Abstract
Expression of the c-Myc proto-oncoprotein is tightly regulated in normal cells. Phosphorylation at two conserved residues, threonine58 (T58) and serine62 (S62), regulates c-Myc protein stability. In cancer cells, c-Myc can become aberrantly stabilized associated with altered T58 and S62 phosphorylation. A complex signalling cascade involving GSK3β kinase, the Pin1 prolyl isomerase, and the PP2A-B56α phosphatase controls phosphorylation at these sites. We report here a novel role for the tumour suppressor scaffold protein Axin1 in facilitating the formation of a degradation complex for c-Myc containing GSK3β, Pin1, and PP2A-B56α. Although knockdown of Axin1 decreases the association of c-Myc with these proteins, reduces T58 and enhances S62 phosphorylation, and increases c-Myc stability, acute expression of Axin1 reduces c-Myc levels and suppresses c-Myc transcriptional activity. Moreover, the regulation of c-Myc by Axin1 is impaired in several tested cancer cell lines with known stabilization of c-Myc or loss of Axin1. This study provides critical insight into the regulation of c-Myc expression, how this can be disrupted in three cancer types, and adds to our knowledge of the tumour suppressor activity of Axin1.
Keywords: Axin1, c-Myc, GSK3β, Pin1, PP2A-B56α
Introduction
c-Myc is a transcription factor that regulates the expression of numerous genes involved in the regulation of cellular proliferation, growth, apoptosis, and differentiation (Dang et al, 1999). Normal cell function requires proper regulation of c-Myc expression, which occurs at multiple levels through transcriptional, translational, and post-translational mechanisms. Recent research has identified a complex signalling pathway that controls c-Myc protein expression at the post-translational level through sequential and reversible phosphorylation at two highly conserved sites, threonine58 (T58) and serine62 (S62), that regulate ubiquitination and 26S proteasomal turnover of c-Myc (reviewed in Sears, 2004). Specifically, S62 phosphorylation increases c-Myc protein stability, whereas T58 phosphorylation stimulates its ubiquitination and degradation by the SCFFbw7 complex (Sears et al, 2000; Welcker et al, 2004; Yada et al, 2004). A number of reports have identified GSK3β, the prolyl isomerase Pin1, and protein phosphatase 2A (PP2A) with the associated B56α regulatory subunit as critical components in c-Myc protein turnover as GSK3β mediates T58 phosphorylation and Pin1 and PP2A-B56α cooperate to dephosphorylate S62 (Sears et al, 2000; Gregory et al, 2003; Yeh et al, 2004; Arnold and Sears, 2006).
Axin1 is a multi-domain scaffold protein that coordinates several different protein complexes that are involved in regulating Wnt, TGFβ, SAPK/JNK, and p53 signalling (Zeng et al, 1997; Zhang et al, 1999; Rui et al, 2004; Liu et al, 2006). Thus far Axin1 has been characterized to be a tumour suppressor and numerous gene mutations have been identified throughout AXIN1 from a number of different cancers (Salahshor and Woodgett, 2005). These mutations most likely compromise the ability of Axin1 to form complexes with DVL, MEKK, GSK3α/β, PP2A, APC, β-catenin, and even Axin1 with itself (summarized in Salahshor and Woodgett, 2005). It has been shown in some cases of hepatocellular carcinomas (HCCs) harbouring mutations in AXIN1 that re-introduction of wild-type Axin1 expression increases apoptosis (Satoh et al, 2000), suggesting that Axin1 may be an important molecular target in HCC as well as other cancers with compromised Axin1 function.
We now report an additional regulatory role for Axin1 in negatively controlling c-Myc protein levels at the post-translational level. Axin1 facilitates the interaction of c-Myc with GSK3β, PP2A, and Pin1, stimulates c-Myc ubiquitin-mediated degradation, and inhibits c-Myc transcriptional activity. Interestingly, several cancer cell lines with increased c-Myc stability have impaired formation of the Axin1–c-Myc degradation complex. Altogether, our results add to our understanding of the regulation of c-Myc expression and provide a new mechanism for the tumour suppressor activity of Axin1.
Results
Axin1 associates with c-Myc, GSK3, PP2A-B56, and Pin1
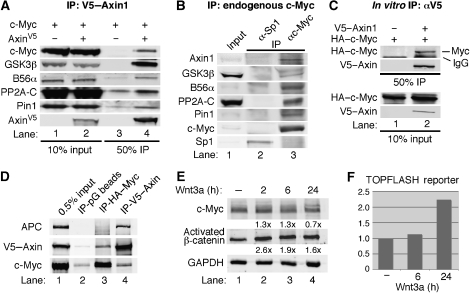
Owing to the high level of regulation and rapid turnover of c-Myc protein, we hypothesized that some of the proteins involved in regulating c-Myc protein turnover might be organized into a complex by associating with a scaffolding protein. We focused our attention on the scaffold protein Axin1, as it had been shown to associate with GSK3α/β and PP2A-B56α (Behrens et al, 1998; Fagotto et al, 1999; Li et al, 2001). To examine whether Axin1 associates with c-Myc, we transiently transfected 293 cells with c-Myc and Axin1 expression vectors and found that c-Myc co-immunoprecipitated with Axin1, and that endogenous GSK3β, B56α, PP2A-C (catalytic subunit), and Pin1 also co-immunoprecipitated with Axin1 (Figure 1A, lane 4). To further examine the association between c-Myc and Axin1, we performed the reverse co-immunoprecipitation and found that Axin1 co-immunoprecipitated with c-Myc (Supplementary Figure 1). Additionally, we found that endogenous Axin1, GSK3β, B56α, PP2A-C, and Pin1 co-immunoprecipitated with endogenous c-Myc, but not another endogenous transcription factor, Sp1 (Figure 1B, lane 3 versus 2). Lastly, immunoprecipitation of in vitro translated Axin1 co-precipitated in vitro translated c-Myc (Figure 1C, lane 2). Altogether, these findings demonstrate that Axin1 and c-Myc associate, which can be detected at endogenous levels along with endogenous GSK3β, PP2A-B56α, and Pin1.
Figure 1.
Axin1 associates with c-Myc along with GSK3β, PP2A-B56α, and Pin1. (A) V5–Axin1 co-immunoprecipitates c-Myc and proteins that stimulate c-Myc degradation. 293 cells were co-transfected with expression plasmids for V5–Axin1 and c-Myc as indicated. Cells were lysed in co-IP buffer and subject to α-V5 immunoprecipitation (IP). Input lysate and immunoprecipitated proteins were detected by western blotting as indicated. (B) Axin1 and c-Myc interact at endogenous levels. 293 cells were lysed in co-IP buffer and subject to IP with αc-Myc or α-Sp1 (control) as indicated. Input and immunoprecipitated proteins were detected by western blotting as indicated. (C) In vitro synthesized Axin1 and c-Myc interact. V5–Axin1 and c-Myc were produced by in vitro transcription/translation. Indicated reactions were suspended in co-IP buffer and subject to α-V5 IP, and input and immunoprecipitated proteins were detected by western blot as indicated. (D) APC associates with Axin1, but not with c-Myc. 293 cells were co-transfected with HA–c-Myc and V5–Axin1, cells were lysed in co-IP buffer, and the lysate was split into thirds. Protein G beads, α-HA, or α-V5 were used for immunoprecipitation, and input and IP proteins were visualized by western blot as indicated. (E) Wnt signalling does not affect c-Myc expression levels. 293T cells were transfected with expression plasmid for c-Myc and starved in 0.2% FBS for 48 h. Cells were then treated with 80 ng/ml Wnt3a for indicated times and c-Myc, endogenous activated β-catenin (anti-active β-catenin; Millipore), and GAPDH were detected by western blot. Fold-change relative to lane 1 and GAPDH is indicated. (F) Activation of the β-catenin/TCF-responsive luciferase reporter plasmid, TOPFLASH, upon stimulation with Wnt3a. 293T cells were co-transfected with TOPFLASH and CMV-β-gal. Cells were starved in 0.2% FBS for 48 h and then treated with 80 ng/ml Wnt3a for indicated times. Luciferase activity was measured and normalized to β-gal activity.
