Abstract
The steroid hormone oestrogen can signal through several receptors and pathways. Although the transcriptional responses mediated by the nuclear oestrogen receptors (ER) have been extensively characterized, the changes in gene expression elicited by signalling through the membrane-associated ER GPR30 have not been studied. We show here for ER-negative human breast cancer cells that the activation of GPR30 signalling by oestrogen or by hydroxytamoxifen (OHT), an ER antagonist but GPR30 agonist, induces a transcription factor network, which resembles that induced by serum in fibroblasts. The most strongly induced gene, CTGF, appears to be a target of these transcription factors. We found that the secreted factor connective tissue growth factor (CTGF) not only contributes to promote proliferation but also mediates the GPR30-induced stimulation of cell migration. These results provide a framework for understanding the physiological and pathological functions of GPR30. As the activation of GPR30 by OHT also induces CTGF in fibroblasts from breast tumour biopsies, these pathways may be involved in promoting aggressive behaviour of breast tumours in response to endogenous oestrogens or to OHT being used for endocrine therapy.
Keywords: oestrogen, G-protein coupled receptor, microarray, signal transduction, tamoxifen resistance
Introduction
The steroid hormone oestrogen binds and activates the oestrogen receptors (ER) α and β, two members of the nuclear receptor superfamily. Activated ERs regulate the transcription of target genes by binding either directly to specific DNA sequences or by tethering to other DNA-bound transcription factors. ERs have been extensively studied at the molecular, cellular, physiological and pathological levels (reviewed by Dahlman-Wright et al, 2006; Deroo and Korach, 2006; Heldring et al, 2007). Tamoxifen and its hydroxylated active form hydroxytamoxifen (OHT) are synthetic ER ligands that compete with the physiological oestrogen 17β-estradiol (E2) for binding. Depending on promoter, cell and signalling context, OHT functions either as a partial agonist or as a partial antagonist. The latter mode has led to its use for endocrine therapy of ERα-positive breast tumours, the proliferation of which can be stimulated by E2 (reviewed by Jordan, 2004).
The early discovery of Filardo et al (2000) that the presence of the completely unrelated transmembrane receptor GPR30 can mediate oestrogen responsiveness of ER-negative breast cancer cells came as a big surprise. GPR30 was later shown to be a genuine ER (Revankar et al, 2005; Thomas et al, 2005). In addition to E2, OHT also functions as a GPR30 agonist (Revankar et al, 2005; Vivacqua et al, 2006a, 2006b). The GPR30 signalling pathway has been studied in a variety of cell lines. GPR30 couples to a trimeric G protein, stimulating the cAMP pathway most likely through a Gαs (Thomas et al, 2005) and Src through Gβγ (Filardo, 2002). Subsequently, Src promotes the shedding of heparin-binding EGF-like growth factor and activation of the EGF receptor (Filardo et al, 2000). This in turn activates a whole series of intracellular signalling events, most notably the activation of mitogen-activated protein kinases (MAPK) Erk1/2, PI3 kinase and phospholipase C (reviewed by Prossnitz et al, 2008). Further cellular responses lie downstream of these signals, including the activation of the gene FOS (Maggiolini et al, 2004).
It is unlikely that the activation of FOS can account for all of the biological effects of GPR30 signalling that have been reported. For example, E2 is able to stimulate the proliferation of breast, thyroid and ovarian carcinomas through GPR30 (Vivacqua et al, 2006b; Albanito et al, 2007, 2008). This effect is clearly independent of ERs and can also be observed with a GPR30-specific ligand, but how GPR30 signalling stimulates proliferation remains unclear. Although the genomic effects of ERα have been extensively studied, and in particular in breast cancer cells (see Carroll and Brown, 2006; Dudek and Picard, 2008 and references therein), the global changes in gene expression triggered by GPR30 signalling are not known. Unlike a transcription factor such as ERα, GPR30 would have to effect these changes indirectly. Nevertheless, GPR30-mediated changes in gene expression patterns have to be considered a specific output of this signal-transduction pathway. Here, we report the transcriptional consequences of GPR30 signalling in human breast cancer cells. The most strongly induced gene suggested a new function of GPR30 signalling in cell migration and proved to be functionally relevant for our understanding of the biological effects of GPR30 signalling.
Results
Gene expression profiling of GPR30 signalling
To determine the changes in gene expression that GPR30 signalling elicits, we chose human SKBr3 breast cancer cells as our model system. These cells lack both ERα and ERβ but express GPR30 and display GPR30 signalling (Filardo et al, 2000; Maggiolini et al, 2004). Despite the absence of other known ERs in SKBr3 cells, we knocked down GPR30 expression with an antisense strategy (Revankar et al, 2005; Vivacqua et al, 2006b) (Supplementary Figure 1A) to ascertain that any observed ligand-induced changes in gene expression are mediated by GPR30. Serum-deprived cells were treated for only 1 h with E2 or OHT to capture the primary responses. It should be pointed out here, that the OHT concentration (10 μM) used for induction is comparable to the micromolar OHT concentrations that are reached in breast tissue of patients undergoing tamoxifen therapy (Kisanga et al, 2004).
The mRNA levels of a total of 175 genes were induced by at least 1.3-fold by one of the treatments by comparison with uninduced control cells (Supplementary Figure 1B). At this point, we decided that we would only consider those genes as potential GPR30 target genes that fulfilled the following stringent criteria: at least 1.3-fold induction by both E2 and OHT, and at least a 1.3-fold reduction of the OHT response by antisense-mediated GPR30 knockdown. These criteria defined 36 genes as GPR30 target genes (Figure 1; Supplementary Table 1). In total, 19 of these 36 genes were induced by more than two-fold by OHT. Within the short time frame of the treatment, no gene was significantly repressed according to the same criteria (data not shown).
Figure 1.
Colour-coded map of hierarchically clustered gene expression profiles. For each gene and condition, the colour indicates the ratio of the values obtained for the treated and untreated samples (as listed in Supplementary Table 1). GPR30 KD, GPR30 knockdown.
We then undertook a Q-PCR experiment with the same RNA samples for a panel of genes to validate the microarray results and to obtain more quantitative data. Qualitatively, GPR30-mediated induction could be confirmed for all of them, although, not surprisingly, larger quantitative differences were obtained by Q-PCR (Figure 2). The gene encoding the connective tissue growth factor (CTGF, also known as CCN2) proved to be induced 15- to 16-fold by OHT and E2. It is a technical limitation of microarray analyses that some genes with a relatively modest induction fall through the cracks. This is the case, for example, for JUN. Our short list of 36 genes contains the genes FOS and FOSB (Supplementary Table 1). These encode components of the heterodimeric transcription factor AP1. Surprisingly, our gene list contains none of the genes, such as JUN, that encode heterodimeric partner proteins of Fos proteins. Although JUN did not pass the third stringent criterion (reduction by at least 1.3-fold in the GPR30 knockdown sample) in the microarray analysis, it easily passed all criteria for a GPR30 target gene in the Q-PCR experiment, including a two-fold induction by both ligands (Figure 2). We therefore include JUN as a GPR30 target gene and consider it very likely that there are other false negatives in the microarray data.
