Abstract
The regulation of endosome dynamics is crucial for fundamental cellular functions, such as nutrient intake/digestion, membrane protein cycling, cell migration and intracellular signalling. Here, we show that a novel lipid raft adaptor protein, p18, is involved in controlling endosome dynamics by anchoring the MEK1–ERK pathway to late endosomes. p18 is anchored to lipid rafts of late endosomes through its N-terminal unique region. p18−/− mice are embryonic lethal and have severe defects in endosome/lysosome organization and membrane protein transport in the visceral endoderm. p18−/− cells exhibit apparent defects in endosome dynamics through perinuclear compartment, such as aberrant distribution and/or processing of lysosomes and impaired cycling of Rab11-positive recycling endosomes. p18 specifically binds to the p14–MP1 complex, a scaffold for MEK1. Loss of p18 function excludes the p14–MP1 complex from late endosomes, resulting in a downregulation of the MEK–ERK activity. These results indicate that the lipid raft adaptor p18 is essential for anchoring the MEK–ERK pathway to late endosomes, and shed new light on a role of endosomal MEK–ERK pathway in controlling endosome dynamics.
Keywords: endosome dynamics, late endosome, lipid rafts, MAPK pathway, MEK1
Introduction
The regulation of intracellular membrane dynamics is crucial for eukaryotic cells to maintain cell homoeostasis and properly respond to extracellular stimuli. The endosome system, which consists of early, recycling and late endosomes, has a central function in membrane dynamics by sorting and delivering cargos to the plasma membrane, trans-Golgi network or lysosomes (Trowbridge et al, 1993; Pfeffer, 2003; Russell et al, 2006). The endosome system is involved in a variety of cell functions that include nutrient intake/digestion, membrane receptor recycling/degradation, recovery of membrane lipids/proteins, antigen presentation, maintenance of cell polarity, cell migration and intracellular signalling (Dudu et al, 2004; Miaczynska et al, 2004; Trombetta and Mellman, 2005; Emery and Knoblich, 2006; Jones et al, 2006). The system can also be utilized by various pathogens to bud in and from cells (Gruenberg and van der Goot, 2006).
In addition to the function as a distribution centre, it has been proposed that the endosome system serves as an intracellular signalling station (Miaczynska et al, 2004). For example, a branch of MAPK signalling pathway is localized to late endosomes through a specific scaffold complex for MEK1, p14–MP1, and the endosome localization of the MEK1–ERK pathway is required for full activation of ERKs in a later phase of signalling (Teis et al, 2002). Further, ablation of endosomal p14–MP1–MEK1 signalling results in severe defects in tissue homoeostasis accompanied by aberrant subcellular distribution and trafficking of late endosomes (Teis et al, 2006). A mutation in human p14 causes a primary immunodeficiency syndrome with defects in endosome/lysosome biogenesis (Bohn et al, 2007). These findings indicate that the p14–MP1–MEK1 signalling pathway on late endosomes is involved in the regulation of endosome/lysosome biogenesis and dynamics. However, the mechanisms underlying the specific localization of the p14–MP1 complex to late endosomes as well as the endosomal functions of the p14–MP1–MEK1 pathway remain unsolved.
The mechanisms of endosome dynamics have been extensively analysed for epidermal growth factor receptor (EGFR) signalling. Concurrent with activation by ligand binding, EGFR is internalized by endocytosis through clathrin-dependent or -independent pathways, the latter including lipid raft- and caveolae-mediated routes (Kirkham and Parton, 2005; Sigismund et al, 2005). Lipid rafts are dynamic cholesterol-enriched microdomains in plasma and intracellular membranes, and are proposed to serve as signalling platforms by facilitating protein–protein interactions (Simons and Toomre, 2000; Anderson and Jacobson, 2002; Helms and Zurzolo, 2004). Caveolae are flask-shaped invaginations of plasma membrane lipid rafts that contain the integral membrane protein caveolin (Rothberg et al, 1992). EGFR internalized by the clathrin-dependent pathway is recycled back to the plasma membrane through recycling endosomes, whereas EGFR internalized through lipid raft/caveolae-dependent pathway is sorted from early endosomes to the intraluminal vesicles of late endosomes, leading to lysosomal degradation (Raiborg et al, 2002; Sachse et al, 2002; Pullikuth et al, 2005; van der Goot and Gruenberg, 2006). Previously, we showed that EGFR is predominantly distributed to lipid rafts in the PC12 pheochromocytoma cell line, and that perturbation of lipid rafts attenuates the downregulation of EGFR signalling (Kasai et al, 2005). These observations supported the important role of lipid rafts in EGFR trafficking through the endosome system. Potential roles of lipid rafts/caveolae have also been demonstrated for other membrane trafficking processes, including endocytosis, exocytosis and vesicle formation (van der Goot and Harder, 2001; Helms and Zurzolo, 2004; Hanzal-Bayer and Hancock, 2007). However, the functions of lipid rafts in the processes of such membrane dynamics are thoroughly unknown.
To address the functions of lipid rafts in EGFR trafficking through the endosome system, we have explored raft proteins that are distributed to intracellular membrane compartments. By LC-MS/MS analysis of potential raft proteins from EGF-stimulated PC12 cells, we identified a novel adaptor-like protein of 18 kDa (termed p18) that is predominantly localized to late endosomes. Analyses of p18 knockout mice and cells show that p18 is indispensable for mouse development, and has a key function in endosome dynamics by anchoring the MEK1–ERK pathway to lipid rafts of late endosomes.
Results
Identification of p18 from detergent-resistant membranes of EGF-stimulated PC12 cells
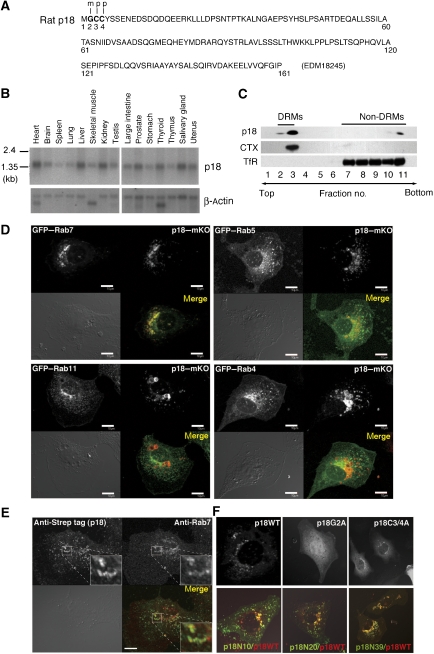
To identify raft proteins distributed in intracellular membrane compartments, we prepared detergent-resistant membrane (DRM) fractions from EGF-stimulated PC12 cells, and analysed potential raft proteins by LC-MS/MS (Supplementary Figure S1A). Among the proteins identified, we focused on the hypothetical protein MGC72560 of about 18 kDa (termed p18; Figure 1A), based on its following unique features. p18 has putative myristoylation and palmitoylation sites in its N terminus (Figure 1A); these modifications are known to function as lipid raft localization signals (Anderson and Jacobson, 2002; Kabouridis, 2006). p18 is highly conserved in vertebrates, but has no defined functional domain (Supplementary Figure S1B). p18 is ubiquitously expressed (Figure 1B), suggesting a general role of p18 in cellular function. Furthermore, a molecule equivalent to p18 was previously identified by proteomic analyses as a substrate of oncogenic v-Src (Rush et al, 2005) and as a secretory granule-associated protein (Brunner et al, 2007), raising a possibility that p18 is involved in intracellular signalling.