The adenomatous polyposis coli (APC) gene product has a critical function in the recruitment and turnover of β-catenin on the Axin1 scaffold protein (Xing et al, 2003). We examined whether APC also has a function in the recruitment of c-Myc to Axin1. 293 cells were transfected with V5-tagged Axin1 and HA-tagged c-Myc. Immunoprecipitation of V5–Axin1 co-immunoprecipitated APC as expected (Figure 1D, lane 4). In contrast, immunoprecipitation of HA–Myc co-immunoprecipitated Axin1, but not APC (Figure 1D, lane 3). In addition, shRNA knockdown of APC does not affect c-Myc or P-T58-Myc levels in conditions in which Axin1 knockdown does, and increasing APC expression does not affect the ability of c-Myc to complex with Axin1 (Dr Bruno Amati, European Institute of Oncology, Italy, personal communication). Lastly, because Wnt signalling inhibits β-catenin turnover by the Axin–APC complex, we tested the effects of Wnt signalling on ectopic c-Myc expression levels. We did not observe a significant induction of c-Myc under conditions where we did observe an increase in activated β-catenin and in β-catenin/TCF-mediated transcription consistent with other reports in the literature (Staal et al, 2002) (Figure 1E and F). Taken together, these data suggest that the Axin1–c-Myc complex is most likely distinct from the Wnt-regulated Axin1–β-catenin complex involving APC. Additionally, c-Myc is primarily nuclear and as such would be expected to associate with a nuclear pool of Axin1.
Axin1 knockdown increases c-Myc levels and decreases its association with GSK3, PP2A, and Pin1
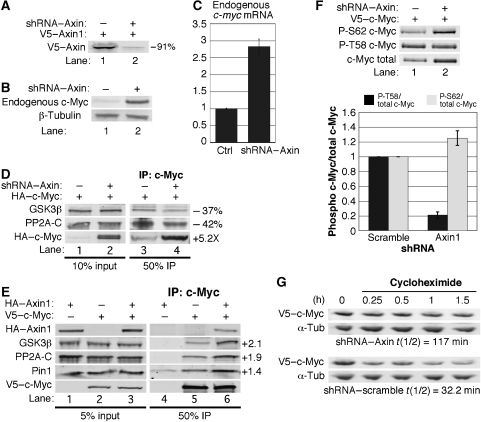
We generated shRNA that efficiently knocks down Axin1 expression (Figure 2A). Knockdown of Axin1 increases endogenous c-Myc protein levels in low-serum growth conditions (Figure 2B). Endogenous c-myc mRNA levels are also increased with Axin1 knockdown (Figure 2C), presumably due to increased β-catenin activity with Axin1 knockdown as β-catenin/TCF has been shown to transcriptionally activate the c-myc gene (He et al, 1998). To focus our research on potential post-transcriptional regulation of c-Myc by Axin1, we examined ectopically expressed c-Myc, driven by the constitutively active CMV enhancer in the following experiments. We found that Axin1 knockdown significantly increased expression of CMV-driven c-Myc (Figure 2D, lower panel, lane 2 versus 1). Moreover, despite a significant increase in immunoprecipitated c-Myc with Axin1 knockdown (5.2-fold), approximately 40% less of both GSK3β and PP2A-C co-immunoprecipitated with c-Myc as compared with control (Figure 2D, compare lane 4 with 3). Quantitation of the amount of GSK3β and PP2A-C co-precipitated relative to the amount of c-Myc indicated an 88% decrease in the association of these proteins with c-Myc upon Axin1 knockdown. This result suggests that c-Myc, which accumulates in the absence of Axin1, does not efficiently associate with GSK3β or PP2A. To address whether this could be partially due to limiting GSK3β and/or PP2A, we tested the effects of ectopic Axin1 expression on the association of c-Myc with GSK3β, PP2A, and Pin1. As shown in Figure 2E, ectopic expression of Axin1 increased the amount of GSK3β, PP2A, and Pin1 that co-immunoprecipitated with c-Myc approximately two-fold (Figure 2E, lane 6 versus 5). Additionally, the association of GSK3β and PP2A with Axin1 is enhanced under low-serum culture conditions where c-Myc protein levels are reduced (Sears et al, 1999) (Supplementary Figure 2, lanes 4 and 6 versus 2).
Figure 2.
Axin1 facilitates the association of c-Myc with GSK3β and PP2A, and negatively regulates c-Myc protein stability. (A) shRNA-mediated knockdown of Axin1. 293 cells were co-transfected with V5–Axin1 and either vector expressing scramble or Axin1-targeted shRNA. Extracts were normalized for transfection efficiency and V5–Axin1 was detected by western blotting and quantified using the LI-COR imager software. (B) Knockdown of Axin1 increases endogenous c-Myc protein expression. Axin1 or scramble shRNA expression vectors were transfected into 293 cells. Cells were maintained in low serum for 48 h. Endogenous c-Myc and β-tubulin were detected by western blot. (C) Knockdown of Axin1 increases c-myc RNA levels. 293tr–shAxin1 cells were treated with or without Dox to induce the expression of Axin1 shRNA under low-serum conditions, and c-myc mRNA expression was analysed by qRT–PCR. (D) Axin1 knockdown reduces the interaction of c-Myc with GSK3β and PP2A. 293 cells were co-transfected with HA–c-Myc and either scramble or Axin1 shRNA. HA–c-Myc was immunoprecipitated and input and immunoprecipitated proteins were detected by western blotting. The change in expression between lane 4 versus 3 was quantified using the LI-COR software. (E) The interaction of c-Myc with GSK3β and PP2A is enhanced by increased Axin1 expression. V5–c-Myc was immunoprecipitated from 293 cells co-transfected with V5–c-Myc and/or HA–Axin1 as indicated. Input and immunoprecipitated proteins were detected by western blot and the fold increase in expression between lane 6 versus 5 was quantified using the LI-COR software. (F) Axin1 knockdown inhibits T58 phosphorylation and increases S62 phosphorylation. 293 cells were co-transfected with V5–c-Myc and either scramble or Axin1 shRNA. Lysates were normalized for transfection efficiency and phosphorylated c-Myc was detected by phospho-specific antibodies. Total c-Myc on the same blots was detected by α-V5 and a representative blot is shown. The signal in lane 2 versus 1 was quantified using the LI-COR software and three separate experiments are graphed showing average change with standard deviation (s.d.) for P-T58/total c-Myc and P-S62/total c-Myc. (G) Axin1 knockdown increases c-Myc protein stability. 293 cells were co-transfected with V5–c-Myc and either scramble or Axin1 shRNA, maintained in low serum, and then treated with cycloheximide and collected at the indicated time points. c-Myc half-life was determined from western blots and quantified using the LI-COR software for V5–c-Myc protein levels using α-tubulin as a control.
Knockdown of Axin1 decreases T58 phosphorylation, increases S62 phosphorylation, and increases c-Myc protein stability
As our data demonstrate that Axin1 is important for c-Myc association with GSK3β and PP2A, and GSK3β phosphorylates T58, whereas PP2A-B56α dephosphorylates S62, we examined the effects of knockdown of Axin1 on c-Myc phosphorylation at T58 and S62. To accurately quantify the level of P-T58 and P-S62 relative to total c-Myc, we dual probed our western blots with α-V5 for total c-Myc and phospho-specific antibodies for P-T58 or P-S62 levels. Upon Axin1 knockdown, we observed an increase in the P-S62 signal, but a decrease in the P-T58 signal (Figure 2F, lane 2 versus 1). Quantifying the ratio of P-T58 to total c-Myc revealed a substantial decrease in the amount of T58 phosphorylated c-Myc with Axin1 knockdown (Figure 2F, graph). In contrast, the signal for P-S62 increased relative to total c-Myc, resulting in a modest increase in S62 phosphorylated c-Myc compared with control (Figure 2F, graph). We also examined whether Axin1 knockdown affects c-Myc protein stability. 293 cells were transfected with CMV-driven c-Myc and either shRNA to Axin1 or scramble control. Following cycloheximide treatment to inhibit protein synthesis, we found that knockdown of Axin1 substantially increased c-Myc protein stability to 117 min, as compared with scramble control shRNA with a 32-min half-life for c-Myc (Figure 2G). Thus, Axin1 post-translationally regulates c-Myc protein stability consistent with its effects on c-Myc T58 and S62 phosphorylation and its effects on the ability of c-Myc to associate with GSK3β, PP2A, and Pin1.
