation and recombination stochastic events). Using this function, the sample could be grouped into 154 parent-child pairs, 107 sibling pairs, avuncular or equivalent 137, first cousins or equivalent 363, first cousins once removed or equivalent 1861, second cousins or equivalent 8962. The mean IBD sharing between all possible pairs of individuals was 0.003 (min = 0, max = 0.61).
Genome-wide associations between anthropometric phenotypes (adjusted for age and sex) and SNP markers were analyzed using the “mmscore” function of the GenABEL R statistical package (38), using an additive model. This score test for family-based association accounts for pedigree structure and allows unbiased estimations of SNP allelic effect (39). No correction for multiple testing was applied. The relationship matrix used in this analysis was generated by the identity-by-state (ibs) function of GenABEL, which used ibs genotype sharing to determine the realized pair-wise kinship coefficient similarly to the PLINK genome function. Minor allele frequencies were reported for the identified SNPs. Weighted genomic inbreeding coefficient was used in the inbreeding estimation (23). All identified SNPs that reached significance at the level of P < 10−5 were visualized using HaploView software (MIT/Harvard Broad Institute, MA, USA).
Results
Genetic variants associated with height, weight, and body mass index
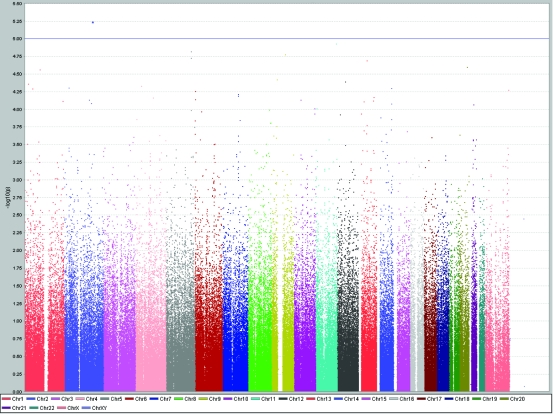
A total of 898 examinees were genotyped and included in the analysis. Descriptive statistics for the investigated traits are presented in Table 1. The average inbreeding coefficient was 0.013 ± 0.025. Initial genome-wide association study revealed two SNPs associated with body height at the level of P < 10−5 on chromosomes 1 and 5. The variant at chromosome 1 (P = 4.81x10−6) did not have a gene in proximity (±100 kilobases). The variant on chromosome 5 (P = 9.77x10−6) implicated a gene ionotropic glutamate receptor AMPA1 (GRIA1) (Table 2, Figure 2).
Table 1.
Descriptive statistics (mean ± standard deviation, SD) of the investigated traits of the examinees (n = 898) from the Korčula island
| Trait | Mean ± SD | Range |
|---|---|---|
| Height (mm) | 1680.9 ± 90.8 | 1410.0-1970.0 |
| Weight (kg) | 79.1 ± 14.2 | 49.5-166.6 |
| Body mass index (kg/m2) | 27.9 ± 4.1 | 16.6-53.8 |
| Brachial circumference (mm) | 333.4 ± 45.0 | 200.0-944.0 |
| Hip circumference (mm) | 1040.6 ± 78.6 | 748.0-1563.0 |
| Waist circumference (mm) | 941.4 ± 121.5 | 620.0-1385.0 |
Table 2.