Figure 1.
Identification of p18 as a late endosomal protein. (A) Primary structure of rat p18. Potential myristoylation and palmitoylation sites are indicated by m and p, respectively. Amino-acid numbers are shown under the primary sequence. (B) Northern blot analysis of p18 transcripts in mouse tissues. Control β-actin blots are also shown. (C) MEFs were treated with Triton X-100 and separated into DRM and non-DRM fractions on a sucrose gradient. Each fraction was immunoblotted with anti-p18, CTX and anti-transferrin receptor (TfR) antibodies. (D) Colocalization of GFP–Rab7, GFP–Rab5, GFP–Rab11 and GFP–Rab4 with p18–mKO in p18−/− cells (Supplementary Figure S3). Scale bar: 10 μm. (E) Double immunostaining of p18rev cells (Supplementary Figure S3) with anti-Strep tag and anti-Rab7 antibodies. Higher magnification images of the indicated areas are shown in the insets. (F) Localization of wild-type p18 and p18 mutants (p18G2A and p18C3/4A) in p18−/− cells (upper panels). Localization of p18 deletion mutants (p18N10, p18N20 and p18N39) was evaluated by co-transfection with wild-type p18 (lower panels). Merged images are shown.
p18 is localized to lipid rafts of late endosomes
Membrane fractionation on a sucrose gradient confirmed the predominant localization of p18 to DRM fractions in mouse embryonic fibroblasts (MEFs) (Figure 1C). Immunostaining of PC12 cells showed that p18 is localized to intracellular endosome-like vesicles positive for the raft marker CTX (cholera toxin B-subunit) (Supplementary Figure S1C). Intracellular localization of p18 was assessed by analysing the distributions of fluorescent fusion proteins using Rab family GTPases, Rab7, Rab5 and Rab4/11, as markers of late endosomes, early endosomes and recycling endosomes, respectively (Feng et al, 1995). The results showed that p18 monomer Kusabira-Orange (mKO) could be colocalized with green fluorescent protein (GFP)–Rab7-positive late endosomes, but not with Rab5-positive early or Rab4/11-positive recycling endosomes (Figure 1D). The localization of p18 to late endosomes was further confirmed by double immunostaining of endogenous Rab7 and strep-tagged p18 (Figure 1E). Higher magnification images showed that Rab7 and p18 were localized to the same vesicles but in different membrane compartments.
The specificity of p18 localization was examined by analysing the localization of a series of mutant p18–GFP fusions. p18 mutants with amino-acid substitutions at the potential acylation sites (p18G2A and p18C3/4A) were excluded from late endosomes and became widely distributed in the cytoplasm (Figure 1F, upper panels). Co-expression analyses using wild-type p18 and p18–GFP deletion mutants showed that the N-terminal 20 amino-acid residues are sufficient for the late endosome localization (Figure 1F, lower panels). These results suggest that p18 is selectively localized to the lipid rafts of late endosomes through its N-terminal unique structure.
p18 knockout mice show severe defects in intracellular organelle organization in the visceral endoderm
To address the function of p18, we generated p18 knockout mice by homologous recombination in embryonic stem cells (Figure 2A; Supplementary Figure S2). The mutant embryos died due to growth retardation at the egg cylinder stage at around embryonic day (E) 7 (Supplementary Figure S2D). At E6.5, p18 is expressed throughout the entire embryo, but it is relatively abundant in the visceral endoderm (VE), which constitutes the outer tissue layer of the pregastrula embryo (Figure 2B). Even at the tissue level, localization of p18 to endosome-like vesicles was observed (Figure 2B, inset). The mutant embryos had no detectable expression of p18 protein (Figure 2B, right). At E7, the mutant embryos underwent growth arrest (Figure 2C, upper row) and defects became evident in the VE (Figure 2C, lower row).
Figure 2.
Phenotypes of p18 knockout mice. (A) Schematic diagram of the p18 gene and targeting vector containing the Neo-resistance gene (neo) as a positive selection marker and the thymidine kinase gene (tk) as a negative selection marker. Locations of PCR primer sequences designed for screening (S1–N1, G3–S2, N2–S2 and N2–S3) and genotyping (G1–G2 and G1–N1) are shown. Details are described in the legend to Supplementary Figure S2. (B) Immunohistochemical analysis of control (cont) and mutant (mut) E6.5 embryos using anti-p18 antibody. Scale bar: 50 μm. Insets show enlarged views of the white boxes. (C) Haematoxylin and eosin (HE)-stained sections of E7 embryos. Lower panels show enlarged views of the white boxes in upper panels. The visceral endoderm (VE) in the mutant embryo and the corresponding visceral yolk sac in the control embryo are indicated. Scale bar: 100 μm in upper panels, 10 μm in lower panels. (D) Immunohistochemical analysis of control and mutant E7 embryos using anti-LAMP-1 antibody. (E) Electron microscopy sections showing E6.5 VE. Boxed areas are enlarged in lower panels. Red asterisks in the control section indicate large lysosomal structures (Anderson and Jacobson, 2002; Zheng et al, 2006), and yellow arrowheads in the mutant section indicate small lysosomal structures with amorphous substances. Scale bar: 5 μm. (F) Immunohistochemical analysis of control and mutant E7 embryos using anti-cubilin (upper) and anti-megalin (lower) antibody. Differential interference contrast (DIC) images are shown in the right panels. Scale bar: 10 μm.
In control embryos, the VE cells were regularly arranged and huge lysosomal structures positive for LAMP-1 (Zheng et al, 2006), so-called giant lysosomes, were observed in the apical side of the cells (Figure 2C and D). In mutant VE, however, the apical surface was expanded, and relatively small LAMP-1-positive vesicles accumulated beneath the apical membrane. Electron microscopy confirmed that the surface membrane of the mutant VE is markedly extended and invaginates into the cytoplasm (Figure 2E). Giant lysosomal structures were detected in the supranuclear region of control cells; however, small lysosomal structures with amorphous substances were abundant in the cytoplasm of mutant cells (Figure 2E). In the mutant VE, accumulation of tubular endosome-related organelles was also observed beneath the apical membranes (Figure 2E). Furthermore, the apical surface scavenger receptors cubilin and megalin (Christensen and Birn, 2002) were predominantly intracellular in the mutant VE cells (Figure 2F). These observations indicate that p18 loss greatly affects organization or biogenesis of endosome-/lysosome-related organelles and the transport of membrane proteins, which are required for the nutrient incorporation and processing. Thus, it is likely that p18 loss abrogates the critical functions of VE, that is, nutrient uptake, digestion and delivery to epiblasts (Bielinska et al, 1999), which may lead to the embryonic lethality.
p18-deficient cells exhibit defects in endosome dynamics through perinuclear compartments
To elucidate the cellular function of p18, we established multiple p18+/+, p18+/− and p18−/− cell lines from littermate embryos (Supplementary Figure S3). However, morphological and functional features of p18−/− cells substantially varied depending upon cell lines, probably due to the difference in the cell lineages. Thus, to precisely examine the function of p18, we established p18rev cells by re-introducing strep-tagged p18 into a p18−/− cell line and used as a control cell line (Supplementary Figure S3).