Figure 2.
Q-PCR validation of a subset of GPR30-regulated genes. ‘Fold induction' denotes the ratio of the values obtained for the treated and untreated samples. Error bars indicate standard deviations of measurements of triplicate samples. GPR30 KD, GPR30 knockdown.
CTGF is a GPR30 target gene
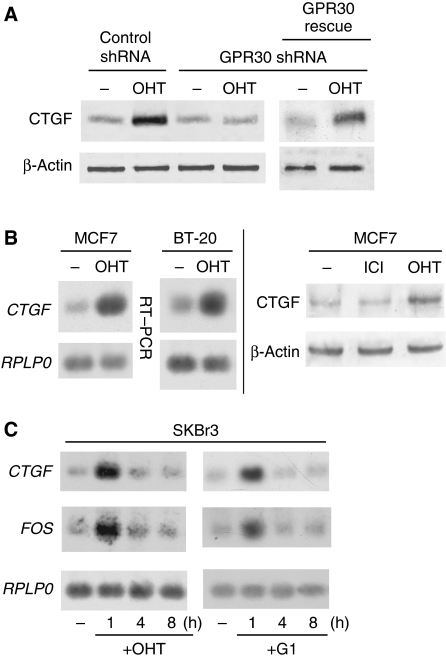
CTGF is by far the gene most strongly induced by E2 or OHT. We performed an immunoblot analysis to determine whether the dramatic induction seen at the mRNA level leads to increased CTGF protein expression in SKBr3 cells. Figure 3A shows that this is the case and that this increase can be blunted by an shRNA-mediated knock down of GPR30. The requirement for GPR30 and the specificity of the GPR30 knockdown are further emphasized by the fact that the co-transfection of an shRNA-resistant version of GPR30 (‘GPR30 rescue') restores the response. The increase at the protein level might seem modest, but note that only cell-associated proteins, and not proteins already released into the medium, were immunoblotted. We further explored the generality of this response with other cell lines and the GPR30-specific ligand G-1 (Bologa et al, 2006) (see Figure 3B and C). CTGF is induced by OHT in the human breast cancer cell lines MCF7 and BT-20, which are ERα positive and negative, respectively. The induction is seen both at the mRNA and protein levels, and it is not elicited by the antioestrogen ICI 182′780 (ICI) (Figure 3B), at least not in MCF7 cells under our experimental conditions. Importantly, the OHT induction of CTGF in MCF7 cells is independent of ERα as it can still be observed when ERα is knocked down (Supplementary Figure 2). The time course experiment confirms the activation of the CTGF and FOS genes by OHT, and shows an identical activation by G-1. Induction at the mRNA level is transient in that it is clearly observed after 1 h but has subsided 3 h later. Note that the microarray analysis was performed with RNA from cells treated for 1 h.
Figure 3.
Induction of CTGF mRNA and protein in a variety of breast cancer cell lines. (A) Immunoblot analysis of CTGF expressed by SKBr3 cells. Cells were transfected with shRNA constructs and the GPR30 rescue vector, and treated with OHT as indicated. (B) Immunoblots of semiquantitative RT–PCR products (‘RT–PCR') and CTGF protein (rightmost panel). (C) Time course of CTGF and FOS induction; semiquantitative RT–PCR analysis of SKBr3 cells treated with OHT or G1. β-Actin and the RPLP0 mRNA served as internal standards for the immunoblot and RT–PCR experiments, respectively.
Mediators of the transcriptional response to GPR30 signalling
The GPR30-mediated activation of target genes must be indirect. Previous analyses had indicated that GPR30 leads to the activation of Erk1/2 (Filardo et al, 2000; Maggiolini et al, 2004). MAPK can activate transcription factors such as the serum response factor (SRF) and members of the ETS family by direct phosphorylation (see for example, Posern and Treisman, 2006; Gutierrez-Hartmann et al, 2007). Moreover, it has been pointed out that the increase in cAMP elicited by GPR30 signalling could be expected to activate CREB (Prossnitz et al, 2008). These factors in turn activate the expression of the second tier of transcription factors such as c-Fos, FosB, c-Jun, EGR1, ATF3, C/EBPδ and NR4A2 (Nurr1). In addition to FOS, which we already knew to be activated by GPR30 signalling (Maggiolini et al, 2004), the genes for the aforementioned second tier transcription factors are all in our list of GPR30-induced genes (complemented with JUN from the Q-PCR experiment).
As a first step towards elucidating the signalling and transcription factor network that might underlie the transcriptional response to GPR30 signalling, we downloaded 5 kb of upstream sequences (relative to the start sites of transcription) for 34 of the 36 target genes of Supplementary Table 1. We scanned them for the presence of common sequence motifs and compared those with the known DNA-binding sequences of the TRANSFAC database to identify the corresponding transcription factors. The results of these analyses are displayed in Figure 4A for CTGF and for the complete set of target genes in Supplementary Table 2. SRF is by far the most over-represented transcription factor with EGR2, CREB and ATF among the runners up. Overall, the results of this bioinformatic analysis are entirely compatible with the aforementioned activation scheme.
Figure 4.
Transcriptional control of CTGF induction. (A) Transcription factor map of CTGF promoter region. Sites with factors indicated above the promoter line are over-represented at least two-fold in all GPR30 target genes (see Supplementary Table 2). AP1 and some additional factors reported in the literature are indicated below the line (see Leask and Abraham, 2006). The shorter line indicates the 2-kb fragment present in the CTGF luciferase reporter construct. The start sites of transcription (TSS) and translation (ATG) are indicated. (B) Immunoblot analysis of CTGF from SKBr3 cells expressing a dominant-negative version of c-Fos (dn-Fos). (C) E2 activation of a CTGF luciferase reporter gene co-transfected with shRNA constructs into SKBr3 cells as indicated. For each pair of samples, the values of the uninduced one were set to 100%. Error bars indicate standard deviations of normalized luciferase activities of triplicate samples.