A summary of single-nucleotide polymorphisms (SNP) that showed association with anthropometric traits that reached levels of genome-wide significance of P < 10−5*
| Trait | SNP | Chromo-some | Position | P value ( × 10−6) | Minor allele frequencies | β | Standard error (β) | Position | Gene |
|---|---|---|---|---|---|---|---|---|---|
| Height | rs7513590 | 1 | 5070572 | 4.81 | 0.104 | -0.316 | 0.069 | no gene | no genes +/− 100kb |
| Height | rs12658202 | 5 | 152959747 | 9.77 | 0.453 | 0110 | 0.025 | intronic | glutamate receptor, ionotropic, AMPA 1 (GRIA1) |
| Weight | rs7792939 | 7 | 99045812 | 3.59 | 0.148 | 0.255 | 0.055 | 7kb 5′ | zinc finger protein 498 (ZNF498) |
| Weight | rs7157940 | 14 | 93632946 | 3.13 | 0.458 | 0.118 | 0.025 | 5′ UTR | hypothetical protein LOC122509 (FAM14B) |
| Body mass index | rs7590983 | 2 | 174070595 | 5.76 | 0.045 | 0.462 | 0.102 | no gene | no genes +/− 100kb |
| Brachial circumference | rs1863080 | 2 | 36379940 | 7.82 | 0.102 | 0.307 | 0.069 | 56kb 5′ | cysteine-rich motor neuron 1 (CRIM1) |
| Brachial circumference | rs7723398 | 5 | 52264862 | 9.68 | 0.222 | 0.191 | 0.043 | intronic | integrin alpha 1 precursor (ITGA1) |
| Brachial circumference | rs32056 | 5 | 156006717 | 9.07 | 0.093 | -0.325 | 0.073 | intronic | delta-sarcoglycan (SGCD) |
| Brachial circumference | rs157350 | 5 | 156072147 | 3.70 | 0.105 | -0.311 | 0.067 | intronic | delta-sarcoglycan (SGCD) |
| Brachial circumference | rs7792939 | 7 | 99045812 | 4.90 | 0.148 | 0.255 | 0.056 | 7kb 5′ | zinc finger protein 498 (ZNF498) |
| Brachial circumference | rs201789 | 13 | 49884119 | 6.20 | 0.279 | 0.170 | 0.038 | intronic | XTP6 |
| Hip circumference | rs157350 | 5 | 156072147 | 6.08 | 0.105 | -0.306 | 0.068 | intronic | delta-sarcoglycan (SGCD) |
| Hip circumference | rs7792939 | 7 | 99045812 | 5.73 | 0.148 | 0.256 | 0.056 | 7kb 5′ | zinc finger protein 498 (ZNF498) |
| Waist circumference | rs7158173 | 14 | 64715003 | 3.93 | 0.471 | -0.114 | 0.025 | 76kb 5′ | MAX |
| Waist circumference | rs4787483 | 16 | 29792948 | 2.10 | 0.343 | 0.164 | 0.035 | intronic | Seizure-related 6 homologue (mouse)-like 2 isoform (SEZ6L2) |
*The table summarizes SNPs, their positions on the chromosomes, P values, minor allele frequencies, effect size and direction (expressed as β and standard error of β), effect allele, and implicated gene.
Figure 2.
Genome-wide association study of height using Haploview software, showing peaks on chromosomes 1 and 5 reaching genome-wide significance level of P < 10−5.
Two further SNPs were associated with weight, one belonging to hypothetical protein LOC122509 (FAM14B) gene on chromosome 14, while the other was within zinc finger protein 498 gene (ZNF498) on chromosome 7 (Table 2, Figure 3). The P value for the former variant was 3.13 × 10−6 and 3.59 × 10−6 for the latter.
Figure 3.
Genome-wide association study of weight using Haploview software, showing peaks on chromosomes 7 and 14 reaching genome-wide significance level of P < 10−5.
Body mass index was associated with a single SNP on the chromosome 2 that was not associated with any gene (Table 2, Figure 4), with a P value of 5.76 × 10−6.
Figure 4.
Genome-wide association study of body mass index using Haploview software, showing a peak on chromosome 2 reaching genome-wide significance level of P < 10−5.
Genetic variants associated with brachial, hip, and waist circumference
Brachial circumference showed association with 6 SNPs at chromosomes 2, 5, 7, and 13. Two of the SNPs (rs157350 and rs7792939) implicated delta-sarcoglycan gene (SGCD). The SNP with the stronger effect (rs157350) of P = 3.70 × 10−6 was also associated with the hip circumference (P = 6.08 × 10−6). Both brachial and hip circumference showed associations with the SNP rs7792939 in zinc finger protein 498 (ZNF498) on chromosome 7 (Table 2, Figure 5 and 6), with significance of P = 4.90 × 10−6 and P = 5.73 × 10−6, respectively. The same SNP showed strong association with body weight. Other variants associated with brachial circumference included rs1863080 in cysteine-rich motor neuron 1 (CRIM1) gene on chromosome 2 (P = 7.82 × 10−6), rs7723398 in integrin alpha 1 precursor (ITGA1) gene on chromosome 5 (P = 9.68 × 10−6), and rs201789 in XTP6 gene on chromosome 13 (P = 6.20 × 10−6).