Ectopic Axin1 expression increases the ubiquitination and degradation of c-Myc
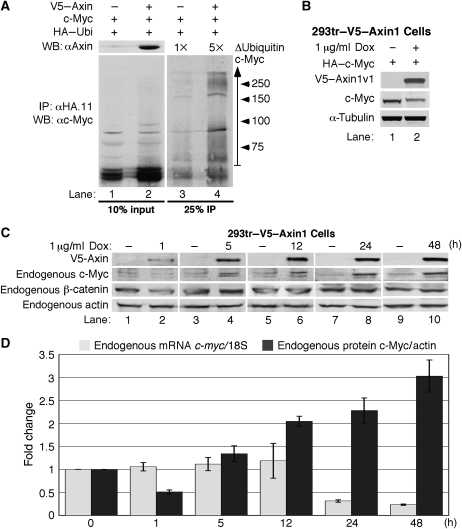
Given the effect of Axin1 expression on c-Myc protein stability, we tested whether Axin1 affects c-Myc ubiquitination. Cells were transfected as indicated in Figure 3A and treated with MG132/MG115 for 4 h prior to collection to prevent turnover of multi-ubiquitinated c-Myc. Ubiquitinated proteins were immunoprecipitated and c-Myc was detected by western blotting. We observed an approximate five-fold increase in ubiquitinated forms of c-Myc with ectopic expression of Axin1 (Figure 3A, lane 4 versus 3). Although our findings support a model where Axin1 coordinates the association of GSK3β, PP2A-B56α, and Pin1 with c-Myc and thus facilitates c-Myc degradation, c-Myc input levels in our co-immunoprecipitation experiments are often not substantially affected by co-expression of Axin1 (see Figure 2E, bottom panel, lanes 2 and 3). This is most likely due to altered stoichiometry of components for complex formation, as the overexpression of scaffold proteins, including Axin1 have been reported to result in dominant-negative effects (Lee et al, 2003). We therefore generated a stable 293tr cell line with tetracycline/doxycycline (Dox)-inducible V5–Axin1 expression to more carefully control the amount and time of Axin1 expression. As shown in Figure 3B, 4 h of Dox treatment induced expression of Axin1 in the 293tr–V5–Axin1 cells (lane 2 versus 1), and this expression of Axin1 decreased ectopic c-Myc protein levels consistently by 60% as compared with control (lane 2 versus 1 and see Figure 5D).
Figure 3.
Acute expression of Axin1 increases c-Myc ubiquitination and downregulates c-Myc protein expression. (A) Axin1 expression increases c-Myc ubiquitination. 293 cells were co-transfected with expression plasmids for c-Myc, HA–ubiquitin, and V5–Axin as indicated. Cells were placed in low-serum media and treated with proteasome inhibitors. Ubiquitinated proteins were immunoprecipitated with α-HA. Input proteins and immunoprecipitated c-Myc were detected by western blotting as indicated. Ubi-Myc (vertical arrow) was quantified by the LI-COR software and the fold change between lane 4 versus 3 is shown. (B) Acute expression of Axin1 suppresses c-Myc expression. Stable 293tr–V5–Axin cells were transfected with HA–c-Myc and then treated with Dox for 4 h as indicated. V5–Axin1 and HA–c-Myc were detected by western blot. (C) Endogenous c-Myc expression is suppressed by acute Axin1 expression. 293tr–V5–Axin cells were treated with or without Dox for the indicated times and cells were collected for western blot analysis as indicated. (D) Acute Axin1 expression suppresses endogenous c-Myc protein prior to affecting c-myc mRNA levels. c-Myc protein levels from (C) were quantified and adjusted to actin. Cells from (C) were also subject to quantitative RT–PCR for c-myc mRNA and 18S rRNA (control). Average fold change for c-Myc protein/actin and c-myc mRNA/18S relative to paired control are graphed as indicated from three independent experiments±s.d.
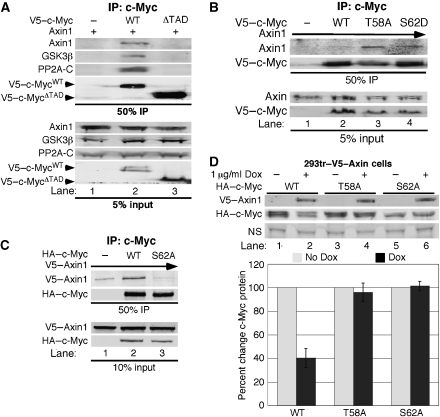
Figure 5.
Axin1 associates with the transactivation domain of c-Myc dependent on S62 phosphorylation. (A) The transactivation domain of c-Myc is required for its interaction with Axin1. 293 cells were co-transfected with expression plasmids for Axin1 and either V5–empty, V5–c-MycWT, or V5–c-MycΔTAD and then subjected to immunoprecipitation with αV5. Input and immunoprecipitated proteins were detected by western blot as indicated. (B) Constitutive S62 phosphorylation increases the association of c-Myc with Axin1. 293 cells were co-transfected with Axin1 and either V5–empty, V5–c-MycWT, V5–c-MycT58A, or V5–c-MycS62D. α-V5 co-IPs were washed stringently with a buffer containing 300 mM NaCl. Input and immunoprecipitated proteins were detected by western blot as indicated. (C) Phosphorylation at S62 is required for c-Myc to associate with Axin1. 293 cells were co-transfected with Axin1 and either HA–empty, HA–c-MycWT, or HA–c-MycS62A. α-HA co-IPs were washed stringently as above. Input and immunoprecipitated proteins were detected by western blot. (D) T58 phosphorylation is required for Axin1-mediated c-Myc turnover. 293tr–V5–Axin cells were transfected with HA–c-MycWT, HA–c-MycT58A, or HA–c-MycS62A and then treated with Dox for 4 h as indicated. V5–Axin1 and HA–c-Myc were detected by western blot (representative shown with nonspecific band loading control (NS)), quantified using the LI-COR software, and averages±s.d. from three separate experiments were graphed.
Acute expression of Axin1 reduces endogenous c-Myc protein levels independent of its effects on c-myc transcription
Our results have demonstrated that Axin1 can negatively regulate ectopic c-Myc protein levels driven from a constitutive enhancer. However, we also examined the effect of Axin1 overexpression on endogenous c-Myc protein levels and found that acute expression of Axin1 in the 293tr–V5–Axin1 cells consistently reduced endogenous c-Myc protein levels by approximately 50% (Figure 3C and D, lane 2 versus 1). We saw no effect of Dox treatment on endogenous c-Myc levels in parental 293tr cells that do not ectopically express Axin1 (Supplementary Figure 3). As a positive control for Axin1 post-translational function on a known target protein, we assessed endogenous β-catenin protein levels and found that β-catenin was also consistently reduced by approximately 50% under the same conditions (Figure 3C, lane 2 versus 1). We also examined c-myc mRNA levels in the above experiments by quantitative RT–PCR analysis as β-catenin/TCF can transcriptionally activate the c-myc gene (He et al, 1998). We found no effect on c-myc mRNA expression levels with activation of Axin1 out to 12 h (Figure 3D, grey bars). However, after 24 h of ectopic Axin1 expression, c-myc mRNA levels were reduced. Although short-term ectopic Axin1 expression reduced endogenous c-Myc protein expression without affecting its mRNA levels, longer term expression resulted in increased c-Myc protein levels, and β-catenin levels were also no longer reduced (Figure 3C, lanes 4, 6, 8, and 10). Endogenous c-Myc was consistently more sensitive to these ‘dominant-negative' effects of increased Axin1 expression compared with ectopic c-Myc, which was degraded by higher Axin 1 expression with 4 h of Dox treatment (compare Figure 3B and C). The discrepancy in c-Myc protein versus c-myc mRNA expression suggests that long-term Axin1 overexpression in 293 cells stoichiometrically reduces productive degradation complexes, preventing efficient c-Myc and β-catenin degradation. Altogether, these results demonstrate that Axin1 can negatively regulate endogenous c-Myc protein expression independent of its effects on the β-catenin/TCF transcriptional regulation of c-myc mRNA expression.