Binding sites for AP1, of which c-Fos can be a component, are also highly represented, although not over-represented by more than two-fold, in promoters of GPR30 target genes (data not shown). For CTGF (Figure 4A), whose upstream sequences contain AP1 sites, we experimentally verified the role of c-Fos. The expression of a dominant-negative variant of c-Fos in SKBr3 cells abolishes the induction of CTGF by OHT or E2 (Figure 4B). To assess whether 5′ flanking sequences of the CTGF gene would be sufficient to mediate the GPR30 response, we used a reporter gene containing a 2 kb CTGF promoter fragment upstream of the luciferase-coding region (Chaqour et al, 2006; see Figure 4A). Upon transfection into SKBr3 cells, this reporter gene could be induced more than two-fold with E2 in a GPR30-dependent manner (Figure 4C). The response to OHT, which appears to be a stronger inducer of GPR30 signalling, could not be determined. The prolonged exposure of the cells to OHT, which this transactivation assay requires, turned out to be too toxic for SKBr3 cells. As observed for the induction of endogenous CTGF protein, AP1 turned out to be important for induction of the CTGF reporter gene, and this observation could be extended to ETS and SRF (Supplementary Figure 3A). A preliminary survey of signalling mediators that are required for the induction of CTGF highlights the role of the EGF receptor (see also Filardo et al, 2000) and the MAPK signalling cascade (Supplementary Figure 3B), mirroring our previous findings related to the induction of FOS (Albanito et al, 2008). In contrast to FOS, the induction of CTGF further depends on actin dynamics, a well-known regulator of SRF activity (Sotiropoulos et al, 1999). These results are compatible with the notion that these signalling pathways mediate the GPR30-induced activation of these transcription factors leading to the activation of CTGF and possibly other target genes.
GPR30 signalling promotes migration through CTGF induction
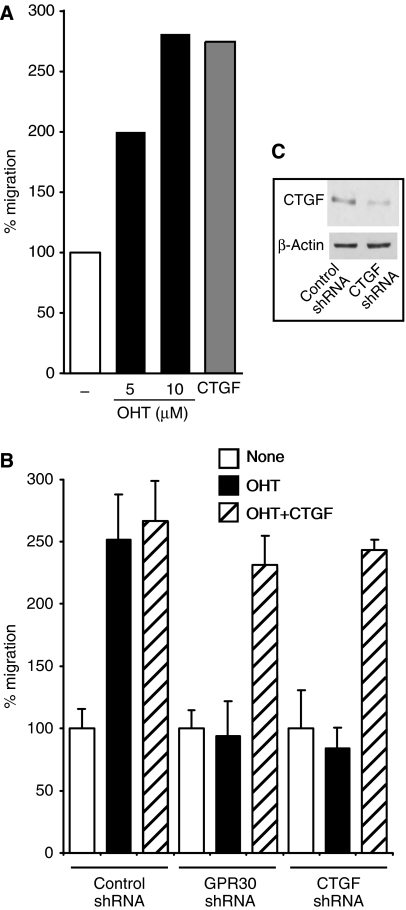
We next wondered what the biological significance of the potent induction of CTGF by GPR30 signalling might be. As CTGF had already been reported to be both sufficient and necessary for the stimulation of migration of other breast cancer cell lines (Chen et al, 2007), we considered the possibility that GPR30 signalling might promote migration through the induction of CTGF. The migration of SKBr3 cells was analysed with a Boyden chamber migration assay. With this assay, the number of SKBr3 cells that are able to migrate through a polycarbonate filter during a 3-h treatment period is counted. Figure 5A demonstrates that both OHT and CTGF stimulate the migration of SKBr3 cells more than two-fold. In the following experiment, we determined whether the stimulation of migration by OHT is indeed mediated by GPR30 signalling and the induction of CTGF. The transient knockdown of GPR30 or CTGF expression using transfected shRNA constructs completely abolishes the stimulation of migration by OHT (Figure 5B). The addition of CTGF to the medium of cells, in which GPR30 or CTGF are knocked down, rescues their migration defect. This result along with an immunoblot confirming the knockdown of CTGF at the protein level (Figure 5C) attests to the specificity of the RNA interference experiment. Thus, OHT stimulates the migration of SKBr3 cells through GPR30, and the GPR30-dependent induction of CTGF expression is necessary for this stimulatory effect. The fact that added CTGF stimulates migration to a similar extent as OHT, both in control and GPR30 knockdown cells, indicates that the GPR30-mediated induction of CTGF is also sufficient for this stimulation.
Figure 5.
CTGF-dependent stimulation of cell migration by GPR30 signalling. (A) OHT and CTGF added to the medium induce the migration of SKBr3 cells. Bar graph shows a representative experiment with means of duplicate samples, standardized to the untreated control set to 100%. (B) Transient knockdown of GPR30 or CTGF in SKBr3 cells blocks OHT stimulation of migration, and CTGF added to the medium restores it. Bar graph shows a representative experiment with means of triplicate samples, standardized to the respective untreated controls set to 100%. Error bars show standard deviations. (C) Immunoblot illustrating the extent of CTGF knockdown of a typical experiment.
A possible role for CTGF in promoting proliferation
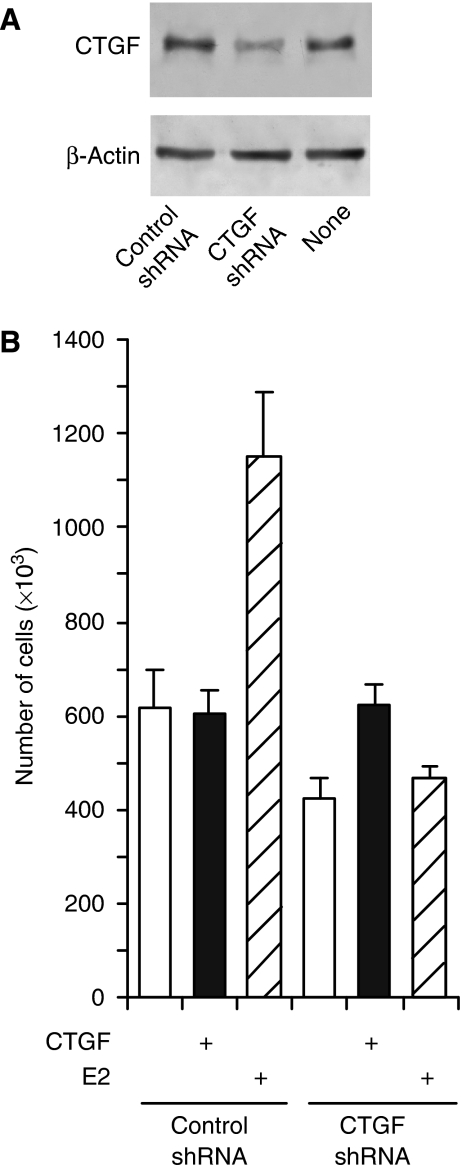
We have previously reported that GPR30 signalling stimulates the proliferation of cell lines, including SKBr3 cells, representing a variety of different carcinomas (Vivacqua et al, 2006b; Albanito et al, 2007, 2008) and mouse spermatogonia (Sirianni et al, 2008). It is very likely that the activation of growth-related transcription factors, such as the ones mentioned above, constitutes a cell-autonomous proliferative stimulus. In addition, primary or secondary GPR30 target genes that encode secreted factors such as CTGF might stimulate proliferation in an autocrine or paracrine manner. Indeed, there is evidence in the literature for a proliferative effect of CTGF on a variety of cell types (Rachfal and Brigstock, 2005; Leask and Abraham, 2006; De Winter et al, 2008), but its effects on normal or cancerous breast epithelial cells are less clear and possibly more complex (see below and Discussion). Pilot experiments indicated that CTGF added to the medium does not stimulate the proliferation of SKBr3 cells (data not shown; see also Figure 6). Although this finding is compatible with the observation that CTGF does not stimulate the proliferation of MCF7 cells (Chen et al, 2007), we noticed that SKBr3 cells grow poorly when CTGF is stably knocked-down below the basal level by a virally transduced shRNA construct (Figure 6). Remarkably, this defect can be corrected by adding CTGF to the medium. In contrast, unlike wild-type SKBr3 cells (Albanito et al, 2008), CTGF knockdown cells are resistant to the proliferative stimulus of E2. These results indicate that CTGF can have a proliferative effect on such breast carcinoma cells under certain conditions, and that CTGF is required for the proliferative response to E2.
