Abstract
Human RECQ helicases have been linked to distinct clinical diseases with increased cancer rates and premature ageing. All RECQ proteins, except RECQ4, have been shown to be functional helicases. Mutations in RECQ4 lead to Rothmund–Thomson syndrome (RTS), and mouse models reveal that the conserved helicase motifs are required for avoidance of RTS. Furthermore, the amino (N) terminus of RECQ4 shares homology with yeast DNA replication initiation factor, Sld2, and is vital for embryonic development. Here, in contrast to previous reports, we show that RECQ4 exhibits DNA helicase activity. Importantly, two distinct regions of the protein, the conserved helicase motifs and the Sld2-like N-terminal domain, each independently promote ATP-dependent DNA unwinding. Taken together, our data provide the first biochemical clues underlying the molecular function of RECQ4 in DNA replication and genome maintenance.
Keywords: DNA repair, DNA replication, genome stability, RECQ, Rothmund–Thomson syndrome
Introduction
The highly conserved RecQ family proteins represent a subset of the helicase superfamily II (SFII) and contain seven characteristic motifs important for ATP binding and hydrolysis. Genetic studies in lower organisms have shown that RecQ is required for efficient induction of the SOS response and suppression of illegitimate recombination leading to DNA rearrangements (Hanada et al, 1997). Interestingly, Escherichia coli and yeast contain only one RecQ protein, whereas there are at least five RECQ proteins in humans: RECQ1, BLM, WRN, RECQ4 and RECQ5.
Mutations in different RECQ homologues are linked to distinct clinical syndromes (for review, Mohaghegh and Hickson, 2002; Bachrati and Hickson, 2008). So far, BLM has been implicated in Bloom syndrome (BS), and individuals with BS have a high risk of cancer predisposition. Mutations in the WRN gene have been associated with Werner syndrome, and individuals with this disease show symptoms associated with premature ageing. Mutations in another member of the RECQ helicase family, the RECQ4 gene, have been associated with Rothmund–Thomson syndrome (RTS), RAPADILINO and Baller–Gerold syndrome. RTS patients, similar to BS patients, exhibit various physical and mental developmental abnormalities. These individuals also show signs of premature ageing, such as the early development of cataracts, loss of hair and osteosarcoma.
Human RECQ4 is a 1208 amino-acid polypeptide, and its conserved helicase domain lies between amino acids 450 and 830. Both the N and C termini of RECQ4 contain unique sequences distinctive from other RECQ family helicases. Indeed, the first 200 aa of human RECQ4 shares homology with the yeast replication initiation factor, Sld2, and this sequence similarity led to observations showing that RECQ4 is required for replication initiation (Sangrithi et al, 2005; Matsuno et al, 2006). The implication of a role for RECQ4 in replication initiation suggests that the N-terminal domain of RECQ4 is vital for cell growth. Mouse models indicate that this is the case; for example, disruption at exons 5–8 of the RECQ4 gene (corresponding to the N terminus upstream of the conserved helicase domain) results in embryonic lethality between day 3.5 and 6.5 (Ichikawa et al, 2002). On the other hand, mice with disruptions of exons 9–13 (corresponding to the further downstream SFII helicase domain) are viable (Mann et al, 2006). Nevertheless, even though the conserved helicase domain of RECQ4 may not be required for viability, deletions of the RECQ4 helicase domain are sufficient to cause severe growth retardation, cancer predisposition and premature ageing reminiscent of the human RTS phenotype (Hoki et al, 2003; Mann et al, 2005).
Although the genetic studies demonstrate that the conserved helicase domain of RECQ4 has a vital function in preventing RTS and cancer predisposition, it is surprising that helicase activity was not observed with recombinant RECQ4 purified from E. coli (Macris et al, 2006). Similarly, endogenous RECQ4 purified from HeLa cell extracts failed to unwind DNA substrates in vitro (Yin et al, 2004). These observations raise the question whether RECQ4 is a functional DNA helicase, and the absence of this biochemical activity has been a hindrance to our understanding of the molecular basis of RTS and its role in normal development.
Here, for the first time, we show that recombinant RECQ4 protein purified from E. coli is an active DNA helicase. RECQ4 is capable of unwinding DNA structures, including splayed arms, bubbles and blunt-end duplex DNA. Surprisingly, domain analyses uncovered two distinct ATP-binding and DNA unwinding activities within RECQ4 protein. One of these ATP-dependent DNA dissociation activities maps to the conserved helicase motifs, whereas the second activity is intrinsic to the N terminus of the RECQ4. We find that the Sld2-like domain is not crucial for ATP binding but is required for the efficient DNA unwinding activity contributed by the RECQ4 N terminus. Our biochemical data, together with the mouse models, suggest that RECQ4 may have distinct functions involving different domains of the RECQ4 in cell proliferation, genome maintenance and cancer prevention.