Axin1 expression reduces c-Myc-dependent transcription from the E2F2 promoter
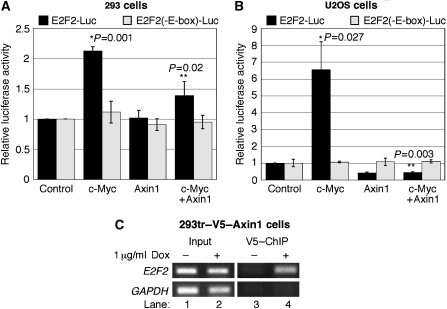
We assessed the effect of Axin1 expression on c-Myc-dependent activation of the E2F2 promoter. As shown in Figure 4A, ectopically expressed c-Myc increased E2F2-driven luciferase activity by two-fold from a luciferase reporter plasmid containing consensus c-Myc-binding E-box elements (Sears et al, 1997). This modest increase in c-Myc-dependent transcription is consistent with numerous previous reports and is dependent upon intact c-Myc-binding sites as the E2F2(-E-box)-Luc showed no change in luciferase activity (Figure 4A). Ectopic Axin1 expression alone had no observable effect on E2F2 promoter activity in 293 cells (Figure 4A). However, co-expression of Axin1 with c-Myc significantly reduced c-Myc-dependent E2F2 promoter activation in these cells (Figure 4A). We observed similar results in U2OS cells except that c-Myc expression alone caused a more robust activation of the E2F2 promoter (Figure 4B). This is most likely due to the deletion of ARF in these cells, as ARF has been shown to inhibit c-Myc transcriptional activity (Qi et al, 2004; Amente et al, 2006). We observed a consistent reduction in basal E2F2 promoter activity in U2OS cells with Axin1 expression alone and a dramatic reduction with co-expression of c-Myc (Figure 4B, third and fourth column sets). As Axin1 expression did not affect the Myc-binding site mutant (-E-box), it is most likely that Axin1 is affecting the activity of endogenous c-Myc as well as ectopic c-Myc on the E2F2 promoter in U2OS cells. Next, we assessed whether Axin1 could be found at the endogenous E2F2 promoter. 293tr–V5–Axin1 cells were stimulated as indicated in Figure 4C with Dox to induce V5–Axin1 expression. Chromatin immunoprecipitation (ChIP) with anti-V5 antibody revealed that Axin1 was present on the E2F2 promoter detected with primers spanning the Myc-binding sites (Figure 4C, lane 4). This finding suggests that Axin1 may suppress c-Myc-dependent transcription at c-Myc target gene promoters.
Figure 4.
Axin1 expression suppresses c-Myc-dependent transcription. (A) c-Myc-dependent activation of the E2F2 promoter is inhibited by Axin1 expression in 293 cells. 293 cells were co-transfected with expression plasmids for β-gal, c-Myc, Axin1, and either E2F2-Luc (containing wild-type E-boxes) or E2F2(-E-box)-Luc (containing mutant E-boxes) as indicated. Luciferase activity was measured and adjusted for transfection efficiency by β-gal assay and three separate experiments were graphed with averages±s.d. Statistical significance is indicated by P-values between c-Myc and control (*), and c-Myc+Axin and c-Myc (**). (B) Axin1 potently inhibits c-Myc activation of the E2F2 promoter in U2OS cells. The same as (A) except U2OS cells were used. (C) Axin1 is present at the c-Myc transcriptional target E2F2 gene promoter. 293tr–V5–Axin1 cells were treated with Dox as indicated and α-V5 conjugated to protein A beads was used for ChIP (see Materials and methods) followed by PCR for the E2F2 promoter region and GAPDH control.
Axin1 associates with the transactivation domain of c-Myc dependent on S62 phosphorylation
We examined whether the transactivation domain (TAD) of c-Myc, which includes the T58 and S62 phosphorylation sites, is important for its association with Axin1. We found that Axin1 co-immunoprecipitated with c-MycWT, but not c-Myc lacking the TAD (c-MycΔTAD) (Figure 5A, lane 2 versus 3). Likewise, GSK3β and PP2A-C also failed to associate with c-MycΔTAD, consistent with a role for Axin1 in coordinating their association with c-Myc. We also examined how the phosphorylation status of c-Myc at T58 and S62 would affect its association with Axin1. To test the effects of constitutive phosphorylation at S62, we utilized two phosphorylation mutants we have previously characterized, the c-MycS62D phosphorylation mimic and c-MycT58A, which has high S62 phosphorylation levels due to the inability of PP2A to dephosphorylate this mutant at S62 (Lutterbach and Hann, 1994; Chang et al, 2000; Sears et al, 2000; Yeh et al, 2004; Arnold and Sears, 2006). To challenge the interaction of Axin1 with the different forms of c-Myc, immunoprecipitates were washed stringently. We found that Axin1 associated more strongly with both c-MycS62D and c-MycT58A compared with c-MycWT (Figure 5B, lanes 3 and 4 versus 2). We also found that Axin1 did not associate with c-Myc lacking S62 phosphorylation, c-MycS62A, compared with c-MycWT (Figure 5C, lane 3 versus 2). These findings suggest that S62 phosphorylation is most likely important for the recruitment and/or retention of c-Myc to the Axin1 degradation complex.
Axin1-mediated c-Myc degradation requires T58 phosphorylation
We tested the ability of Axin1 to negatively regulate the expression of the c-MycT58A and c-MycS62A phosphorylation mutants. Unlike c-MycWT, which showed a consistent decrease of 60% with induction of Axin1 (Figure 5D, lane 2 versus 1 and graph), we found no effect of Axin1 expression on c-MycT58A or c-MycS62A protein levels (Figure 5D, lanes 3–6 and graph). The inability of Axin1 expression to affect c-MycS62A levels can be explained by its lack of interaction with Axin1 (Figure 5C). In contrast, Axin1 showed increased association with c-MycT58A (Figure 5B), but did not stimulate its degradation, suggesting a substrate-trapping phenomenon. Consistent with this, T58 phosphorylation is required for recognition by Fbw7, and the c-MycT58A mutant is resistant to turnover by the SCFFbw7 ubiquitin ligase machinery (Welcker et al, 2004; Yada et al, 2004). These results suggest that T58 phosphorylation is critical for Axin1-mediated c-Myc turnover.
Multiple domains in Axin1 are important for its association with c-Myc
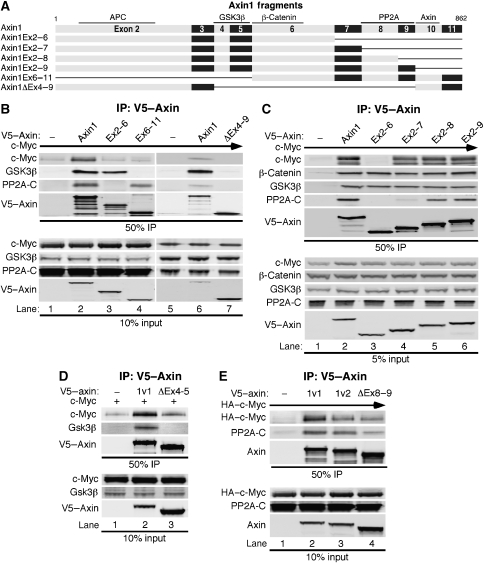
We performed mapping experiments to examine what domain(s) in Axin1 are important for its association with c-Myc. Initially, we tested whether c-Myc bound to either an amino, Axin1Ex2−6, or carboxyl, Axin1Ex6−11, fragment of Axin1 (Figure 6A). We found that c-Myc co-immunoprecipitated with full-length Axin1, but not with either amino or carboxyl fragment (Figure 6B, top panel, lanes 3 and 4 versus 2). We also found that a large internal deletion encompassing exons 4–9 of Axin1 did not bind to c-Myc (Figure 6B, lane 7). We generated three more Axin1 amino-terminal fragments, Axin1Ex2−7, Axin1Ex2−8, and Axin1Ex2−9 (Figure 6A) to sequentially extend the Axin1Ex2−6 fragment that did not associate with c-Myc. Interestingly, we found that just adding exon 7 to the AxinEx2−6 fragment (AxinEx2−7) resulted in the association of c-Myc with Axin1 (Figure 6C, lane 4 versus 3), and this association increased as exons 8 and 9 were sequentially added (lanes 5 and 6). Taken together, these data demonstrate that exon 7 is important for c-Myc binding but that other amino and carboxyl sequences are also involved as c-Myc did not bind Ex6–11 alone and exons 8 and 9 contribute to binding. However, it is important to note that all of these mapping experiments involve c-Myc and Axin1 overexpression. In contrast, endogenous β-catenin bound all amino-terminal fragments of Axin1 examined (Figure 6C, lanes 2–6), which is consistent with previous reports mapping the β-catenin-binding domain to exon 6 (Ikeda et al, 1998; Xing et al, 2003). These results suggest that c-Myc and β-catenin require different regions of Axin1 for association. Immunofluorescent analysis of the various Axin1 fragments demonstrated that none of these mutants were mislocalized relative to full-length Axin1, which showed predominately cytoplasmic and variable amounts of nuclear staining consistent with its known nuclear/cytoplasmic shuttling (Cong and Varmus, 2004) (Supplementary Figure 4).