Figure 6.
CTGF is required for the proliferative stimulation of SKBr3 by E2. (A) CTGF immunoblot of stable CTGF knockdown SKBr3 cells. They are compared with SKBr3 cells stably transduced with an unrelated control shRNA and untransfected wild-type SKBr3 cells. (B) Proliferation assay with SKBr3 cells stably transduced with shRNA constructs and treated with CTGF or E2 as indicated. Bar graph shows means of triplicate samples with standard deviations.
Contribution to GPR30 signalling from the stroma
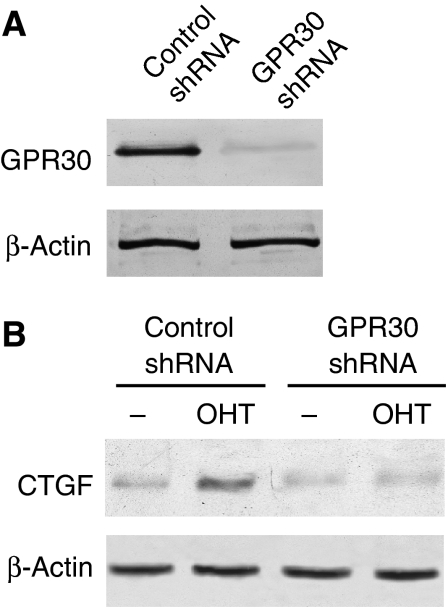
Given that secreted factors regulating proliferation and migration need not only be made by the breast carcinoma cells themselves, we examined the expression and signalling of GPR30 in fibroblasts obtained from biopsies of primary breast tumours. GPR30 is clearly expressed in these fibroblasts, and its activation by OHT stimulates the expression of CTGF (Figure 7).
Figure 7.
GPR30 expression and signalling in fibroblasts from breast tumours. The data shown are from a representative experiment of three experiments performed with samples from different patients. (A) Immunoblot analysis showing expression and knock down of GPR30 in transfected fibroblasts. (B) Immunoblot analysis of CTGF expressed by fibroblasts. Cells were transfected with indicated shRNA constructs and treated with OHT as indicated.
Discussion
In the process of characterizing the transcriptional response to GPR30 signalling, we have found a set of target genes that contribute to mediating the proliferative stimulus of GPR30 activators for carcinoma cell lines. Moreover, the prominent induction of CTGF by both E2 and OHT led us to the discovery that GPR30 signalling stimulates cell migration through CTGF. As these responses may occur in breast cancer cells independently of their ERα status as well as in the surrounding stromal cells, these findings may have important implications for our understanding of endocrine resistance in breast cancer.
GPR30 signalling activates a transcription factor network
Our microarray analysis indicated that GPR30 signalling triggers the activation of a network of transcription factors. A first tier, including SRF, members of the ETS family and CREB, is directly activated through post-translational modifications by kinases that are activated by GPR30 signalling. These transcription factors are the ones that transcriptionally activate the expression of a second tier of transcription factors (for example, Fos and Jun proteins, EGR1, ATF3, C/EBPδ and NR4A2). The response is most likely further amplified by GPR30-stimulated post-translational modifications of these proteins and positive feedback loops between many of these transcription factors. GPR30 itself may be part of a positive reinforcement loop as we have found that its expression is induced through MAPK and c-Fos in response to the growth factor EGF (Albanito et al, 2008). It remains to be seen whether these mechanisms, possibly involving other transcription factors mentioned above, also operate in response to GRP30 signalling, which incidentally depends on the EGF receptor (Filardo et al, 2000).
Considering the GPR30 signalling pathway, it is not surprising that the GPR30 response resembles the well-characterized transcriptional ‘immediate early response' of fibroblasts to serum (Iyer et al, 1999). The strong induction of CTGF is consistent with the following: (i) CTGF is known to be activated by a panel of different extracellular signals and its promoter contains binding sites for transcription factors of the ‘immediate early response', notably SRF, ETS, ATF, AP1, and TEAD2/ETF (Leask and Abraham, 2006; Cooper et al, 2007; Figure 4A); (ii) the inhibitory effect of a dominant-negative Fos points to a role for AP1, and additional preliminary functional results suggest that ETS and SRF are also required; and (iii) the activation of MAPK and actin dynamics are essential for the activation of both the full complement of these transcription factors and CTGF.
GPR30 expression and signalling in carcinomas
Activation of GPR30 by oestrogens elicits proliferative responses of breast and other carcinomas (Vivacqua et al, 2006b; Albanito et al, 2007, 2008). Although GPR30 is also expressed in normal breast epithelium, a survey of 321 primary breast tumours showed that 60% maintain GPR30 expression, including half of all ER-negative tumours (Filardo et al, 2006, 2008). Unlike ERα, the expression of which correlates with good prognosis, GPR30 expression was found to correlate very strongly with tumour size, HER-2 expression and distant metastasis. Remarkably, another study on breast cancer failed to find a similar correlation (Kuo et al, 2007), but methodological differences and a smaller cohort compared with the aforementioned studies call for more investigations. In the meantime, it is noteworthy that GPR30 expression was also associated with poor prognosis in a survey of endometrial carcinomas (Smith et al, 2007). Even in GPR30-positive cells, GPR30 is not expressed abundantly at the mRNA and protein levels (data not shown). Rather than monitoring only the expression of GPR30 itself, it might be more informative to combine it with measurements of the expression of a set of GPR30 target genes. Hence, our results highlight a set of GPR30 target genes that might both provide a signature for GPR30 signalling and a mechanistic underpinning of the aforementioned phenomena.