Figure 5.
Genome-wide association study of brachial circumference using Haploview software, showing peaks on chromosomes 2, 5, 7, and 13 reaching genome-wide significance level of P < 10−5.
Figure 6.
Genome-wide association study of hip circumference using Haploview software, showing peaks on chromosomes 5 and 7 reaching genome-wide significance level of P < 10−5.
Finally, waist circumference was associated with two SNPs, one on chromosome 14 (rs7158173) belonging to the MAX gene (P = 3.83 × 10−6) and the other on chromosome 16 (rs4787483) belonging to seizure-related 6 homologue (mouse)-like 2 isoform gene (SEZ6L2; P = 2.10 × 10−6) (Table 2, Figure 7).
Figure 7.
Genome-wide association study of waist circumference using Haploview software, showing peaks on chromosomes 14 and 16 reaching genome-wide significance level of P < 10−5.
Discussion
The results of this study suggest several potential candidate genes involved in determining height, weight, and body mass index, as well as the hip, waist, and brachial circumference. We identified novel genetic associations with 6 human anthropometric traits using an isolate population. In isolated populations, genome-wide association studies can be particularly useful because of several potential advantages. First, allele frequencies of some important variants may be skewed from those found in the general population due to a combination of population genetic effects such as founder effect, genetic drift, and inbreeding (2). This could dramatically increase the power of genome-wide association study to detect otherwise rare variants of reasonably large effects on phenotypes of interest, if their frequency is unusually increased in an isolate population, a suggestion that has been proposed for some isolated Croatian Adriatic islands (22). In addition, accepted methods of computation of formal threshold for genome-wide statistical significance that corrects for multiple testing performed for hundreds of thousands of genetic markers may be overly stringent in isolate populations because of their population genetic properties (22). Increased linkage disequilibrium in such populations makes many of the markers linked so that many of the performed tests are dependent on each other (linkage disequilibrium works against the assumption of the independence between all performed tests) (31), which in turn mathematically lowers the threshold for genome-wide significance for an unknown but substantial amount. Lowering the significance threshold means greater power for the studies performed in isolate populations. Due to the two mechanisms described above (2,22), it would not be unexpected that genetic variants underlying complex traits are identified in human isolate populations rather than in outbred population by using genome-wide scans of comparable density and much larger sample sizes (5,8).
In recent years, genome-wide association studies have proven their worth as an extremely powerful tool in identifying genetic variants underlying many human diseases and complex traits (40). Hundreds of genetic variants are presently being discovered by large consortia of scientists in pioneering efforts to use this reliable approach to provide hypothesis-free insights into the genetic architecture of complex human phenotypes (36). Our study succeeded in identifying 11 genetic loci that reached significance level of P < 10−5. Although this does not make all of them necessarily genome-wide significant in formal turns, we believe that they should be carefully considered for replication in other populations and further functional studies due to combination of factors described above.
The gene AMPA1 (GRIA1) implicated in height in our study has also been implicated in stimulus-reward learning in mice (41,42) and possibly in bipolar disorder and schizophrenia in humans (43,44), but possible mechanisms remain obscure, and the finding has yet to be replicated. It has not been associated with human height to date. However, human height has been strongly inversely associated with the risk of schizophrenia (45), making this finding potentially interesting for further replication and functional follow-up, as GRIA1 gene could be a potential common factor contributing to both phenotypes.
It has recently been revealed that SCGD codes aminomutase catalyzing a-tyrosine to beta-tyrosine (46). The gene has been hypothesized to have a role in cardiomyopathy and muscular dystrophy (47). Furthermore, we have not been able to find information on the possible role and function of ZNF498 gene. Thus, both genes represent good candidates for further functional studies to clarify their possible role in determining body shape and size.
CRIM1, implicated in brachial circumference, is an interesting candidate gene. It encodes a putative transmembrane protein with multiple cysteine-rich (CR) domains known to have bone morphogenetic proteins binding activity, and is usually associated with vertebrate central nervous system development and organogenesis (48). It has been suggested that it regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface (49). Interestingly, several reports linked it to control of body size (50,51), thus making it a promising candidate for further follow-up studies.