Results
ATP-dependent DNA unwinding by RECQ4
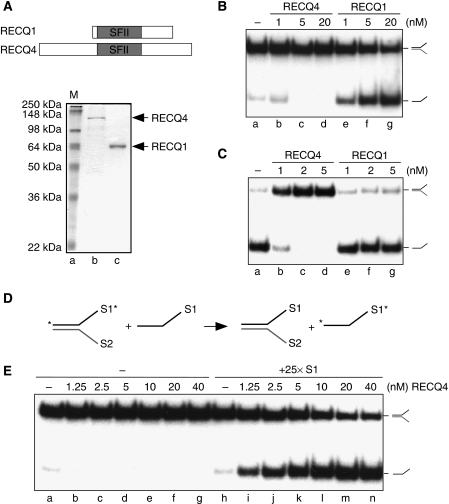
To determine whether RECQ4 exhibits DNA helicase activity, recombinant human RECQ4 was overexpressed and purified from E. coli (Figure 1A). RECQ4 was analysed and compared with RECQ1 for the presence of DNA helicase activities with 32P-labelled splayed-arm substrates, made by annealing oligos S1 and S2. In accord with previous observations, RECQ1 exhibits a robust DNA unwinding activity, whereas RECQ4 fails to unwind the splayed-arm DNA under the same reaction conditions (Figure 1B; Cui et al, 2003; Macris et al, 2006). RECQ helicases possess DNA annealing activities that antagonize their helicase activities but can be overcome by human replication protein A (RPA) through both the stabilization of the ssDNA product and by direct protein–protein interactions (for review, Bachrati and Hickson, 2008; Liu and West, 2008). We found that RECQ4 DNA annealing activity is significantly stronger than that observed with RECQ1, and that the presence of RPA had no effect when added to RECQ4 helicase reactions (Figure 1C and data not shown; Macris et al, 2006).
Figure 1.
Biochemical activities of human RECQ4. (A) (Upper) Schematic diagram of RECQ1 and RECQ4 helicases. The conserved SFII helicase domain is shown in grey. (Lower) Visualization of recombinant RECQ1 and RECQ4 proteins purified from E. coli by SDS–PAGE and Coomassie blue staining. (B) Helicase activities of the recombinant RECQ1 and RECQ4 proteins. Dissociations of the 32P-labelled splayed-arm structures were carried out using indicated amounts of proteins. 32P-Labelled single-stranded DNA products were visualized by autoradiography following neutral PAGE. (C) DNA strand annealing activities of the recombinant RECQ1 and RECQ4 proteins. DNA annealing of 32P-labelled S1 and unlabelled complementary S2 oligos were carried out using the indicated amounts of proteins. The annealed 32P-labelled splayed-arm products were visualized as described in (B). (D) Scheme of DNA helicase assay in the presence of excess of unlabelled S1 oligos. DNA unwinding allows the separation of 32P-labelled S1 (indicated with an asterisk) and unlabelled S2 to ssDNA. DNA annealing promotes the hybridization between the excess amounts of unlabelled S1 with S2, allowing the stabilization of the single-stranded 32P-labelled S1. (E) Helicase activities of the recombinant RECQ4 in the presence and absence of unlabelled S1 oligos. 32P-Labelled dissociated products were visualized by autoradiography following neutral PAGE.
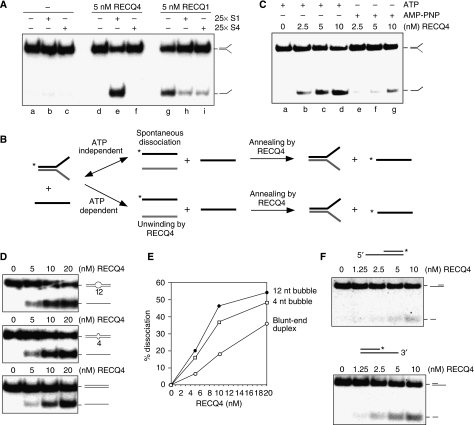
It is possible that the failure to observe RECQ4 DNA unwinding activity in vitro is due to its strong DNA annealing activity, which promotes rapid re-association of the ssDNA products. To test this possibility, we carried out RECQ4 helicase assays in the presence of an excess of unlabelled oligonucleotide S1, which might be expected to prevent re-annealing of the dissociated 32P-labelled S1 and S2 products (Figure 1D). In contrast to the reactions containing no S1 (Figure 1E, lanes b–g), we found that addition of a 25-fold excess of unlabelled S1 revealed an efficient DNA unwinding activity that resulted in the dissociation of the splayed-arm substrate (Figure 1E, lanes i–n). The stimulatory effect was observed only using a complementary oligonucleotide (S1), as it was not seen with non-complementary oligo S4 (Figure 2A, compare lanes e and f). Under the same reaction conditions, excess ssDNA, regardless of the sequence, inhibited unwinding by RECQ1, possibly by competing with the splayed-arm substrates for RECQ1 binding (Figure 2A, lanes g–i).
Figure 2.