GPR30 target genes and cancer
Our experiments with SKBr3 cells demonstrate that CTGF is necessary for the stimulation of proliferation and migration by GPR30 signalling. Although CTGF is sufficient to stimulate migration, steady-state levels of CTGF might normally be sufficient to sustain a basal level of proliferation. The stimulation of proliferation by GPR30 signalling might arise from the combined induction of CTGF with multiple other GPR30 target genes. How the highly transient activation of GPR30 target genes that we have observed at the mRNA level is converted into this long-term response remains to be further analysed. Perhaps not a single, but repeated pulses of GPR30 signalling may be sufficient to produce relatively persistent higher levels of key proteins such as CTGF.
CTGF defines the cystein knot family of proteins along with Cyr61 (also known as CCN1) and Nov. Note that CYR61 is also induced by GPR30 signalling in SKBr3 cells, albeit much less than CTGF (Figures 1 and 2; Supplementary Table 1). Cyr61 and CTGF bind integrins and heparan sulphate-containing proteoglycans. CTGF also binds receptors or co-receptors for other signalling molecules such as Wnt, and CCN proteins even bind growth factors and cytokines themselves. These findings have led to the notion that CCN proteins exert their biological functions by modifying the action of other signals and the interactions with the extracellular matrix (reviewed by Rachfal and Brigstock, 2005; Leask and Abraham, 2006; De Winter et al, 2008; Holbourn et al, 2008). This may explain why the effects of CCN proteins are exquisitely dependent on cell type and experimental conditions, and why they may at times be seemingly contradictory. For example, low concentrations of CTGF can promote angiogenesis, but high concentrations inhibit the angiogenic effects of VEGF (Inoki et al, 2002). The evidence for an implication of CTGF in breast cancer is equally confusing. Although we have observed stimulatory effects of CTGF with SKBr3 cells, CTGF was shown to mediate TGFβ-induced apoptosis of MCF7 cells (Hishikawa et al, 1999). Nevertheless, CTGF was found to be part of a gene expression signature of osteolytic metastatic variants of the ER-negative breast cancer cell line MDA-MB-231, and to contribute to metastasis upon overexpression (Kang et al, 2003). A recent study confirmed this association and notably demonstrated that the overexpression of the genome organizer protein SATB1, which is overexpressed by aggressive breast cancer cells, induces CTGF expression (Han et al, 2008). In contrast, in a survey of 122 human breast tumours, Cyr61 and CTGF were found to be oppositely correlated with poor outcome, being high and low, respectively (Jiang et al, 2004). Elevated Cyr61 and CTGF levels have been reported to be characteristic of a number of other carcinomas (Bleau et al, 2005; Rachfal and Brigstock, 2005; Deng et al, 2007; Liu et al, 2008; Mullis et al, 2008). Whether the ERα status of breast tumours and cell lines may influence the effects of CCN proteins and account for some of the apparent discrepancies remains to be investigated.
There is yet another set of GPR30 target genes, the expression of which may contribute to breast cancer progression. Of the 11 most strongly induced genes, 5 are metallothionein genes. These include four members of the MT1 gene cluster and one, MT2A, from another chromosome. Interestingly, high expression of metallothioneins in breast tumours correlates with poor outcome, perhaps owing to their protective and proliferative functions (Bay et al, 2006).
Relevance to tamoxifen resistance of breast cancer and to physiology
Tamoxifen is used clinically for the endocrine treatment of ERα-positive breast cancer. How and why many of these tumours ultimately become resistant to this therapy remain poorly understood (reviewed by Herynk and Fuqua, 2007; Riggins et al, 2007). On the basis of our findings, we would suggest that GPR30 continues to mediate stimulatory oestrogen signals for proliferation and migration even in tumours that lose ERα expression. Indeed, GPR30 may become a risk factor in patients that are treated with tamoxifen as OHT is a potent GPR30 agonist. Thus, for a subset of breast tumours as well as other carcinomas, perhaps independently of ERα status, activation of GPR30 might actually promote poor outcome by stimulating tissue invasion and remodelling through CTGF. GPR30 antagonists, which are not yet available, could be of great therapeutic value. As our results with primary fibroblasts indicate, the stroma could contribute to relaying stimulatory signals of oestrogens or OHT to breast carcinoma cells, again independently of ERα status. It has been increasingly recognized that there is a decisive interplay between cancer cells and stroma (Bhowmick and Moses, 2005; Finak et al, 2008). Future studies should therefore include the evaluation of GPR30 levels in the cancer-associated stroma.
Although our transcriptome analysis was carried out with a breast cancer cell line, its results, which we have partially confirmed with primary fibroblasts, may be of heuristic value for understanding other biological GPR30 functions. The induction of a network of transcription factors and target genes such as CTGF, CYR61 and metallothioneins may be relevant to the wide range of GPR30 functions currently being unravelled in thymic atrophy (Wang et al, 2008a), mechanical hyperalgesia (Kuhn et al, 2008), liver injury (Hsieh et al, 2007), in the hypothalamus (Qiu et al, 2006; Brailoiu et al, 2007; Canonaco et al, 2008), for primordial follicle formation (Wang et al, 2008b), and for the proliferation of urothelial cells (Teng et al, 2008) and osteoblasts (Teplyuk et al, 2008).
Materials and methods
Cell culture
SKBr3 breast cancer cells were maintained in RPMI-1640 without phenol red supplemented with 10% fetal bovine serum (FBS). MCF7 and BT-20 breast cancer cells were cultured in Dulbecco's modified Eagle's medium and MEM, respectively, supplemented with 10% FBS. At 24 h prior to induction experiments, MCF7 and BT-20 cells were switched to a medium without serum and without phenol red. Lentiviruses were produced by transient co-transfection of lentiviral shRNA constructs with the VSV-G envelope plasmid pMD2G and the packaging plasmid psPAX2 (gifts from Dr D Trono; see http://tronolab.epfl.ch) into 293T cells. SKBr3 cells with a stable knockdown of CTGF were generated by lentiviral transduction of an shRNA construct (see below). In parallel, we obtained cells stably transduced with an unrelated control shRNA. To this end, cells were consecutively infected twice overnight, washed twice and subjected to a selection with 3 μg/ml puromycin for at least 24 h to eliminate the small fraction of non-infected cells.