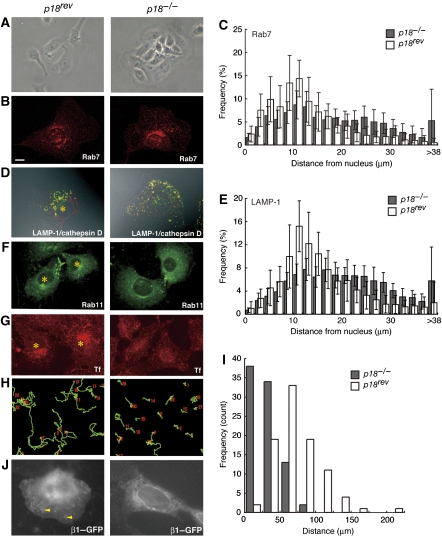
The p18rev cells show mesenchymal cell-like features and grow dispersed in low-density cultures, whereas p18−/− cells have an epithelial cell-like morphology and grow as compact colonies (Figure 3A). Consistent with the in vivo observations, p18−/− cells showed apparent defects in the intracellular distribution of endosome-related organelles (Figure 3). In control p18rev cells, the majority of Rab7-positive late endosomes accumulates in perinuclear region, whereas in p18−/− cells, some populations of late endosomes significantly exhibit a scattered distribution pattern (Figure 3B and C). In p18rev cells, relatively large LAMP-1/cathepsin D-positive lysosomal structures accumulates in the perinuclear region, whereas smaller sized lysosomal structures are diffusely distributed in the peripheral cytoplasm of p18−/− cells (Figure 3D and E; Supplementary Figure S4) and some parts of signals for LAMP-1 and cathepsin D are detected in distinct vesicles (Figure 3D). These observations demonstrate that p18 loss affects the processing of lysosome-related organelles, such as lysosome fusion with late endosome and their intracellular transports.
Figure 3.
Analysis of p18−/− cells. (A) Phase-contrast images of growing p18rev (left) and p18−/− (right) cells. (B) Immunofluorescence analysis of Rab7-positive late endosomes in p18rev and p18−/− cells. (C) Semiquantitative analysis of the distribution of Rab7-positive late endosomes in the cells. The frequency distributions were measured in distances from the nucleus (n=17 cells). χ2 independence test; P<0.001. (D) Immunofluorescence analysis of LAMP-1 (green) and cathepsin D (red) in p18rev and p18−/− cells. Merged images are shown. Asterisk indicates the location of PNRC. (E) Semiquantitative analysis of the distribution of LAMP-1-positive lysosomes in the cells (Supplementary Figure S4). The frequency distributions were measured in distances from the nucleus (n=17 cells). χ2 independence test; P<0.001. (F) Immunofluorescence analysis of Rab11-positive endosomes in p18rev and p18−/− cells. (G) Fluorescence images of transferrin-Alexa 594 (Tf) incorporated into p18rev and p18−/− cells. (H) Time-lapse imaging of cell migration of p18−/− and p18rev cells. Representative tracks for 16 h are shown. (I) Histograms of the migration distance for 4 h of 25 μm windows are shown for p18rev and p18−/− cells. (J) Time-lapse imaging of integrin β1–GFP (β1–GFP) in p18−/− and p18rev cells. QuickTime movies are provided in Supplementary Movies S1 and S2.
It is known that a variety of membrane receptors accumulate in the so-called perinuclear recycling compartment (PNRC), prior to being returned to the plasma membrane through the Rab11 pathway (Mohrmann and van der Sluijs, 1999; Caswell and Norman, 2006). Indeed, Rab11-positive recycling endosomes accumulates in PNRC of p18rev cells (Figure 3F). However, the Rab11-positive endosomes are dispersedly distributed in the cytoplasm of p18−/− cells (Figure 3F). Transferrin receptor, which is also known to be transported to PNRC through Rab11-dependent mechanism (Mohrmann and van der Sluijs, 1999), cannot accumulate in PNRC either (Figure 3G). These features of p18−/− cells suggest that p18 loss affects Rab11-mediated ‘long-loop' transport/recycling through PNRC (Jones et al, 2006). p18−/− cells also showed apparent defects in cell migration, with substantially reduced migration velocities (Figure 3H and I). During cell migration, β1 integrins are known to be recycled back to the plasma membrane through Rab11-positive PNRC (Caswell and Norman, 2006). Time-lapse analysis of integrin β1–GFP showed that p18rev cells actively transport integrin β1 to the cell periphery and focal adhesions, whereas in p18−/− cells, integrin β1 constitutively accumulates in perinuclear organelles (Figure 3J; Supplementary Movie S1 and S2). From these observations, it is postulated that p18 can also participate in the cycling of the endosome system through PNRC.
Identification of the p14–MP1 complex as a binding partner of p18
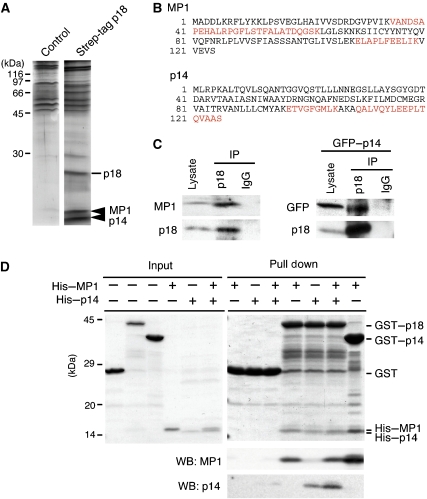
To elucidate the mechanism of p18 function, we searched for p18-binding partners. When the strep-tagged p18 was purified from MEFs using a Streptactin column, a doublet protein of around 14 kDa was co-purified with p18 (Figure 4A). LC-MS/MS analyses of these protein bands revealed that the doublet constitutes a complex of p14 and MP1 (Figure 4A and B; Supplementary Figure S5A and B). The p14–MP1 complex was previously identified as a scaffold for MEK1 in late endosomes, and was implicated in the biogenesis and trafficking of endosomes/lysosomes (Schaeffer et al, 1998; Wunderlich et al, 2001; Teis et al, 2002, 2006). Interestingly, the defects caused by p14 deficiency appeared quite similar to those caused by the loss of p18: p14−/− embryos manifest severe developmental defects by E8.5, endosomes/lysosomes are displaced to the cell periphery in p14−/− MEFs (Teis et al, 2006), and cathepsin D-positive endosomes/lysosomes are widely distributed throughout the cytoplasm of p14-deficient patient cells (Bohn et al, 2007). Similar defects in the distribution of endosomes/lysosomes was also observed in MEK1−/− cells (Teis et al, 2006). These observations strongly suggest the functional link between p18 and the p14–MP1–MEK1 signalling pathway on late endosomes.