ITGA1, another gene implicated in brachial circumference, is an equally interesting candidate gene. It is a part of integrin collagen receptor locus on human chromosome 5q11.2 (52). Thus, it is involved in the early remodelling of osteoarthritic cartilage and plays an essential role in the regulation of mesenchymal stem cell proliferation and cartilage production (53). Further replication of our finding and follow-up functional studies will be necessary to confirm its regulation of brachial circumference and interactions with other genes.
Unlike the previous candidate genes, no plausible information is currently available on possible roles and functions of XTP6, SEZ6L2, and MAX genes that would help us understand their possible role in determining human body size and shape.
The results presented here suggest that some identified genes were implicated in more than one trait, suggesting that they might be responsible for the genetic action on the common underlying property for these traits. Other genes, such as CRIA1 and ITGA1, represent very promising and biologically plausible functional candidates.
The shortcomings of this study primarily include the potential low statistical power, which is a consequence of the limited population size encountered in any genetic isolate. Studies performed in isolated populations should always seek replication to ensure that the findings are indeed representative of wider general human populations and not limited to specific circumstances of a particular isolate. Finally, variants identified in this study all require functional follow-up and replication in other populations in order to establish their true significance in determination of human body size and shape.
Acknowledgment
A. M. is the Editor in Chief, I. R. is the Editor for the International Health Issues, and Z. B. is the Cover Page Editor of the Croatian Medical Journal. S. J. is the Dean of the University of Split School of Medicine, one of the owners of the journal. To ensure that any possible conflict of interest has been addressed, this article was reviewed according to best practice guidelines of international editorial organizations.
This work was supported by the grant 108-1080315-0302 to IR and several other grants from the Croatian Ministry for Science, Education, and Sport to Croatian co-authors. It was also supported by the grants from the Medical Research Council UK to HC, AFW, and IR; and European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) to HC. The authors collectively thank to very large number of individuals for their individual help in organizing, planning and carrying out the field work related to the project and data management.”
References
- 1.Vesalius, A. De humanis corporis fabrica libri septem. Basileae [Basel]: Ex officina Joannis Oporini; 1543.
- 2.Hartl DL, Clark AG. Principles of population genetics. 4th edition. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- 3.Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–41. doi: 10.1016/S0169-5347(02)02489-8. [DOI] [Google Scholar]
- 4.Falconer DS, Mackay TFC. Quantitative genetics. Harlow (UK): Pearson, Prentice Hall; 1996. [Google Scholar]
- 5.Wright A, Charlesworth B, Rudan I, Carothers A, Campbell H. A polygenic basis for late-onset disease. Trends Genet. 2003;19:97–106. doi: 10.1016/S0168-9525(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 6.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–90. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 7.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson A, Marroni F, Hayward C, Franklin CS, Kirichenko AV, Jonasson I, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18:373–80. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang TL, Xiong DH, Guo Y, Recker RR, Deng HW. Comprehensive association analyses of IGF1, ESR2, and CYP17 genes with adult height in Caucasians. Eur J Hum Genet. 2008;16:1380–7. doi: 10.1038/ejhg.2008.113. [DOI] [PubMed] [Google Scholar]
- 10.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 12.Peeters MW, Beunen GP, Maes HH, Loos RJ, Claessens AL, Vlietinck R, et al. Genetic and environmental determination of tracking in subcutaneous fat distribution during adolescence. Am J Clin Nutr. 2007;86:652–60. doi: 10.1093/ajcn/86.3.652. [DOI] [PubMed] [Google Scholar]