Generation of primary fibroblast cells from breast cancer tissues
Breast cancer specimens were collected from primary tumours of patients who signed informed consent and underwent surgery before any pharmacological treatment. Following tumour excision, small pieces (1–2 mm diameter) were placed in a digestion solution (400 IU collagenase, 100 IU hyalurodinase and 20% FBS in Hank's balanced salt solution; all components from Sigma) containing antibiotics and antimycotics (Sigma) and incubated at 37°C with 5% CO2 for 5–6 h. After centrifugation at 90 g for 2 min, the supernatant containing fibroblast cells was centrifuged at 500 g for 8 min, then resuspended and cultured in RPMI-1640 medium supplemented with 20% FBS, antibiotics and antimycotics.
Plasmids
The GPR30 knockdown for the microarray analysis was carried out with the plasmid GPR30/AS (Revankar et al, 2005) using the related empty expression vector pRK5 (Schall et al, 1990) as a negative control. The CTGF luciferase reporter plasmid p(−1999/+36)-luc (Chaqour et al, 2006), which is based on the backbone of vector pGL3-basic (Promega), was a gift from Dr B Chaqour. The plasmid A-FOS, which encodes a c-Fos mutant that heterodimerizes with c-Fos dimerization partners but does not allow DNA binding (Gerdes et al, 2006), was obtained from Dr C Vinson. We have previously reported the characteristics and the evaluation of an shRNA construct to knock down the expression of GPR30 and of an unrelated shRNA control construct (Albanito et al, 2008). The shRNA construct for CTGF was obtained from the same supplier (Open Biosystems; www.Biocat.de). It has clone ID TRCN0000061950 and is based on the same lentiviral expression vector pLKO.1 as the other shRNA constructs. The targeting strand generated from the CTGF shRNA construct is TAGTACAGCGATTCAAAGATG. The expression vector for Flag-tagged human GPR30 has been described (Albanito et al, 2008). It was used to generate the GPR30 rescue vector containing silent mutations in the shRNA-targeted sequence: codons 293–297 were changed to CCG TGT AAA CAA AGT (changes underlined).
Microarray analysis
SKBr3 cells were transiently transfected either with the empty expression vector pRK5 or with the plasmid GPR30/AS using the FuGENE 6 transfection reagent (Roche) at 3 μl/μg DNA (5 μg DNA per 10 cm dish) in a medium with only 1% FBS. At 36 h after transfection, cells were switched to serum-free medium for 12 h before induction with 100 nM E2 or 10 μM OHT for 1 h. Total RNA from triplicate samples was extracted with Trizol (Invitrogen) according to the manufacturer's instructions. RNA quality was assessed with a BioAnalyzer (Bio-Rad). All RNA samples were stored at −80°C until required for further processing. Expression profiles were determined with HumanWG-6 v2.0 BeadChip microarrays (Illumina) at the genomics platform of the University of Geneva. The data have been submitted to GEO (access code GSE11567).
Promoter sequence analysis
Sequence retrieval: 5000 nucleotide sequences from upstream of the transcription start sites of the 34 genes in Supplementary Table 1 with a RefSeq accession number were retrieved from the UCSC genome browser (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/bigZips/upstream5000.fa.gz). Similarly, a large set of promoter sequences, to be used as a background/control sequences, was retrieved from http://ani.embl.de/trawler. Transcription factor identification: TRANSFAC Professional 11.4 was downloaded from Biobase (http://www.biobase-international.com) and installed locally on a computer running Ubuntu. The set of all vertebrate transcription factor-binding sites was retrieved from matrix.dat in TRANSFAC. For these factors, a number of profiles were generated with matrix similarity ranging from 0.8 to 1.00 and core similarity ranging from 0.8 to 1.00. The profile minFP to minimize false positives was generated from MATCH on the website. MATCH was used locally to identify transcription factor-binding sites in the set of promoter sequences with the profile minFP. Thereafter, F-MATCH was used with a cutoff P-value of 0.001 to identify over-represented transcription factor-binding sites. These transcription factor-binding sites were sorted by their relative over-representation in the target sequence set. To retrieve the predicted sites for 5000 bp of CTGF upstream sequences, the profile with 0.95/0.95 matrix/core similarity was used.
PCR analyses
Quantitative real-time PCR analysis was performed for the triplicate RNA samples used for the microarray experiment. To identify appropriate control genes for standardization, the analysis was performed with three candidates that had proved useful in many other gene expression analyses (TFRC, ALAS1 and TBP). As TFRC and ALAS1 varied the least across all sample, all measurements were normalized to the geometric mean of these two control genes. Semiquantitative RT–PCR was carried out as described (Maggiolini et al, 1999) with total RNA from cells induced for 1 h. FOS, CTGF and the internal control RPLP0 (also known as 36B4) cDNAs yielded bands of 420, 392 and 408 bp with 20, 20 and 10 PCR cycles, respectively. The primers are listed in Supplementary Table 3.
Immunoblotting
Total protein extracts were prepared from cells after a 2-h induction. For inhibition experiments with shRNAs or the dominant-negative c-Fos variant, cells were transiently transfected in a medium without serum and induced 24 h later. CTGF, β-actin and GPR30 were revealed by immunoblotting with the goat polyclonal antiserum sc-14939, the mouse monoclonal antibody sc-8432 (Santa Cruz Biotechnology) and the rabbit polyclonal antiserum LS-A1183 (MBL-Eppendorf), respectively.
Reporter gene assay
The CTGF luciferase construct was co-transfected with a Renilla luciferase expression plasmid as an internal transfection control into SKBr3 cells as described above for the microarray analysis. Induction with 100 nM E2 was carried out overnight. Luciferase activities of triplicate samples were measured with the Dual-Luciferase Reporter Assay system (Promega).
Migration and proliferation assays
Migration assays were performed with SKBr3 cells in triplicate using Boyden chambers (Costar Transwell, 8 μm polycarbonate membrane). For knockdown experiments, cells were transfected with shRNA constructs directed against GPR30 or CTGF or with an unrelated shRNA construct (3 μg DNA with FuGENE 6 at a 3 μl to 1 μg DNA ratio in 60 mm plates) in standard growth medium. After 24 h, they were seeded in the upper chambers. OHT or CTGF was added to the medium without serum in the bottom wells. After 3 h, cells on the bottom side of the membrane were fixed and counted. Human CTGF was purchased from MBL and added at 100 ng/ml. For proliferation assays, SKBr3 cells with a stable CTGF (or control) knockdown were cultured in a medium supplemented with 5% charcoal-treated FBS. Where indicated, CTGF was added each day. Cells were counted at day 6.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Acknowledgments
We are grateful to Drs B Chaqour, E Prossnitz, H Sato, D Trono and C Vinson for gifts of reagents, Dr R Treisman for the SRF shRNA sequence and to Dr K Strub for a critical reading of the paper. We are indebted to Dr S Abonante and Dr F Romeo for providing the breast cancer specimens. We thank the team of the genomics platform of the NCCR Frontiers-in-Genetics at the University of Geneva for help with microarrays. Funding for this project was provided by the Associazione Italiana Ricerca sul Cancro (AIRC) and Ministero dell'Università e Ricerca to MM, the Fondation Medic, the Swiss National Science Foundation and the Canton de Genève to DP.