Figure 6.
The interaction of c-Myc with Axin1 requires exon 7 and is facilitated by the GSK3β- and PP2A-binding domains. (A) Schematic diagram of Axin1-coding exons and deletion mutants with known binding domains indicated. (B) Both amino- and carboxyl regions in Axin1 are required for c-Myc association. 293 cells were co-transfected with expression plasmids for c-Myc and either V5–empty, V5–Axin1, V5–AxinEx2−6, V5–AxinEx6−11, or V5–AxinΔEx4−9. Cells were lysed and subjected to α-V5 IP. Input and immunoprecipitated proteins were detected by western blot as indicated. (C) Exon 7 of Axin1 is required for the association of c-Myc. The same as (B) except different Axin1 fragments were used as indicated. (D) The GSK3β-binding domain in Axin1 is important for c-Myc association. The same as (B) except V5–AxinΔEx4−5 was used. (E) The PP2A-binding domain in Axin1 contributes to c-Myc association. The same as (B) except the splice variant, V5–Axin1v2 missing exon 9, and V5–AxinΔEx8−9 were used.
Analysis of the association of GSK3β and PP2A-C with the amino-terminal Axin1 fragments demonstrated that endogenous GSK3β bound to all of them (Figure 6B, lanes 2, 3, and 6, and 6C, lanes 2–6), consistent with previous reports that GSK3β associates within exons 4 and 5 (Itoh et al, 1998). In contrast, the C subunit of PP2A associated with full-length Axin1 and the carboxyl Axin1Ex6−11 fragment (Figure 6B, lanes 2 and 4), but not Axin1Ex2−6 (lane 3). It also associated with Axin1Ex2−8 and more strongly with Axin1Ex2−9 (Figure 6C, lanes 5 and 6), consistent with a reported binding region for PP2A within exons 8 and 9 (Hsu et al, 1999).
GSK3 and PP2A facilitate the association of c-Myc with Axin1
As both amino and carboxyl domains in Axin1 appear important for c-Myc association, we examined the effect of removing the GSK3β- and PP2A-binding domains from Axin1 on c-Myc interaction. Removal of exons 4 and 5 in Axin1 prevented its interaction with GSK3β (Figure 6D, lane 3) and we also found c-Myc association to be substantially reduced (Figure 6D, lane 3 versus 2). The naturally occurring splice variant of Axin1 (Axin1v2), in which exon 9 is spliced out, also showed reduced association with c-Myc as well as PP2A-C, and this was exacerbated by the removal of both exons 8 and 9 (Figure 6E, lanes 2–4). Taken together, our results demonstrate that multiple components facilitate Axin1–c-Myc complex formation.
Cancer cell lines with deletion, mutation, or compromised Axin1 function show impaired c-Myc regulation
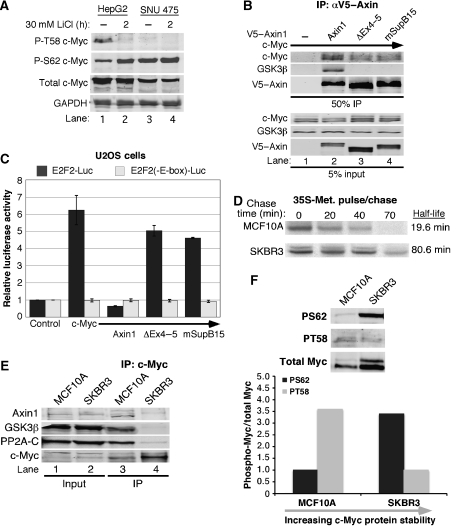
Approximately 10% of hepatocellular carcinomas (HCCs) harbour missense mutations or deletions in the AXIN1 gene, and loss of heterozygosity at the AXIN1 locus is often observed (Taniguchi et al, 2002). One such HCC cell line, SNU475 showed loss of exons one and two, and no AXIN1 transcript was detected (Satoh et al, 2000). In contrast, the HCC cell line HepG2 is wild type for Axin1. We examined the phosphorylation status of c-Myc in both the HepG2 and SNU475 cell lines. As shown in Figure 7A, c-Myc is phosphorylated at T58 in HepG2 cells but not in SNU475 cells, consistent with the role of Axin1 in promoting the interaction between c-Myc and GSK3β (compare lanes 1 and 3). Short treatment with lithium chloride, an inhibitor of GSK3β, reduced T58 phosphorylation and increased S62 phosphorylation in the HepG2 cells (lane 2 versus 1), consistent with the inter-relationship between phospho-T58 and phospho-S62 levels (Yeh et al, 2004). S62 phosphorylation was constitutively elevated in the SNU475 cells (lanes 3 and 4). The reduced phospho-T58 and elevated phospho-S62 observed in the SNU475 cells relative to the HepG2 cells is similar to the change in c-Myc phosphorylation observed with Axin1 knockdown (Figure 2F).
Figure 7.
Disruption of Axin1 regulation of c-Myc in human cancer cell lines. (A) Phosphorylation status of c-Myc in hepatocellular carcinomas. Cells were treated with 30 mM LiCl for the indicated time, lysed in SDS–PAGE loading buffer, and detected by western blot as indicated. (B) Decreased association of c-Myc with an Axin1 mutant identified in the SupB15 ALL cell line. 293 cells were co-transfected with expression plasmids for c-Myc and either V5–empty, V5–Axin1, V5–AxinΔEx4−5, or V5–AxinmSupB15 Cells were lysed and subjected to α-V5 IP. Input and immunoprecipitated proteins were detected by western blot as indicated. (C) Axin1 mutants lacking GSK3β binding no longer suppress c-Myc-dependent transcription. 293 cells were co-transfected with β-gal, c-Myc, Axin1, V5–AxinΔEx4−5, and V5–AxinmSupB15 and either E2F2-Luc or E2F2(-E-box)-Luc, as indicated. Luciferase activity was measured and adjusted for β-gal and three separate experiments were graphed with averages±s.d. (D) Increased c-Myc stability in SKBR3 breast cancer cells. SKBR3 or control MCF10A cells were pulse-labelled with 35S-methionine and then chased for the indicated times with media containing excess unlabelled methionine. Endogenous 35S-c-Myc was immunoprecipitated from lysates with equal radioactive counts and detected by SDS–PAGE radiography. 35S-c-Myc was quantified by phosphorimager and c-Myc half-life was calculated with Excel. (E) Association of c-Myc with Axin1, GSK3β, and PP2A is impaired in SKBR3 breast cancer cells. Endogenous c-Myc was immunoprecipitated from MCF10A and SKBR3 cells. Input and immunoprecipitated proteins were detected by western blot as indicated. (F) c-Myc exhibits high S62 and low T58 phosphorylation in a breast cancer cell line that shows impaired interaction between c-Myc and Axin1. Lysates from MCF10A and SKBR3 were immuno-blotted with T58 or S62 phospho-specific and total c-Myc antibodies. Visualization and quantitation were carried out with the LI-COR imaging system and the ratio of phospho-Myc to total Myc is graphed.