Figure 4.
Identification of the p14–MP1 complex as p18-binding proteins. (A) Silver staining of p18-binding proteins purified from control MEFs and strep-tagged p18-expressing MEFs. Arrowheads, MP1 and p14 identified by LC-MS/MS. (B) Amino-acid sequences of MP1 and p14. Peptide sequences obtained by LC-MS/MS are indicated in red. (C) p18 was immunoprecipitated from normal or GFP–p14-expressing MEFs, and immunoblotted with anti-MP1 and anti-p18 (left) or anti-GFP and anti-p18 (right) antibodies. As input controls, 16% of total cell lysate was analysed. (D) In vitro binding assays among p18, p14 and MP1. As input controls, 7% of GST, GST–p18, His–MP1 and His–p14 used for pull down assay were separated by SDS–PAGE and stained with Coomassie brilliant blue (CBB, left panel). GST, GST–p18 or GST–p14 was incubated with His–MP1 and/or His–p14, and the complexes were pulled down with glutathione beads, followed by staining with CBB and immunoblotting with anti-MP1 and anti-p14 (right panels). Locations of GST, GST–p18, His–MP1 and His–p14 are indicated.
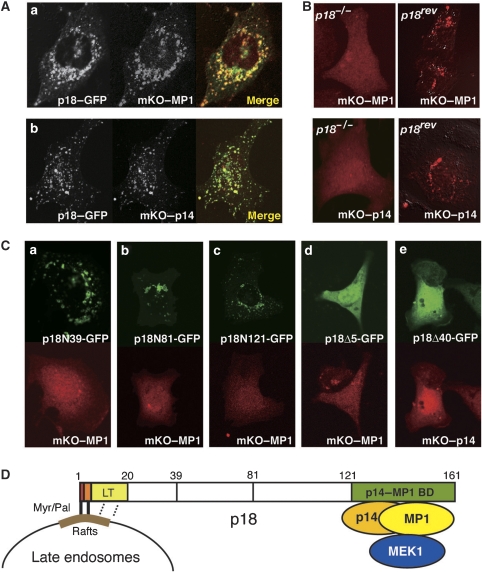
We confirmed the interaction between p18 and the p14–MP1 complex by immunoprecipitation from MEFs (Figure 4C) and in vitro pull-down assays using recombinant proteins (Figure 4D). The pull-down assay revealed that p18 could directly interact with both MP1 and p14. Analyses using fluorescent fusion proteins showed striking colocalization between p18 and MP1 (Figure 5Aa), and between p18 and p14 (Figure 5Ab). Notably, in p18−/− cells, either MP1 or p14 could not localize to late endosomes (Figure 5B), indicating an essential role of p18 for late endosome localization of the p14–MP1 complex. Analyses of p18 mutants showed that deletion mutants containing the N-terminal portion of p18 (p18N39, N81 and N121) could localize to late endosomes, but failed to anchor the p14–MP1 complex (Figure 5Ca–c; Supplementary Figure S5C). Also, p18 mutants that lack endosome-anchoring regions (p18Δ5 and p18Δ40) could not anchor the p14–MP1 complex to late endosomes (Figure 5Cd and e). These results indicate that the N- and C-terminal portions of p18 are specifically employed for the binding to late endosomes and the p14–MP1 complex, respectively (Figure 5D).
Figure 5.
Interaction between p18 and the p14–MP1 complex. (A) Colocalization of p18–GFP and mKO–MP1 (a) or mKO–p14 (b) in MEFs. (B) Intracellular localization of mKO–MP1 (upper) and mKO–p14 (lower) in p18−/− (left) and p18rev (right) cells. (C) p18−/− cells were co-transfected with GFP fusions with p18 deletion mutants containing aa 1–39 (p18N39, a), 1–81 (p18N81, b), 1–121 (p18N121, c), 6–161 (p18Δ5, d) and 41–161 (p18Δ40, e), and mKO–MP1 or mKO–p14. Intracellular distribution of each construct is shown. (D) A schematic model of the interaction domains of p18. Myr, myristate; Pal, palmitate; LT, late endosome targeting domain; p14–MP1 BD, p14–MP1-binding domain.
p18 serves as an essential scaffold for the p14–MP1–MEK1 pathway on late endosomes
To further confirm the function of p18 as a scaffold for the p14–MP1–MEK1 complex, we examined the impact of p18 loss on MEK–ERK activity. In p18−/− cells cultured under growth conditions (10% serum), the steady-state activities of MEK and ERK were significantly reduced as compared with those in p18rev cells (Figure 6A and B). Under lower serum conditions (0.1%), EGF-dependent activation of MEK was significantly suppressed in p18−/− cells (Figure 6C and D). In this experiment, the effect of p18 loss on ERK activity was not evident, probably due to the activation of MAP kinase phosphatase by EGF stimulation (Kondoh and Nishida, 2007). These results suggest that p18 serves as a functional component of the MEK–ERK pathway by anchoring the p14–MP1–MEK complex to late endosomes.
Figure 6.
Effects of p18 loss on the MEK–ERK activity. (A) Total cell lysates from p18−/− and p18rev cells grown under normal growth conditions were immunoblotted with the indicated antibodies. (B) Mean±s.d. of the relative activities of MEK (left) and ERK (right) in p18rev (n=4) and p18−/− (n=4) cells are shown. ***P<0.001 by Student's t-test. (C) p18−/− and p18rev cells cultured in the presence of 0.1% serum were treated with EGF (100 ng/ml). Cell lysates prepared at the indicated time points were immunoblotted with the indicated antibodies. (D) Means±s.d. of the relative activity of MEK in p18rev (n=4, white) and p18−/− (n=4, grey) cells are shown. Differences between groups were analysed by using repeated measures analysis of variance (ANOVA); P=0.0013. Differences at the indicated time points were analysed by Student's t-test; **P<0.01; ***P<0.001. (E) Cell lysates prepared from p18−/− and p18rev cells treated with EGF for the indicated times were immunoblotted with the indicated antibodies. Caveolin was detected as a control of DRMs. (F) Means±s.d. of the relative activity of ERK in DRM fractions from p18rev (n=4, white) and p18−/− (n=4, grey) cells are shown. Differences between groups were analysed by using repeated measures ANOVA; P=0.0008. Differences at the indicated time points were analysed by Student's t-test; **P<0.01; ***P<0.001. (G) p18rev was treated with U0126 (10 μM) for the indicated periods, and was co-stained with anti-LAMP-1 and anti-cathepsin D antibodies. Merged images are shown. An image of p18−/− cell is also shown.