References
- Albanito L, Madeo A, Lappano R, Vivacqua A, Rago V, Carpino A, Oprea TI, Prossnitz ER, Musti AM, Ando S, Maggiolini M (2007) G protein-coupled receptor 30 (GPR30) mediates gene expression changes and growth response to 17β-estradiol and selective GPR30 ligand G-1 in ovarian cancer cells. Cancer Res 67: 1859–1866 [DOI] [PubMed] [Google Scholar]
- Albanito L, Sisci D, Aquila S, Brunelli E, Vivacqua A, Madeo A, Lappano R, Pandey DP, Picard D, Mauro L, Andò S, Maggiolini M (2008) EGF induces GPR30 expression in estrogen receptor-negative breast cancer cells. Endocrinology 149: 3799–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay BH, Jin R, Huang J, Tan PH (2006) Metallothionein as a prognostic biomarker in breast cancer. Exp Biol Med 231: 1516–1521 [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Moses HL (2005) Tumor–stroma interactions. Curr Opin Genet Dev 15: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Perbal B (2005) CCN proteins and cancer: two to tango. Front Biosci 10: 998–1009 [DOI] [PubMed] [Google Scholar]
- Bologa CG, Revankar CM, Young SM, Edwards BS, Arterburn JB, Kiselyov AS, Parker MA, Tkachenko SE, Savchuck NP, Sklar LA, Oprea TI, Prossnitz ER (2006) Virtual and biomolecular screening converge on a selective agonist for GPR30. Nat Chem Biol 2: 207–212 [DOI] [PubMed] [Google Scholar]
- Brailoiu E, Dun SL, Brailoiu GC, Mizuo K, Sklar LA, Oprea TI, Prossnitz ER, Dun NJ (2007) Distribution and characterization of estrogen receptor G protein-coupled receptor 30 in the rat central nervous system. J Endocrinol 193: 311–321 [DOI] [PubMed] [Google Scholar]
- Canonaco M, Giusi G, Madeo A, Facciolo RM, Lappano R, Canonaco A, Maggiolini M (2008) A sexually dimorphic distribution pattern of the novel estrogen receptor G-protein-coupled receptor 30 in some brain areas of the hamster. J Endocrinol 196: 131–138 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Brown M (2006) Estrogen receptor target gene: an evolving concept. Mol Endocrinol 20: 1707–1714 [DOI] [PubMed] [Google Scholar]
- Chaqour B, Yang R, Sha Q (2006) Mechanical stretch modulates the promoter activity of the profibrotic factor CCN2 through increased actin polymerization and NF-kappaB activation. J Biol Chem 281: 20608–20622 [DOI] [PubMed] [Google Scholar]
- Chen P-S, Wang M-Y, Wu S-N, Su J-L, Hong C-C, Chuang S-E, Chen M-W, Hua K-T, Wu Y-L, Cha S-T, Babu MS, Chen C-N, Lee P-H, Chang K-J, Kuo M-L (2007) CTGF enhances the motility of breast cancer cells via an integrin-αvβ3-ERK1/2-dependent S100A4-upregulated pathway. J Cell Sci 120: 2053–2065 [DOI] [PubMed] [Google Scholar]
- Cooper SJ, Trinklein ND, Nguyen L, Myers RM (2007) Serum response factor binding sites differ in three human cell types. Genome Res 17: 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA (2006) International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev 58: 773–781 [DOI] [PubMed] [Google Scholar]
- De Winter P, Leoni P, Abraham D (2008) Connective tissue growth factor: structure–function relationships of a mosaic, multifunctional protein. Growth Factors 26: 80–91 [DOI] [PubMed] [Google Scholar]
- Deng YZ, Chen PP, Wang Y, Yin D, Koeffler HP, Li B, Tong XJ, Xie D (2007) Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem 282: 36571–36581 [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS (2006) Estrogen receptors and human disease. J Clin Invest 116: 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek P, Picard D (2008) Genomics of signaling crosstalk of estrogen receptor α in breast cancer cells. PLoS ONE 3: e1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo EJ (2002) Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. J Steroid Biochem Mol Biol 80: 231–238 [DOI] [PubMed] [Google Scholar]
- Filardo EJ, Graeber CT, Quinn JA, Resnick MB, Giri D, DeLellis RA, Steinhoff MM, Sabo E (2006) Distribution of GPR30, a seven membrane-spanning estrogen receptor, in primary breast cancer and its association with clinicopathologic determinants of tumor progression. Clin Cancer Res 12: 6359–6366 [DOI] [PubMed] [Google Scholar]
- Filardo EJ, Quinn JA, Bland KI, Frackelton AR Jr (2000) Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol Endocrinol 14: 1649–1660 [DOI] [PubMed] [Google Scholar]
- Filardo EJ, Quinn JA, Sabo E (2008) Association of the membrane estrogen receptor, GPR30, with breast tumor metastasis and transactivation of the epidermal growth factor receptor. Steroids 73: 870–873 [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14: 518–527 [DOI] [PubMed] [Google Scholar]
- Gerdes MJ, Myakishev M, Frost NA, Rishi V, Moitra J, Acharya A, Levy MR, Park S-w, Glick A, Yuspa SH, Vinson C (2006) Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res 66: 7578–7588 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Hartmann A, Duval DL, Bradford AP (2007) ETS transcription factors in endocrine systems. Trends Endocrinol Metab 18: 150–158 [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T (2008) SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452: 187–193 [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA (2007) Estrogen receptors: how do they signal and what are their targets. Physiol Rev 87: 905–931 [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA (2007) Estrogen receptors in resistance to hormone therapy. Adv Exp Med Biol 608: 130–143 [DOI] [PubMed] [Google Scholar]
- Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Luscher TF, Fujii T (1999) Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J Biol Chem 274: 37461–37466 [DOI] [PubMed] [Google Scholar]
- Holbourn KP, Acharya KR, Perbal B (2008) The CCN family of proteins: structure–function relationships. Trends Biochem Sci 33: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Yu HP, Frink M, Suzuki T, Choudhry MA, Schwacha MG, Chaudry IH (2007) G protein-coupled receptor 30-dependent protein kinase A pathway is critical in nongenomic effects of estrogen in attenuating liver injury after trauma-hemorrhage. Am J Pathol 170: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y (2002) Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J 16: 219–221 [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283: 83–87 [DOI] [PubMed] [Google Scholar]