We have previously characterized a number of leukaemia cell lines as well as bone marrow samples from paediatric patients with acute lymphoblastic leukaemia (ALL) and found that c-Myc protein stability was significantly increased in the majority of tested samples (Malempati et al, 2006; O'Neil et al, 2007). In particular, c-Myc protein half-life was increased to 70 min in the pre-B ALL cell line, SupB15, as compared with another leukaemia cell line, HL60, which showed a normal turnover rate for c-Myc of 20 min. Moreover, c-Myc association with GSK3β was disrupted in the SupB15 cell line as compared with the HL60 cell line, although the mechanism for this was unknown at the time of publication. We have now sequenced AXIN1 mRNA in the SupB15 cell line. Strikingly, we found an in-frame deletion that removes amino acids 373–418, which includes most of the GSK3α/β-binding domain. This is the first report of an Axin1 mutation in a blood cancer. We assessed the ability of the mutant form of Axin1 from the SupB15 cells to associate with c-Myc. We found that c-Myc did not associate with Axin1mSupB15 as robustly as wild-type Axin1 (Figure 7B, lane 4 versus 2). We also found that GSK3β did not associate with this mutant at all (lane 4). These findings most likely explain the impaired association of c-Myc with GSK3β in the SupB15 cells, as well as the increased stability and decreased T58 phosphorylation observed in this cell line (Malempati et al, 2006). We also examined the effect of Axin1mSupB15 on c-Myc-dependent transcription and found that neither Axin1ΔEx4−5 nor Axin1mSupB15 significantly suppressed c-Myc-dependent E2F2-driven luciferase activity as did full-length Axin1 (Figure 7C). This result most likely reflects both the reduced association of these Axin1 mutants with c-Myc and an important role for GSK3β in Axin1's ability to suppress c-Myc-dependent transcription.
Although there are a number of reported mutations in Axin1 from solid tumours, only non-coding or silent mutations have been detected in breast cancers (Webster et al, 2000; Salahshor and Woodgett, 2005). We sequenced AXIN1 mRNA from several breast cancer cell lines in which we found c-Myc to have increased stability and similarly found no AXIN1-coding sequence mutations. We examined the formation of the Axin1–c-Myc degradation complex in a control mammary epithelial cell line, MCF10A, in which c-Myc is unstable with a 20 min half-life (Figure 7D, top panel) and found that immunoprecipitation of endogenous c-Myc readily co-precipitated endogenous Axin1, GSK3β, and PP2A-C (Figure 7E, lane 3). We compared this to a breast cancer cell line, SKBR3, in which c-Myc has increased stability to 80 min (Figure 7D, bottom panel) and consistently observed that immunoprecipitation of endogenous c-Myc from the breast cancer SKBR3 cells co-immunoprecipitated substantially less Axin1, GSK3β, and PP2A-C (Figure 7E, lane 4). We examined whether this reduced association with Axin1 affected c-Myc phosphorylation in the SKBR3 cells. We found dramatically increased phospho-S62 and decreased phospho-T58 in the SKBR3 cells compared with MCF10A control cells (Figure 7F). This is again consistent with the phosphorylation status of c-Myc in the SNU475 and SupB15 cell lines (Malempati et al, 2006), and with knockdown of Axin1 (Figure 2F). This shows that in the absence of mutations in either c-Myc or Axin1, the association of c-Myc with Axin1, as well as GSK3β and PP2A can still become disrupted leading to deregulation of c-Myc T58 and S62 phosphorylation and stability.
Discussion
The regulation of c-Myc expression at the post-translational level involves a number of proteins, which sequentially modify the phosphorylation status of c-Myc on the conserved T58 and S62 residues (Sears, 2004; Escamilla-Powers and Sears, 2007). To date, we have focused on the signalling pathways that activate or repress the activity of these proteins. It is now becoming clear that signalling pathways are regulated by the coordination of proteins into complexes by multi-domain scaffold proteins. We previously characterized critical roles for GSK3β, PP2A-B56α, and Pin1 in promoting c-Myc protein turnover (Sears et al, 2000; Yeh et al, 2004; Arnold and Sears, 2006). We report here that the scaffold protein Axin1 facilitates the association of GSK3β, PP2A-B56α, and Pin1 with c-Myc, which negatively regulates c-Myc protein expression and transcriptional activity. Our findings expand our understanding of the regulation of c-Myc and provide a new important target for the tumour suppressor activity of Axin1.
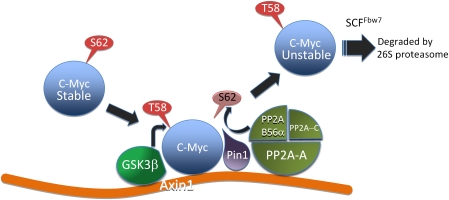
Formation of the Axin1-c-Myc degradation complex and sequential steps in c-Myc destruction
Previous work showed that initial S62 phosphorylation not only transiently stabilizes c-Myc but also primes the subsequent phosphorylation of T58 by GSK3β that targets c-Myc for ubiquitination and proteolysis (Lutterbach and Hann, 1994; Sears et al, 2000). We show here that S62 phosphorylation also promotes the association of c-Myc with Axin1 (see proposed model in Figure 8). Moreover, degradation of c-Myc and its release from Axin1 appear to require subsequent T58 phosphorylation, consistent with the reported requirement of T58 phosphorylation for SCFFbw7 recognition and poly-ubiquitination of c-Myc targeting it to the 26S proteasome (Welcker et al, 2004; Yada et al, 2004).
Figure 8.
Model showing sequential steps in c-Myc degradation and the role of Axin1 as a scaffold in this process. Initial phosphorylation of S62 allows the association of c-Myc with Axin1. Axin1-associated GSK3β can then phosphorylate T58. Pin1 recognizes P-T58 and isomerizes proline 63 from cis to trans. Axin1-associated PP2A-B56α can then dephosphorylate S62, allowing c-Myc recognition by Fbw7 and subsequent proteasomal degradation.
Although we cannot conclude from our experiments that the interaction between c-Myc and Axin1 is direct, our mapping experiments with Axin1 indicate that exon 7 of Axin1 is critical for at least ectopic c-Myc recruitment. Moreover, binding sites for GSK3α/β and PP2A facilitated c-Myc association with Axin1, suggesting that the formation of the Axin1-mediated degradation complex for c-Myc is a coordinated process. Consistent with this, we have previously reported that knockdown, knockout, or inhibition of PP2A-B56α, Pin1, or GSK3β, respectively, all increase c-Myc protein stability and expression to a similar degree as knockdown of Axin1 shown here (Sears et al, 2000; Yeh et al, 2004; Arnold and Sears, 2006). It is most likely that other proteins are involved in the Axin1–c-Myc degradation complex. For example, there may be an adapter protein to recruit SCFFbw7, analogous to WTX that brings in the SCFβTRCP E3 ligase to efficiently ubiquitinate β-catenin (Major et al, 2007). This could explain why we do not readily detect Fbw7 in a complex with Axin1 and c-Myc.
We have shown earlier that mitogenic signalling transiently stabilizes c-Myc as a cell exits quiescence and enters the cell cycle (Sears et al, 1999). Consistent with this, the effect of Axin1 knockdown on c-Myc expression is most apparent under low-serum culture conditions, and our preliminary experiments indicate that the association of GSK3β and PP2A with Axin1 is reduced with high serum cell culture conditions compared with low-serum conditions (Supplementary Figure 2). Although the mechanism of this regulation is unknown, it has been shown earlier that the mitogenic-stimulated kinases MEKK1 and GSK3β competitively associate with Axin1 resulting in a ‘switch' of Axin1-mediated complexes (Zhang et al, 2001).
Another important consideration for future research regarding Axin-mediated regulation of c-Myc expression is to assess whether a homologue of Axin1, Axin2, has any role in regulating c-Myc expression. In contrast to the ubiquitous and constitutive expression of Axin1, Axin2 is reported to have tissue-specific expression (Zeng et al, 1997). Axin2 expression has also been shown to be induced upon Wnt signalling as part of a negative feedback pathway in which Axin2 coordinates a degradation complex for β-catenin (Leung et al, 2002; Lustig et al, 2002). Whether Axin2 has a similar function in c-Myc regulation will certainly be explored and carefully examined in future experiments.
Mutation and/or impaired Axin1 function in human cancer cells can contribute to altered T58 and S62 phosphorylation and increased c-Myc protein stability
Multiple AXIN1 mutations have been observed in solid tumours, often occurring in the binding sites for GSK3α/β and PP2A (Salahshor and Woodgett, 2005). Such mutations are most likely to contribute to altered phosphorylation and aberrant stabilization of c-Myc in human cancer. Indeed, loss of Axin1 expression in the SNU475 HCC cell line was associated with loss of T58 phosphorylation and elevated S62 phosphorylation, a phosphorylation ratio that favours increased stability (Malempati et al, 2006). Additionally, we have identified an in-frame deletion disrupting most of the GSK3α/β-binding domain in Axin1 in a leukaemia cell line in which c-Myc was shown earlier to have increased stability and lack association with GSK3β (Malempati et al, 2006). This is the first report of an AXIN1 mutation in a blood cancer and future studies will include sequencing of AXIN1 in additional leukaemia cell lines and in primary leukaemia patient samples.