The contribution of lipid rafts to the p18–p14–MP1–MEK1 pathway was next examined by separating DRMs from EGF-stimulated cells. Immunoblot analysis showed that the localization of MP1 and p14 to DRMs is mostly dependent on the presence of p18 (Figure 6E). MEK and ERK were also significantly concentrated in DRMs of p18rev cells, although a smaller fraction of MEK–ERK was detected in DRMs even in the absence of p18 (Figure 6E; Supplementary Figure S6). This p18-independent DRM localization of MEK–ERK might be due to the presence of some other scaffolds for the MEK–ERK pathway in DRMs (Kolch, 2005; Sacks, 2006). It should also be noted that MEK and ERK in DRMs tended to decrease following EGF stimulation (Supplementary Figure S6). This is consistent with the previous finding that MP1 preferentially binds to an inactive form of MEK (Schaeffer et al, 1998). Indeed, the activated form of MEK was undetectable in DRMs (data not shown). Nonetheless, the amount of activated ERK in DRMs was significantly increased in the presence of p18 (Figure 6F), presumably because the activated ERK could interact with some DRM components. These observations suggest that the activation of p18-dependent MEK–ERK pathway takes place on the lipid raft platform of late endosomes.
We finally examined the role of the MEK–ERK activity in the membrane dynamics regulated by p18. In p18rev cells, we observed that lysosomes positive for LAMP-1 and cathepsin D accumulated in the perinuclear compartments (Figure 3D). When p18rev cells were treated with a specific MEK inhibitor U0126, LAMP-1-positive lysosomes were time-dependently scattered throughout the cytoplasm and became smaller in size, resulting in aberrant lysosomal distribution as observed in p18−/− cells (Figure 6G). This indicates that the p18–MEK–ERK activity is required for lysosome processing.
Discussion
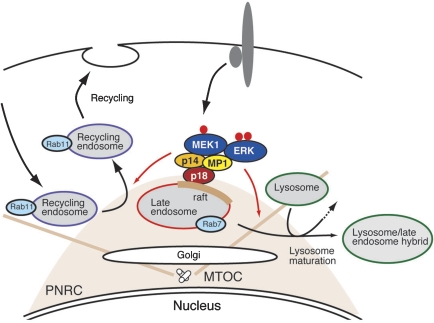
In this study, we analysed functions of a novel adaptor protein, p18, which is potentially localized to lipid rafts of late endosomes. In vivo and in vitro analyses revealed that p18 serves as an essential anchor for the p14–MP1–MEK–ERK pathway in late endosomes, and is involved in controlling endosome dynamics, including lysosome processing and endosome cycling through PNRC (Figure 7).
Figure 7.
The p18–MEK–ERK pathway in the intracellular organelle dynamics. A schematic model of p18 function. p18 serves as a lipid raft anchor for p14–MP1–MEK1 signalling components on late endosomes. The loss of p18 function or the inhibition of MEK activity causes defects in membrane dynamics, including Rab11-mediated endosome recycling and lysosome processing. From these observations, it is suggested that the p18–MEK–ERK pathway anchored to intracellular lipid rafts takes part in controlling intracellular membrane dynamics potentially by regulating organelle interactions and/or transports along cytoskeletons. MTOC, microtubule organizing centre; PNRC, perinuclear recycling compartment.
We identified p18 as a potential component of lipid rafts. The predominant distribution of p18 to DRMs suggested its potential localization to lipid rafts. However, it is currently accepted that DRMs do not necessarily correspond to lipid rafts and that the DRM separation method is insufficient for the identification of lipid raft-associated proteins (Lichtenberg et al, 2005; Hancock, 2006). Thus, to verify the raft localization of p18, we examined intracellular distribution of p18 and its mutants. The cell staining analyses showed that p18 could be colocalized with GM1 ganglioside, a marker of lipid rafts (Harder et al, 1998), and that the N-terminal potential myristoylation and palmitoylation sites, which are known to function as lipid raft localization signals, were required for the late endosome localization of p18. These observations strongly supported the presence of p18 in lipid rafts of late endosomes (Balbis et al, 2007). It is of interest that the N-terminal only 20 residues of p18 are sufficient for specific localization to late endosomes. Lipid raft components that specifically recognize this sequence may contribute to the specific functions of late endosomes. Given that lipid rafts are indeed present in late endosomes, their roles in cellular functions remain as critical issues. A line of evidence has suggested that lipid rafts in intracellular organelles have functions in the recruitment of specific signalling components, membrane curvature, membrane compartmentalization and membrane fusion (Sönnichsen et al, 2000; del Pozo et al, 2005; Balasubramanian et al, 2007; Hanzal-Bayer and Hancock, 2007). Specific roles of cholesterol and phosphatidylinositol phosphates in the lipid rafts have also been implicated in these events. Late endosomes have one of the most dynamic membranes in cells, where formation of intraluminal vesicles (late endosomes come from multivesicular bodies), budding in and off of recycling vesicles and fusion with lysosomes are continuously and actively taking place. Thus, it seems reasonable to speculate that lipid rafts are required for these late endosome-specific membrane dynamics, and the lipid raft proteins, such as p18, have functions in recruiting signalling components involved in regulating membrane dynamics.
Loss of p18 function caused embryonic lethality with growth retardation at early developmental stages (around E7). The most unequivocal defects were observed in the VE, which has critical functions in nutrient uptake, digestion and delivery to support epiblast development (Bielinska et al, 1999). The defects in lysosome maturation and the transport of scavenger receptors suggest that the impaired nutrient transport and/or digestion of maternal materials are the main causes of embryonic lethality. This notion is supported by the observations that the phenotypes of p18−/− embryos are quite similar to the phenotypes of amnionless (Strope et al, 2004) and dab2−/− mice (Maurer and Cooper, 2005), both of which caused growth retardation at early developmental stages due to defective transport of maternal materials through VE. These in vivo observations highlighted the critical role of p18 in regulating endosome dynamics involved in intracellular material transport and lysosome processing. VE cells morphologically and functionally resemble small intestinal epithelial cells (Ezzell et al, 1989), suggesting that p18 might be necessary for the functions of digestive and secretory organs even in adult animals. More interestingly, a mutation in human p14 causes a primary immunodeficiency syndrome that carries defects in endosome–lysosome biogenesis, which is critical for antigen processing (Bohn et al, 2007). Furthermore, defects in lysosome-related organelles have been implicated in various human autosomal recessive disorders (Dell'Angelica, 2007). Thus, the understanding of the function and regulatory features of p18 as a regulator of organelle biogenesis may offer new avenues for therapeutic intervention in these organelle diseases.