- Jiang WG, Watkins G, Fodstad O, Douglas-Jones A, Mokbel K, Mansel RE (2004) Differential expression of the CCN family members Cyr61, CTGF and Nov in human breast cancer. Endocr Relat Cancer 11: 781–791 [DOI] [PubMed] [Google Scholar]
- Jordan VC (2004) Selective estrogen receptor modulation: concept and consequences in cancer. Cancer Cell 5: 207–213 [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massagué J (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549 [DOI] [PubMed] [Google Scholar]
- Kisanga ER, Gjerde J, Guerrieri-Gonzaga A, Pigatto F, Pesci-Feltri A, Robertson C, Serrano D, Pelosi G, Decensi A, Lien EA (2004) Tamoxifen and metabolite concentrations in serum and breast cancer tissue during three dose regimens in a randomized preoperative trial. Clin Cancer Res 10: 2336–2343 [DOI] [PubMed] [Google Scholar]
- Kuhn J, Dina OA, Goswami C, Suckow V, Levine JD, Hucho T (2008) GPR30 estrogen receptor agonists induce mechanical hyperalgesia in the rat. Eur J Neurosci 27: 1700–1709 [DOI] [PubMed] [Google Scholar]
- Kuo WH, Chang LY, Liu DL, Hwa HL, Lin JJ, Lee PH, Chen CN, Lien HC, Yuan RH, Shun CT, Chang KJ, Hsieh FJ (2007) The interactions between GPR30 and the major biomarkers in infiltrating ductal carcinoma of the breast in an Asian population. Taiwan J Obstet Gynecol 46: 135–145 [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119: 4803–4810 [DOI] [PubMed] [Google Scholar]
- Liu LY, Han YC, Wu SH, Lv ZH (2008) Expression of connective tissue growth factor in tumor tissues is an independent predictor of poor prognosis in patients with gastric cancer. World J Gastroenterol 14: 2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiolini M, Donzé O, Picard D (1999) A non-radioactive method for inexpensive quantitative RT–PCR. Biol Chem 380: 695–697 [DOI] [PubMed] [Google Scholar]
- Maggiolini M, Vivacqua A, Fasanella G, Recchia AG, Sisci D, Pezzi V, Montanaro D, Musti AM, Picard D, Andò S (2004) The G protein-coupled receptor GPR30 mediates c-fos up-regulation by 17b-estradiol and phytoestrogens in breast cancer cells. J Biol Chem 279: 27009–27016 [DOI] [PubMed] [Google Scholar]
- Mullis TC, Tang X, Chong KT (2008) Expression of connective tissue growth factor (CTGF/CCN2) in head and neck squamous cell carcinoma. J Clin Pathol 61: 606–610 [DOI] [PubMed] [Google Scholar]
- Posern G, Treisman R (2006) Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16: 588–596 [DOI] [PubMed] [Google Scholar]
- Prossnitz ER, Arterburn JB, Smith HO, Oprea TI, Sklar LA, Hathaway HJ (2008) Estrogen signaling through the transmembrane G protein-coupled receptor GPR30. Annu Rev Physiol 70: 165–190 [DOI] [PubMed] [Google Scholar]
- Qiu J, Bosch MA, Tobias SC, Krust A, Graham SM, Murphy SJ, Korach KS, Chambon P, Scanlan TS, Ronnekleiv OK, Kelly MJ (2006) A G-protein-coupled estrogen receptor is involved in hypothalamic control of energy homeostasis. J Neurosci 26: 5649–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachfal AW, Brigstock DR (2005) Structural and functional properties of CCN proteins. Vitam Horm 70: 69–103 [DOI] [PubMed] [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER (2005) A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307: 1625–1630 [DOI] [PubMed] [Google Scholar]
- Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH (2007) Pathways to tamoxifen resistance. Cancer Lett 256: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall TJ, Lewis M, Koller KJ, Lee A, Rice GC, Wong GH, Gatanaga T, Granger GA, Lentz R, Raab H, Kohr WJ, Goeddel DV (1990) Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 61: 361–370 [DOI] [PubMed] [Google Scholar]
- Sirianni R, Chimento A, Ruggiero C, De Luca A, Lappano R, Andò S, Maggiolini M, Pezzi V (2008) The novel estrogen receptor GPR30 mediates the proliferative effects induced by 17β-estradiol on mouse spermatogonial GC-1 cell line. Endocrinology 149: 5043–5051 [DOI] [PubMed] [Google Scholar]
- Smith HO, Leslie KK, Singh M, Qualls CR, Revankar CM, Joste NE, Prossnitz ER (2007) GPR30: a novel indicator of poor survival for endometrial carcinoma. Am J Obstet Gynecol 196: 386 e381–389 [DOI] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R (1999) Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98: 159–169 [DOI] [PubMed] [Google Scholar]
- Teng J, Wang ZY, Prossnitz ER, Bjorling DE (2008) The G protein-coupled receptor GPR30 inhibits human urothelial cell proliferation. Endocrinology 149: 4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ (2008) Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem 283: 27585–27597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Pang Y, Filardo EJ, Dong J (2005) Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology 146: 624–632 [DOI] [PubMed] [Google Scholar]
- Vivacqua A, Bonofiglio D, Albanito L, Madeo A, Rago V, Carpino A, Musti AM, Picard D, Andò S, Maggiolini M (2006a) 17β-Estradiol, genistein and 4-hydroxytamoxifen induce the proliferation of thyroid cancer cells through the G protein-coupled receptor GPR30. Mol Pharmacol 70: 1414–1423 [DOI] [PubMed] [Google Scholar]
- Vivacqua A, Bonofiglio D, Recchia AG, Musti AM, Picard D, Andò S, Maggiolini M (2006b) The G protein-coupled receptor GPR30 mediates the proliferative effects induced by 17β-estradiol and hydroxytamoxifen in endometrial cancer cells. Mol Endocrinol 20: 631–646 [DOI] [PubMed] [Google Scholar]
- Wang C, Dehghani B, Magrisso IJ, Rick EA, Bonhomme E, Cody DB, Elenich LA, Subramanian S, Murphy SJ, Kelly MJ, Rosenbaum JS, Vandenbark AA, Offner H (2008a) GPR30 contributes to estrogen-induced thymic atrophy. Mol Endocrinol 22: 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Prossnitz ER, Roy SK (2008b) G protein-coupled receptor 30 expression is required for estrogen stimulation of primordial follicle formation in the hamster ovary. Endocrinology 149: 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3