We have also observed increased c-Myc protein stability in several breast cancer cell lines, but this was not associated with any mutations. Instead, we observed that c-Myc was no longer able to associate with Axin1, GSK3β, or PP2A in a breast cancer cell line with more stable c-Myc compared with a mammary epithelial cell line with unstable c-Myc. This was also associated with similarly decreased T58 and increased S62 phosphorylation. Thus, even in the absence of Axin1 mutations, the ability of Axin1 to coordinate a degradation complex for c-Myc can be impaired, leading to altered c-Myc phosphorylation and increased stability. Although the mechanism for this is currently unknown, it clearly suggests that the scaffold function of Axin1 is subject to important regulation.
Axin1 is a key regulator of multiple signalling pathways
Given the important function Axin1 has in other signalling pathways such as Wnt, TGFβ, SAPK/JNK, and p53, the addition of c-Myc as an Axin1 target protein further highlights the critical function Axin1 has in cellular regulation. The expression of Axin1 is very low in cells and it has been reported to be a limiting factor in the turnover of β-catenin (Hart et al, 1998; Lee et al, 2003). Although our data suggest that the Axin1–c-Myc degradation complex does not involve APC and is not influenced by Wnt signalling, and c-Myc would be predicted to interact with nuclear Axin1, it remains an open question as to what extent changes in the expression of one Axin-regulated oncoprotein could influence Axin's regulation of its other targets. Clearly, further research is required to reveal how this important tumour suppressor coordinately regulates multiple oncogenic signalling pathways.
Materials and methods
Plasmids and RNAi
Expression plasmids CMV-empty, CMV-β-gal, CMV-Myc, CMV-HA-Myc, pD40-His/V5-c-Myc, pD40-His/V5-c-MycT58A, pD40-His/V5-c-MycS62A, pD40-His/V5-c-MycS62D, pD40-His/V5-c-MycΔTAD, E2F2-Luc, and E2F2(-E-box)-Luc have been described earlier (Sears et al, 1997; Yeh et al, 2004; Arnold and Sears, 2006). Full-length, human Axin1, Axin1v2, and AxinmSupB15 were PCR amplified with Pfu Ultra (Stratagene) from human liver cDNA library, cDNA from HL60 cells, and cDNA from SupB15 cells, respectively. PCR primers are listed in Supplementary Table 1. PCR products were cloned into pDEST40 mammalian expression vector through Gateway cloning. Axin1Ex6–11, Axin1Ex2–6, AxinEx2–7, AxinEx2–8, and AxinEx2–9 were PCR amplified from pDEST40-Axin1 using Pfu Ultra with primers listed in Supplementary Table 1. AxinΔEx4–5, AxinΔEx4–9, and AxinΔEx8–9 were generated by PCR amplifying two fragments of Axin1, ligated using T4 DNA ligase (Roche) and Gateway cloned into pDEST40 (detailed in Supplementary data). shRNA expression vectors were generated using a target sequence in Axin1, GAGGAAGAAAAGAGAGCCA. Oligos encoding the sense and antisense shRNA sequences were cloned into the pSUPER (OligoEngine) or pENTR-H1/TO (Invitrogen, Carlsbad, CA) shRNA expression vectors.
Cell lines and transfection
HEK-293 and U2OS cells were maintained in DMEM supplemented with 10% characterized fetal bovine serum (FBS), 2 mM L-glutamine, and 1 × penicillin/streptomycin at 37°C and 5% CO2. Plating of cells was done to achieve 60–80% confluency 24 h post-split for transfection. Transfections involving shRNAs were carried out as described earlier (Arnold and Sears, 2006). All other transfections were performed using Metafectene (Biontex, Germany) or HEK-Fectin (Bio-Rad). Total transfected DNA was held constant by the addition of empty control plasmid and included 50 ng of CMV-β-gal to normalize for transfection efficiencies between experimental conditions. Experimental cells were maintained in 2% or 0.2% FBS and L-glutamine for 24–48 h prior to collection.
MCF10A cells were grown in 45% DMEM, 45% F-12 hams, 5% horse serum, 2.5 mM L-glutamine, 20 ng/ml EGF, 10 μg/ml insulin, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 1 × penicillin/streptomycin. SKBR3 and HepG2 cells were grown in DMEM with 10% FBS and 1 × penicillin/streptomycin. SupB15 and SNU475 cells were grown in RPMI with 10% FBS (defined FBS for SupB15), 2 mM L-glutamine, and 1 × penicillin/streptomycin.
Stable 293tr–V5–Axin1 and 293tr–shAxin1 cells were generated as detailed in the Supplementary data. Briefly, HEK-293 cells (ATCC) were infected with lentivirus encoding the tet-repressor, pLenti6/TR (Invitrogen). The best TR expressing clone was then infected with lentivirus expressing V5–Axin1, pLenti4/TO/V5-Dest-Axin1 or transfected with pENTR/H1/TO-shRNA-Axin1. The resulting 293tr–V5–Axin1 or 293tr–shAxin1 cell line was then maintained in DMEM supplemented with 10% defined FBS, 2 mM L-glutamine, 5 μg/ml blasticidin, and 100 μg/ml zeocin. For experiments, 293tr–V5–Axin1 cells were placed in DMEM with 0.2% defined FBS for 24 h prior to the addition of Dox.
In vitro protein expression
T7-driven HA–c-Myc or V5-Axin1 was expressed using the T7 Quick Coupled Transcription/Translation kit (Promega) according to the manufacturer's protocol.
Antibodies
c-Myc antibodies (N262, C-33, and agarose-conjugated C-33), B56α, actin, and Sp1 (PEP-2) are from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA.11 is from Covance (Berkeley, CA). Rabbit HA tag antibody (rabHA), c-Myc (Y69), and APC (Ali 12–28) are from Abcam (Cambridge, MA). PP2A-Cα antibody is from BD Biosciences (San Jose, CA). Mouse V5 antibody is from Invitrogen. β-Catenin and α-tubulin antibodies are from Sigma (St Louis, MO). Generation of the c-Myc serine62 phospho-specific antibody (αP-S62) is described (Escamilla-Powers and Sears, 2007). The threonine58 phospho-specific (αP-T58) and GSK3β antibodies are from Cell Signaling Technology (Beverly, MA). Specificity of the phospho-T58 c-Myc antibody is achieved by blocking with milk (see western blotting for details). The threonine58 phospho-specific antibody used in Figure 7A is from Applied Biological Materials Inc. (BC, Canada). Pin1 antibody is from GenWay Biotech (San Diego, CA). Anti-active β-catenin 8E7 is from Millipore (Temecula, CA).
Co-immunoprecipitation
Lysates were collected in 10 × cell pellet volumes of co-IP buffer: 20 mM Tris, pH 7.5, 12.5% glycerol, 0.2% NP-40, 200 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT plus protease and phosphatase inhibitors (Supplementary data). Lysates were incubated on ice for 20 min, sonicated 10 pulses (output=1 and 10% duty), and cleared by centrifugation at 20K r.c.f. for 10 min at 4°C. Cleared lysate volumes were adjusted for transfection efficiency by β-gal assay. Alternatively, in vitro samples were diluted in 250 μl of co-IP buffer. Immunoprecipitations were carried out with antibodies at the following dilutions, 1:150 dilution of agarose-conjugated αC-33, 1:150 conjugated αSp1, 1:500 dilution αHA.11, or 1:750 dilution of αV5 conjugated to either protein A or G depending upon immunoglobulin type. Three washes with 10 × volumes of co-IP buffer with 1-min incubation during each wash. Where specified, washes were carried out with co-IP buffer containing 300 mM NaCl.