We identified the p14–MP1 complex (Wunderlich et al, 2001) as a specific binding partner of p18. Previously, it was shown that the localization of the MP1–MEK1 complex is mediated by p14 (Teis et al, 2002). However, as p14 has no significant membrane localization signal, some other membrane components have been thought to be required for its membrane anchoring. Crystal structure of the p14–MP1 complex also provided a potential binding surface for as-yet unidentified partners (Kurzbauer et al, 2004; Lunin et al, 2004). Our studies on the p18–p14–MP1 interaction indicate that p18 serves as the critical determinant for the specific localization of the p14–MP1–MEK1 complex to late endosomes. These indicate that p18 represents the missing link among late endosomes, the MEK–ERK pathway and membrane dynamics (Deacon et al, 2005; Dell'Angelica, 2007), and raise a possibility that p18 may serve as a limiting regulatory site. As p18 has been identified as a potential substrate of oncogenic v-Src (Rush et al, 2005) and has potential Ser/Thr phosphorylation sites, it is possible that the function of p18 is regulated downstream of some cell signalling pathways to organize endosome dynamics.
The MEK–ERK pathway is activated by numerous extracellular stimuli, including growth factors, and participates in diverse cellular functions including cell growth, differentiation, transformation and cell migration. To properly sort the diverse functions of the MEK–ERK pathway, several scaffolds, such as KSR (kinase repressor of Ras), β-arrestin, Sef and IQGAP1, have been shown to spatially specify the route of the MEK–ERK pathway in the cells (Kolch, 2005; Sacks, 2006). For example, KSR produces a docking platform at the plasma membrane and modulate the intensity and duration of the MEK–ERK pathway, altering cell fate (Morrison and Davis, 2003). IQGAP1, which interacts with cytoskeletal components, is assumed to link the MEK–ERK pathway to the cytoskeleton (Brown and [45]Sacks, 2006). In this study, we showed that approximately half of the total MEK–ERK activity in cells passed through the p18–p14–MP1 scaffold on late endosomes. This branch of the MEK–ERK pathway may specifically contribute to the regulation of endosome dynamics.
However, the functions of the late endosomal MEK–ERK pathway in endosome dynamics remain to be addressed. We observed that Rab11-mediated endosome cycling was greatly impaired by p18 loss, despite the localization of p18 to late endosomes. This spatial inconsistency suggests an apparent lack of causal link between p18 and the defects caused by p18 loss. However, Rab11-positive recycling endosomes are known to be cycled through PNRC, where late endosomes are accumulated (Caswell and Norman, 2006). Thus, it can be hypothesized that, similar to lysosomes that fuse to late endosomes in the maturation process, Rab11-positive recycling endosomes may be processed by directly interacting with late endosomes in PNRC. The aberrant distribution of recycling endosomes as well as lysosomes observed in p18−/− cells could be a consequence of the lack of proper interaction or fusion with late endosomes. To prove this hypothesis, more detailed analysis of vesicular interaction with late endosomes and identification of specific ERK targets involved in the regulation of the interaction will be necessary. Alternatively, the endosomal p18–MEK–ERK pathway might regulate polarized transport of vesicles and organelles along microtubules and/or actin filaments (Pullikuth and Catling, 2007). It was reported that the crystal structure of MP1/p14 is strikingly similar to the Roadblock/LC7 dynein light chain homodimer (Kurzbauer et al, 2004; Lunin et al, 2004; Ilangovan et al, 2005). Thus, it would also be of great interest to analyse the interaction between the p18-anchored molecules and motor machineries containing dynein or kinesin.
In summary, we presented a line of evidence that the novel adaptor protein p18 on late endosomes has a crucial function in endosome dynamics by serving as an anchor of a branch of the MEK/ERK pathway. Further analysis of the function of the late endosomal MEK/ERK pathway would shed new light on the molecular mechanisms for regulating the endosome dynamics, which is critical for fundamental cellular dynamics.
Materials and methods
Cell culture
Rat pheochromocytoma (PC12) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Nissui) supplemented with 5% fetal bovine serum (FBS; GIBCO Life Technology) and 5% horse serum (GIBCO Life Technology) on dishes coated with collagen type I (BD Biosciences). MEFs, p18+/+, p18+/−, p18−/−, p18rev, C127, HaCaT and MDCK cells were cultured in DMEM supplemented with 10% FBS. p18+/+, p18+/− and p18−/− cell lines were established by introducing SV40 T antigen into primary cultures of embryonic cells from a litter of embryos obtained from an intercross of p18+/− mice. The embryonic stem cell line EGR05 was established from 129S2 mice (Charles River Laboratories) at the Genome Information Research Centre of Osaka University.
DRM fractionation and protein analysis
DRM and non-DRM fractions were separated as described previously (Shima et al, 2003). Briefly, cells were lysed in buffer A (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM sodium orthovanadate, 50 mM NaF, 5 mM 2-mercaptoethanol and protease inhibitors) containing 0.25% Triton X-100 at 4°C. The lysate was separated on a discontinuous sucrose gradient (40–35–5%) by ultracentrifugation at 100 000 g for 16 h at 4 °C. The gradient was separated into 12 fractions collected from the top. The DRM proteins were solubilized in buffer A containing 2% N-octyl-β-D-glucoside (ODG) and 1% Nonidet P40 (NP40). To prepare total cell lysates, cells were lysed in ODG buffer (buffer A containing 5% glycerol, 2% ODG and 1% NP40). Protein content was measured by the Bradford method using bovine serum albumin as the standard. Immunoprecipitation and immunoblotting were performed as described previously (Shima et al, 2003; Yagi et al, 2007). An anti-p18 antibody was generated in rabbits by immunizing with glutathione S-transferase-tagged rat or mouse p18, and was affinity purified using maltose-binding protein-tagged p18. Anti-cathepsin D antibody was prepared as described previously (Koike et al, 2003). Anti-Rab7 was kindly provided by Y Wada (Osaka university). Other antibodies used were obtained commercially: anti-TfR (Zymed), anti-LAMP-1 (1D4B; Developmental Studies Hybridoma Bank), anti-cubilin (Santa Cruz Biotechnology), anti-megalin (Santa Cruz Biotechnology), anti-Rab11 (BD Transduction Laboratories), anti-MP1 (Santa Cruz Biotechnology), anti-GFP (MBL, Nagoya) and anti-β-tubulin (Sigma). Alexa566- and Alexa694-conjugated anti-rabbit IgGs and FITC-conjugated anti-mouse IgG were obtained from Molecular Probes. Peroxidase- and FITC-conjugated CTX were obtained from Sigma.