- 13.Fantuzzi G, Mazzone T. Adipose tissue and atherosclerosis: exploring the connection. Arterioscler Thromb Vasc Biol. 2007;27:996–1003. doi: 10.1161/ATVBAHA.106.131755. [DOI] [PubMed] [Google Scholar]
- 14.Cikim AS, Ozbey N, Orhan Y. Relationship between cardiovascular risk indicators and types of obesity in overweight and obese women. J Int Med Res. 2004;32:268–73. doi: 10.1177/147323000403200306. [DOI] [PubMed] [Google Scholar]
- 15.Casiglia E, Tikhonoff V, Schiavon L, Guglielmi F, Pagnin E, Bascelli A, et al. Skinfold thickness and blood pressure across C-344T polymorphism of CYP11B2 gene. J Hypertens. 2007;25:1828–33. doi: 10.1097/HJH.0b013e32826308a0. [DOI] [PubMed] [Google Scholar]
- 16.Dai F, Keighley ED, Sun G, Indugula SR, Roberts ST, Aberg K, et al. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int J Obes (Lond) 2007;31:1832–42. doi: 10.1038/sj.ijo.0803675. [DOI] [PubMed] [Google Scholar]
- 17.Podolsky RH, Barbeau P, Kang HS, Zhu H, Treiber FA, Snieder H. Candidate genes and growth curves for adiposity in African- and European-American youth. Int J Obes (Lond) 2007;31:1491–9. doi: 10.1038/sj.ijo.0803673. [DOI] [PubMed] [Google Scholar]
- 18.Little BB, Malina RM. Gene-environment interaction in skeletal maturity and body dimensions of urban Oaxaca Mestizo schoolchildren. Ann Hum Biol. 2007;34:216–25. doi: 10.1080/03014460601144011. [DOI] [PubMed] [Google Scholar]
- 19.Bouchard L, Tremblay A, Bouchard C, Pérusse L. Contribution of several candidate gene polymorphisms in the determination of adiposity changes: results from the Québec Family Study. Int J Obes (Lond) 2007;31:891–9. doi: 10.1038/sj.ijo.0803542. [DOI] [PubMed] [Google Scholar]
- 20.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol. 1999;23:531–46. [PubMed] [Google Scholar]
- 21.Rudan I. Inbreeding and cancer incidence in human isolates. Hum Biol. 1999;71:173–87. [PubMed] [Google Scholar]
- 22.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 23.Carothers AD, Rudan I, Kolcic I, Polasek O, Hayward C, Wright AF, et al. Estimating human inbreeding coefficients: comparison of genealogical and marker heterozygosity approaches. Ann Hum Genet. 2006;70:666–76. doi: 10.1111/j.1469-1809.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 24.Rudan I, Smolej-Narancic N, Campbell H, Carothers A, Wright A, Janicijevic B, et al. Inbreeding and the genetic complexity of human hypertension. Genetics. 2003;163:1011–21. doi: 10.1093/genetics/163.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudan I, Biloglav Z, Vorko-Jovic A, Kujundzic-Tiljak M, Stevanovic R, Ropac D, et al. Effects of inbreeding, endogamy, genetic admixture, and outbreeding on human health: a (1001 Dalmatians) study. Croat Med J. 2006;47:601–10. [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell H, Carothers AD, Rudan I, Hayward C, Biloglav Z, Barac L, et al. Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Mol Genet. 2007;16:233–41. doi: 10.1093/hmg/ddl473. [DOI] [PubMed] [Google Scholar]
- 27.Polasek O, Kolcic I, Smoljanovic A, Stojanovic D, Grgic M, Ebling B, et al. Demonstrating reduced environmental and genetic diversity in human isolates by analysis of blood lipid levels. Croat Med J. 2006;47:649–55. [PMC free article] [PubMed] [Google Scholar]
- 28.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudan I, Carothers AD, Polasek O, Hayward C, Vitart V, Biloglav Z, et al. Quantifying the increase in average human heterozygosity due to urbanisation. Eur J Hum Genet. 2008;16:1097–102. doi: 10.1038/ejhg.2008.48. [DOI] [PubMed] [Google Scholar]
- 30.Kolcic I, Vorko-Jovic A, Salzer B, Smoljanovic M, Kern J, Vuletic S. Metabolic syndrome in a metapopulation of Croatian island isolates. Croat Med J. 2006;47:585–92. [PMC free article] [PubMed] [Google Scholar]
- 31.Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–42. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 32.Rudan I, Campbell H, Carothers AD, Hastie ND, Wright AF. Contribution of consanguinuity to polygenic and multifactorial diseases. Nat Genet. 2006;38:1224–5. doi: 10.1038/ng1106-1224. [DOI] [PubMed] [Google Scholar]