Western blotting
Total cell lysates were collected as described earlier (Arnold and Sears, 2006). Lysates from transfected cells were subjected to β-gal assay to normalize for transfection efficiency. Normalized samples were separated by SDS–PAGE and western blotted using the LI-COR Odyssey Infrared Imager system (Lincoln, Nebraska) as described earlier (Arnold and Sears, 2006) and detailed in the Supplementary data. Quantification of western blots was carried out using the LI-COR Odyssey Infrared Imager software version 1.2. Error bars in graphs represent two standard deviations as calculated using Excel (Microsoft, Redmond, WA). t-test analysis (two-tailed distribution and two-sample unequal variance) was carried out using Excel to determine statistical differences as indicated on graphs.
qRT–PCR analysis
RNA was isolated in 1 ml TRIzol reagent (Invitrogen) and DNase treated in 100 mM MgCl2, 10 mM DTT, RNasin (Promega), RNase-free DNase (Roche) for 15 min at 37°C and purified using RNeasy (Qiagen). cDNA was made using M-MLV Reverse Transcriptase (Invitrogen) with oligo dT primers. qRT–PCR analysis was carried out using Taqman primers for c-myc and 18S designed by Applied Biosystems (cat. no. Hs00905030_m1 and Hs99999901_s1) on a 7300 qRT–PCR machine (Applied Biosystems).
Cyclohexamide half-life
Cyclohexamide half-life was carried out as described earlier (Arnold and Sears, 2006). Briefly, transfected cells were split into six plates. 24 h post-splitting, cells were placed in starve media (DMEM supplemented with 0.2% FBS and L-glutamine) for 48 h. Cyclohexamide (100 μg/ml) was added to inhibit protein synthesis and sample collection began 10 min later for western blot analysis.
Ubiquitin assay
293 cells were transfected as indicated and 12 h following transfection cell media were changed to DMEM supplemented with 2% FBS. At 32 h post re-feeding, cells were treated with 10 μm each MG132 and MG115 for 4 h, and collected in RIPA lysis buffer (Supplementary data), sonicated (20 pulses, output=2.5, and 30% duty), and cleared by centrifugation (20K r.c.f. for 10 min at 4°C). Lysate volumes were adjusted based on β-gal activity, input samples were collected, and remaining lysate volume was immunoprecipitated with conjugated αHA.11. Immunoprecipitates were washed 4 × with RIPA buffer and then subjected to western blot analysis.
Luciferase assay
Luciferase assay was carried out as described earlier (Arnold and Sears, 2006). TOPFLASH reporter plasmid is from Millipore (Billerica, MA). Luciferase activity was detected using a Promega Luciferase Assay kit and Berthold luminometer (Bundoora, Australia). Luciferase activity was adjusted for β-gal activity.
35S-Methionine pulse/chase
35S-Methionine pulse/chase was carried out as described earlier (Sears et al, 2000). Briefly, cells were pre-starved of methionine and cystine and labelled in vivo with 600 μCi/ml 35S-methionine/cysteine (PerkinElmer, Boston, MA) in DMEM medium without L-met/L-cys with 10% dialysed FBS for 20–30 min. Cells were then chased with DMEM medium with 10% FBS, 5 mM L-methionine, and 3 mM L-cysteine. Cells were harvested in Ab lysis buffer (Supplementary data). Labelled c-Myc was immunoprecipitated from an equal number of cells and equal radioactive counts for each time point using the c-Myc C33 monoclonal antibody-conjugated beads. Immunoprecipitated c-Myc was separated by SDS–PAGE, and radiolabelled c-Myc was quantified using a phosphorimager. After background subtraction, band intensity was plotted on a log scale for each time point using Microsoft Excel graphing function, and c-Myc half-life was determined from the graph.
ChIP analysis
293tr–V5–Axin1 cells were crosslinked with a 1% formaldehyde solution in phosphate-buffered saline for 10 min and stopped by incubating the cells in 0.125 M glycine for 5 min. ChIP analysis was performed as described (Zeng et al, 2002) using anti-V5 (Invitrogen) coupled to protein A beads. Immunoprecipitated DNA fragments were purified by phenol–chloroform extraction and ethanol precipitation and then analysed by semiquantitative PCR using 2 × Immomix Red from BIOLINE (Randolph, MA) with primers for E2F2 and GAPDH genes (Supplementary data).
Supplementary Material
Supplementary Material
Acknowledgments
We thank members of the Sears lab for a critical reading of this paper. We thank Dr Bruno Amati for helpful comments on this work. We thank Dr Virshup for providing us with several Axin1 expression constructs. HKA received support from the NIH training grant 5-T32-GM08617. This study was supported by the NIH/NCI grant R01-CA100855 and Department of Defense grant BC061306 to RCS.
References
- Amente S, Gargano B, Varrone F, Ruggiero L, Dominguez-Sola D, Lania L, Majello B (2006) p14ARF directly interacts with Myc through the Myc BoxII domain. Cancer Biol Ther 5: 287–291 [DOI] [PubMed] [Google Scholar]
- Arnold HK, Sears RC (2006) Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol 26: 2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W (1998) Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science 280: 596–599 [DOI] [PubMed] [Google Scholar]
- Chang DW, Claassen GF, Hann SR, Cole MD (2000) The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol 20: 4309–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Varmus H (2004) Nuclear–cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci USA 101: 2882–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K (1999) Function of the c-Myc oncogenic transcription factor. Exp Cell Res 253: 63–77 [DOI] [PubMed] [Google Scholar]
- Escamilla-Powers JR, Sears RC (2007) A conserved pathway that controls c-Myc protein stability through opposing phosphorylation events occurs in yeast. J Biol Chem 282: 5432–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, Costantini F (1999) Domains of axin involved in protein–protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol 145: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, Hann SR (2003) Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 278: 51606–51612 [DOI] [PubMed] [Google Scholar]
- Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P (1998) Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol 8: 573–581 [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW (1998) Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Hsu W, Zeng L, Costantini F (1999) Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem 274: 3439–3445 [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J 17: 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Krupnik VE, Sokol SY (1998) Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta-catenin. Curr Biol 8: 591–594 [DOI] [PubMed] [Google Scholar]
- Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW (2003) The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 1: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER (2002) Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J Biol Chem 277: 21657–21665 [DOI] [PubMed] [Google Scholar]
- Li X, Yost HJ, Virshup DM, Seeling JM (2001) Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J 20: 4122–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, Ye Z, Zhang S, Chan SC, Chen YG, Han J, Lin SC (2006) Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J 25: 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol 22: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR (1994) Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 14: 5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, Angers S, Moon RT (2007) Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science 316: 1043–1046 [DOI] [PubMed] [Google Scholar]
- Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, Sears RC (2006) Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 20: 1572–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT (2007) FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 204: 1813–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR (2004) p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431: 712–717 [DOI] [PubMed] [Google Scholar]
- Rui Y, Xu Z, Lin S, Li Q, Rui H, Luo W, Zhou HM, Cheung PY, Wu Z, Ye Z, Li P, Han J, Lin SC (2004) Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J 23: 4583–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahshor S, Woodgett JR (2005) The links between axin and carcinogenesis. J Clin Pathol 58: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y (2000) AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet 24: 245–250 [DOI] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR (1999) Ras enhances Myc protein stability. Mol Cell 3: 169–179 [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR (1997) Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol 17: 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC (2004) The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3: 1133–1137 [PubMed] [Google Scholar]
- Staal FJ, Noort Mv M, Strous GJ, Clevers HC (2002) Wnt signals are transmitted through N-terminally dephosphorylated beta-catenin. EMBO Rep 3: 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, Murphy LM, Nagorney DM, Burgart LJ, Roche PC, Smith DI, Ross JA, Liu W (2002) Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21: 4863–4871 [DOI] [PubMed] [Google Scholar]
- Webster MT, Rozycka M, Sara E, Davis E, Smalley M, Young N, Dale TC, Wooster R (2000) Sequence variants of the axin gene in breast, colon, and other cancers: an analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer 28: 443–453 [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JA, Harper JW, Eisenman RN, Clurman BE (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA 101: 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Kimelman D, Xu W (2003) Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev 17: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI (2004) Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23: 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, Counter CM, Nevins JR, Means AR, Sears R (2004) A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 6: 308–318 [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL III, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F (1997) The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90: 181–192 [DOI] [PubMed] [Google Scholar]
- Zeng SX, Dai MS, Keller DM, Lu H (2002) SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J 21: 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Neo SY, Wang X, Han J, Lin SC (1999) Axin forms a complex with MEKK1 and activates c-Jun NH(2)-terminal kinase/stress-activated protein kinase through domains distinct from Wnt signaling. J Biol Chem 274: 35247–35254 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qiu WJ, Liu DX, Neo SY, He X, Lin SC (2001) Differential molecular assemblies underlie the dual function of Axin in modulating the WNT and JNK pathways. J Biol Chem 276: 32152–32159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material