LC-MS/MS analysis
To identify p18, DRMs were prepared from PC12 cells as described above. For the identification of p18-binding proteins, p18 with a C-terminal Strep tag was stably expressed in MEFs. Protein complexes containing Strep-tagged p18 were purified from total cell lysates using a Strep-Tactin Superflow column (IBA). The protein samples were separated by SDS–PAGE. After visualization by silver staining, protein bands were excised from the gel and digested in situ with Achromobacter protease I (Lys-C) (Wako, Osaka) or Trypsin Gold (Promega). The digested samples were analysed by nanocapillary reversed-phase LC-MS/MS using a C18 column (φ75 mm) on a nanoLC system (Ultimate, LC Packing) coupled to a quadruple time-of-flight mass spectrometer (QTOF Ultima, Waters). Direct injection data-dependent acquisition was performed using one MS channel for every three MS/MS channels and dynamic exclusion for selected ions. Proteins were identified by database searching using Mascot Daemon (Matrix Science).
cDNA cloning and northern blotting
cDNAs for p18, Rab4, Rab5, Rab7, Rab11, MP1 and p14 were obtained by RT–PCR using total RNA from PC12 cells (p18), MEFs (Rab proteins) or mouse brain (MP1 and p14) as templates. cDNA fragments were cloned into the pEXPR-IBA3, pCX4 (a gift from T Akagi), pmKO1-N1 (pEGFP-N1 vector carrying a monomer Kusabira-Orange 1 gene instead of EGFP), pEGFP-C1 and pEGFP-N1 vectors. Site-specific and deletion mutants of p18 were generated by PCR using p18 cDNA as a template. Transient transfection was performed using Lipofectamine 2000. For northern blotting of p18 transcripts in mouse tissues, the entire ORF of mouse p18 cDNA was radiolabelled and hybridized onto multiple tissue blots (MTN and MTNII filters from Clontech). Control β-actin blots were obtained by re-hybridizing the p18 blots.
Cell imaging
To examine the localization of fluorescent proteins, cells seeded onto collagen- or fibronectin-coated cover slips were observed using an Olympus IX81 confocal microscope controlled by Fluoview FV1000 software. For live cell imaging, cells were seeded onto a 35-mm diameter glass-based dish coated with collagen or fibronectin in DMEM (phenol red-free), and monitored using an Olympus IX71 microscope equipped with a CoolSNAP HQ camera (Roper Scientific, Trenton, NJ) controlled by MetaMorph software (Universal Imaging, West Chester, PA). For cell motility analysis, differential interference contrast images were recorded for 20 h at 10-min intervals and the cell locations were traced using the ‘track points' function in MetaMorph.
Immunohistochemistry
Embryos were immersed in a 4% paraformaldehyde–4% sucrose solution in 0.1 M phosphate buffer (PB; pH 7.2), and fixed overnight at 4°C. Samples were embedded in OCT compound (Miles, Elkhart, IN) after cryoprotection with 15 and 30% sucrose solutions, and were cut into 10-μm sections with a cryostat (CM3050; Leica). The sections were placed on silane-coated glass slides and stored at −80 °C until use. The sections were incubated with anti-LAMP-1 (1:100), further incubated with biotinylated goat anti-rat IgG for 1 h, and finally with peroxidase-conjugated streptavidin (Vector Laboratories) for 1 h at RT. Staining for peroxidase and immunofluorescence histochemistry were performed as described previously (Koike et al, 2003; Yagi et al, 2007).
Electron microscopy
Transmission electron microscopy was performed as described previously (Koike et al, 2003). Briefly, embryos were immersed in 2% glutaraldehyde–2% paraformaldehyde buffered with 0.1 M PB (pH 7.2) and fixed overnight (or longer) at 4°C. Embryos were post-fixed with 1% OsO4 in 0.1 M PB, block stained with a 2% aqueous solution of uranyl acetate, dehydrated with a graded series of ethanol and embedded in Epon 812. Ultrathin sections were cut with an ultramicrotome (Ultracut N; Reichert-Nissei), stained with uranyl acetate and lead citrate, and observed with a Hitachi H7100 electron microscope.
Generation of p18 knockout mice
A targeting vector was constructed using pNT1.1 containing the Neo-resistance gene (neo r) as a positive selection marker and the herpes simplex virus 1-thymidine kinase gene as a negative selection marker. A PCR-amplified 1345-bp fragment upstream of the first exon and a 5690-bp fragment encompassing the first intron to the fifth exon were inserted as the short and long arms, respectively. Embryonic stem cells were electroporated with NotI-digested linearized DNA. Two clones that had undergone homologous recombination, as confirmed by PCR analysis on both homologous arms (Supplementary Figure S2), were isolated and injected into C57BL/6 blastocysts, resulting in the birth of male chimaeric mice. These mice were crossed with C57BL/6 females to obtain heterozygous mutants. Mice used in the study were the offspring of crosses between F1 and/or F2 generations. Mice were handled and maintained according to the Osaka University guidelines for animal experimentation.
Supplementary Material
Supplementary Movie S1
Supplementary Movie S2
Supplementary Figures
Acknowledgments
We thank JA Cooper, A Tarakhovski and T Yoshimori for critical reading of the paper, C Oneyama for advices on the gene expression experiment, Y Koreeda and A Kawai for technical support in generating p18 knockout mice, Y Takada for integrin β1 cDNA, T Akagi for pCX4 vector, Y Wada for anti-Rab7 antibody and KW Wong for assistance in editing the paper. LC-MS/MS analysis was performed in the DNA-chip Development Centre for Infectious Diseases (RIMD, Osaka University). This study was supported by grants-in-aid from the Ministry of Education, Culture, Sports Science and Technology––Japan and by The Yasuda Medical Foundation.