- 33.Rudan I, Rudan D, Campbell H, Carothers A, Wright A, Smolej-Narancic N, et al. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40:925–32. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudan I, Smolej-Narancic N, Campbell H, Carothers A, Wright A, Janicijevic B, et al. Inbreeding and the genetic complexity of human hypertension. Genetics. 2003;163:1011–21. doi: 10.1093/genetics/163.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivkovic V, Vitart V, Rudan I, Janicijevic B, Smolej-Narancic N, Skaric-Juric T, et al. The Eysenck personality factors: psychometric structure, reliability, heritability and phenotypic and genetic correlations with psychological distress in an isolated Croatian population. Pers Individ Dif. 2007;42:123–33. doi: 10.1016/j.paid.2006.06.025. [DOI] [Google Scholar]
- 36.Pulanić D, Polašek O, Petrovečki M, Vorko-Jović A, Peričić M, Barać Lauc L, et al. Effects of isolation and inbreeding on human quantitative traits – an example of biochemical markers of hemostasis and inflammation. Hum Biol. 2008;80:513–33. doi: 10.3378/1534-6617-80.5.513. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–85. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mead AN, Stephens DN. Selective disruption of stimulus-reward learning in glutamate receptor gria1 knock-out mice. J Neurosci. 2003;23:1041–8. doi: 10.1523/JNEUROSCI.23-03-01041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mead AN, Brown G, Le Merrer J, Stephens DN. Effects of deletion of gria1 or gria2 genes encoding glutamatergic AMPA-receptor subunits on place preference conditioning in mice. Psychopharmacology (Berl) 2005;179:164–71. doi: 10.1007/s00213-004-2071-8. [DOI] [PubMed] [Google Scholar]
- 43.Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20:687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 44.Kerner B, Jasinska AJ, Deyoung J, Almonte M, Choi OW, Freimer NB. Polymorphisms in the GRIA1 gene region in psychotic bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:24–32. doi: 10.1002/ajmg.b.30780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zammit S, Rasmussen F, Farahmand B, Gunnell D, Lewis G, Tynelius P, et al. Height and body mass index in young adulthood and risk of schizophrenia: a longitudinal study of 1 347 520 Swedish men. Acta Psychiatr Scand. 2007;116:378–85. doi: 10.1111/j.1600-0447.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 46.Li M, Liu W, Li Y. Cloning, expression and characterization of gene sgcD involved in the biosynthesis of novel antitumor lidamycin. Sci China C Life Sci. 2003;46:310–9. doi: 10.1360/03yc9033. [DOI] [PubMed] [Google Scholar]
- 47.Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, et al. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–74. doi: 10.1016/S0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 48.Kolle G, Georgas K, Holmes GP, Little MH, Yamada T. CRIM1, a novel gene encoding a cysteine-rich repeat protein, is developmentally regulated and implicated in vertebrate CNS development and organogenesis. Mech Dev. 2000;90:181–93. doi: 10.1016/S0925-4773(99)00248-8. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson L, Kolle G, Wen D, Piper M, Scott J, Little M. CRIM1 regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface. J Biol Chem. 2003;278:34181–8. doi: 10.1074/jbc.M301247200. [DOI] [PubMed] [Google Scholar]
- 50.Fung WY, Fat KF, Eng CK, Lau CK. crm-1 facilitates BMP signaling to control body size in Caenorhabditis elegans. Dev Biol. 2007;311:95–105. doi: 10.1016/j.ydbio.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Glienke J, Sturz A, Menrad A, Thierauch KH. CRIM1 is involved in endothelial cell capillary formation in vitro and is expressed in blood vessels in vivo. Mech Dev. 2002;119:165–75. doi: 10.1016/S0925-4773(02)00355-6. [DOI] [PubMed] [Google Scholar]
- 52.Lee HJ, Kim SY, Koh JM, Bok J, Kim KJ, Kim KS, et al. Polymorphisms and haplotypes of integrinalpha1 (ITGA1) are associated with bone mineral density and fracture risk in postmenopausal Koreans. Bone. 2007;41:979–86. doi: 10.1016/j.bone.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Cheli Y, Kanaji S, Jacquelin B, Chang M, Nugent DJ, Kunicki TJ. Transcriptional and epigenetic regulation of the integrin collagen receptor locus ITGA1-PELO-ITGA2. Biochim Biophys Acta. 2007;1769:546–58. doi: 10.1016/j.bbaexp.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]