References
- Anderson RG, Jacobson K (2002) A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296: 1821–1825 [DOI] [PubMed] [Google Scholar]
- Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA (2007) Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol 9: 1381–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbis A, Parmar A, Wang Y, Baquiran G, Posner BI (2007) Compartmentalization of signaling-competent epidermal growth factor receptors in endosomes. Endocrinology 148: 2944–2954 [DOI] [PubMed] [Google Scholar]
- Bielinska M, Narita N, Wilson DB (1999) Distinct roles for visceral endoderm during embryonic mouse development. Int J Dev Biol 43: 183–205 [PubMed] [Google Scholar]
- Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schaffer AA, Rathinam C, Taub N, Teis D, Zeidler C, Dewey RA, Geffers R, Buer J, Huber LA, Welte K, Grimbacher B, Klein C (2007) A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med 13: 38–45 [DOI] [PubMed] [Google Scholar]
- Brown MD, Sacks DB (2006) IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol 16: 242–249 [DOI] [PubMed] [Google Scholar]
- Brunner Y, Coute Y, Iezzi M, Foti M, Fukuda M, Hochstrasser DF, Wollheim CB, Sanchez JC (2007) Proteomics analysis of insulin secretory granules. Mol Cell Proteomics 6: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Caswell PT, Norman JC (2006) Integrin trafficking and the control of cell migration. Traffic 7: 14–21 [DOI] [PubMed] [Google Scholar]
- Christensen EI, Birn H (2002) Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol 3: 256–266 [DOI] [PubMed] [Google Scholar]
- Deacon SW, Nascimento A, Serpinskaya AS, Gelfand VI (2005) Regulation of bidirectional melanosome transport by organelle bound MAP kinase. Curr Biol 15: 459–463 [DOI] [PubMed] [Google Scholar]
- del Pozo MA, Balasubramanian N, Alderson NB, Kiosses WB, Grande-Garcia A, Anderson RG, Schwartz MA (2005) Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat Cell Biol 7: 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC (2007) Bad signals jam organelle traffic. Nat Med 13: 31–32 [DOI] [PubMed] [Google Scholar]
- Dudu V, Pantazis P, Gonzalez-Gaitan M (2004) Membrane traffic during embryonic development: epithelial formation, cell fate decisions and differentiation. Curr Opin Cell Biol 16: 407–414 [DOI] [PubMed] [Google Scholar]
- Emery G, Knoblich JA (2006) Endosome dynamics during development. Curr Opin Cell Biol 18: 407–415 [DOI] [PubMed] [Google Scholar]
- Ezzell RM, Chafel MM, Matsudaira PT (1989) Differential localization of villin and fimbrin during development of the mouse visceral endoderm and intestinal epithelium. Development 106: 407–419 [DOI] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A (1995) Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol 131: 1435–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, van der Goot FG (2006) Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol 7: 495–504 [DOI] [PubMed] [Google Scholar]
- Hancock JF (2006) Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol 7: 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer MF, Hancock JF (2007) Lipid rafts and membrane traffic. FEBS Lett 581: 2098–2104 [DOI] [PubMed] [Google Scholar]
- Harder T, Scheiffele P, Verkade P, Simons K (1998) Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol 141: 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JB, Zurzolo C (2004) Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic 5: 247–254 [DOI] [PubMed] [Google Scholar]
- Ilangovan U, Ding W, Zhong Y, Wilson CL, Groppe JC, Trbovich JT, Zuniga J, Demeler B, Tang Q, Gao G, Mulder KM, Hinck AP (2005) Structure and dynamics of the homodimeric dynein light chain km23. J Mol Biol 352: 338–354 [DOI] [PubMed] [Google Scholar]
- Jones MC, Caswell PT, Norman JC (2006) Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol 18: 549–557 [DOI] [PubMed] [Google Scholar]
- Kabouridis PS (2006) Lipid rafts in T cell receptor signalling. Mol Membr Biol 23: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai A, Shima T, Okada M (2005) Role of Src family tyrosine kinases in the down-regulation of epidermal growth factor signaling in PC12 cells. Genes Cells 10: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Kirkham M, Parton RG (2005) Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim Biophys Acta 1746: 349–363 [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S, Waguri S, Momoi T, Kominami E, Peters C, Figura K, Saftig P, Uchiyama Y (2003) Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci 22: 146–161 [DOI] [PubMed] [Google Scholar]
- Kolch W (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 6: 827–837 [DOI] [PubMed] [Google Scholar]
- Kondoh K, Nishida E (2007) Regulation of MAP kinases by MAP kinase phosphatases. Biochim Biophys Acta 1773: 1227–1237 [DOI] [PubMed] [Google Scholar]
- Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, Clausen T (2004) Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci USA 101: 10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg D, Goni FM, Heerklotz H (2005) Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 30: 430–436 [DOI] [PubMed] [Google Scholar]
- Lunin VV, Munger C, Wagner J, Ye Z, Cygler M, Sacher M (2004) The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J Biol Chem 279: 23422–23430 [DOI] [PubMed] [Google Scholar]
- Maurer ME, Cooper JA (2005) Endocytosis of megalin by visceral endoderm cells requires the Dab2 adaptor protein. J Cell Sci 118: 5345–5355 [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Pelkmans L, Zerial M (2004) Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol 16: 400–406 [DOI] [PubMed] [Google Scholar]
- Mohrmann K, van der Sluijs P (1999) Regulation of membrane transport through the endocytic pathway by rabGTPases. Mol Membr Biol 16: 81–87 [DOI] [PubMed] [Google Scholar]
- Morrison DK, Davis RJ (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol 19: 91–118 [DOI] [PubMed] [Google Scholar]
- Pfeffer S (2003) Membrane domains in the secretory and endocytic pathways. Cell 112: 507–517 [DOI] [PubMed] [Google Scholar]
- Pullikuth A, McKinnon E, Schaeffer HJ, Catling AD (2005) The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals. Mol Cell Biol 25: 5119–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullikuth AK, Catling AD (2007) Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell Signal 19: 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG (1992) Caveolin, a protein component of caveolae membrane coats. Cell 68: 673–682 [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101 [DOI] [PubMed] [Google Scholar]
- Russell MR, Nickerson DP, Odorizzi G (2006) Molecular mechanisms of late endosome morphology, identity and sorting. Curr Opin Cell Biol 18: 422–428 [DOI] [PubMed] [Google Scholar]
- Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J (2002) Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell 13: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DB (2006) The role of scaffold proteins in MEK/ERK signalling. Biochem Soc Trans 34: 833–836 [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ (1998) MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281: 1668–1671 [DOI] [PubMed] [Google Scholar]
- Shima T, Nada S, Okada M (2003) Transmembrane phosphoprotein Cbp senses cell adhesion signaling mediated by Src family kinase in lipid rafts. Proc Natl Acad Sci USA 100: 14897–14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S (2005) Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA 102: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M (2000) Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 149: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope S, Rivi R, Metzger T, Manova K, Lacy E (2004) Mouse amnionless, which is required for primitive streak assembly, mediates cell-surface localization and endocytic function of cubilin on visceral endoderm and kidney proximal tubules. Development 131: 4787–4795 [DOI] [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, Klein C, Hess MW, Huber LA (2006) p14–MP1–MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol 175: 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA (2002) Localization of the MP1–MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell 3: 803–814 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Mellman I (2005) Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol 23: 975–1028 [DOI] [PubMed] [Google Scholar]
- Trowbridge IS, Collawn JF, Hopkins CR (1993) Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol 9: 129–161 [DOI] [PubMed] [Google Scholar]
- van der Goot FG, Gruenberg J (2006) Intra-endosomal membrane traffic. Trends Cell Biol 16: 514–521 [DOI] [PubMed] [Google Scholar]
- van der Goot FG, Harder T (2001) Raft membrane domains: from a liquid-ordered membrane phase to a site of pathogen attack. Semin Immunol 13: 89–97 [DOI] [PubMed] [Google Scholar]
- Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA (2001) A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol 152: 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Waguri S, Sumikawa Y, Nada S, Oneyama C, Itami S, Schmedt C, Uchiyama Y, Okada M (2007) C-terminal Src kinase controls development and maintenance of mouse squamous epithelia. EMBO J 26: 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Tang T, Tang N, Kudlicka K, Ohtsubo K, Ma P, Marth JD, Farquhar MG, Lehtonen E (2006) Essential role of RGS-PX1/sorting nexin 13 in mouse development and regulation of endocytosis dynamics. Proc Natl Acad Sci USA 103: 16776–16781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie S1
Supplementary Movie S2
Supplementary Figures