ATP-dependent DNA unwinding activity of human RECQ4. (A) Comparison of the helicase activities of the recombinant RECQ4 and RECQ1 in the presence of unlabelled S1 or a control oligo, S4, with different sequence composition. (B) Schematic diagram of the production of 32P-labelled single-stranded S1 in the DNA dissociation reaction. 32P-end labels are indicated with asterisks. In the ATP-independent event (upper), low level of spontaneous DNA dissociation of splayed arm allows the exchange of 32P-labelled single-stranded S1 with the unlabelled S1, a reaction facilitated by the DNA annealing activity of RECQ4. In the ATP-dependent reaction (lower), DNA dissociation is catalysed by RECQ4 unwinding of splayed arm upon ATP hydrolysis to generate 32P-labelled single-stranded S1, which is then stabilized by the presence of unlabelled S1 to compete for re-annealing back to S2. (C) Helicase activities of the recombinant RECQ4 using splay-arm substrates in the presence of ATP or AMP-PNP. (D) Helicase assays of the recombinant RECQ4 proteins were carried out as described using duplex DNA with 12-nt bubble (upper panel), duplex DNA with 4-nt bubble (middle panel) and blunt-ended duplex DNA (lower panel). (E) Product formation in (D) was quantified by phosphorimaging. Close circle, dissociation product using duplex DNA with 12-nt bubble. Open rectangle, dissociation product using duplex DNA with 4-nt bubble. Open circle, dissociation product using blunt-ended duplex DNA. (F) Helicase activities of the recombinant RECQ4 using either a 32P-labelled 5′ overhang (upper panel) or 3′ single stranded overhang (lower panel).
Next, we wished to exclude the possibility that spontaneous dissociation of the splayed-arm substrate could lead to the capture of the unlabelled S2 strand by excess complementary S1 strand, in a reaction mediated by the ssDNA annealing activity of RECQ4 (Figure 2B, upper panel). To do this, we determined whether DNA unwinding by RECQ4 was an ATP-dependent reaction. Helicase assays were carried out in the presence of ATP, or the non-hydrolysable ATP analogue AMP-PNP, leading us to observe a significantly higher levels of dissociation in the presence of ATP compared with the reactions containing AMP-PNP (Figure 2C). This observation shows that the majority of the dissociated ssDNA products result from ATP-dependent DNA unwinding driven by the RECQ4 helicase (Figure 2B, lower panel). Therefore, RECQ4 is an active ATP-dependent DNA helicase.
Substrate specificity and helicase polarity
In Xenopus extracts, depletion of RECQ4 prevents the subsequent loading of RPA and DNA polymerase α during replication initiation. These results indicate that DNA melting reactions that generate ssDNA at replication origins may be defective in the absence of RECQ4 (Sangrithi et al, 2005; Matsuno et al, 2006). MCM complex is thought to unwind DNA at the origin of replication; however, in vitro studies have shown that MCM complex can only exert an effect upon DNA substrates containing partial ssDNA (You et al, 2003; Bochman and Schwacha, 2008). We have shown that RECQ4 is capable of unwinding a replication fork-like structure (Figure 1E). To determine whether RECQ4 is capable of unwinding additional substrates reminiscent of DNA structures found during replication, we analysed RECQ4 helicase activity on a 61 bp blunt-end duplex DNA containing either a 12-nt bubble, a 4-nt bubble or without a bubble. Unlike most other human RECQ helicases, which require a 3′ single-stranded tail for efficient DNA unwinding, RECQ4 was found to unwind all three DNA substrates (Figure 2D and E), an activity similar to that exhibited by E. coli RecQ helicase (Umezu et al, 1990; Harmon et al, 1999; Harmon and Kowalczykowski, 2001). The ability of RECQ4 to unwind duplex DNA suggests a possible role of RECQ4 in assisting MCM complex in melting DNA at the origin of replication.
To determine the polarity of the helicase activity of RECQ4, we analysed its activity on 3′- and 5′-overhang substrates. As RECQ4 is capable of unwinding blunt-ended duplex DNA (Figure 2D), reactions were carried out for relatively short times (15 min), as prolonged incubation results in the unwinding of both substrates, as observed previously with E. coli RecQ protein (Umezu et al, 1990). As expected, although DNA unwinding was observed with both substrates, RECQ4 showed a preference towards the 3′-overhang structure (Figure 2F, lower panel) compared with the 5′-overhang substrate (Figure 2F, upper panel). These results show that RECQ4 exhibits a 3′ → 5′ polarity.
RECQ4 possesses dual DNA unwinding activities
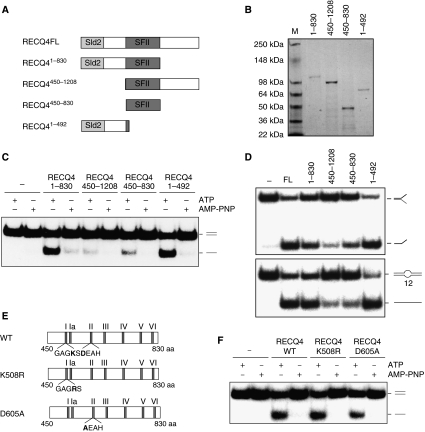
In addition to the conserved helicase domain, all RecQ family helicases contain a RecQ C-terminal (RQC) domain, with the exception of human RECQ4 protein. The RQC domain has been shown to be important for the enzymatic activities for the RecQ family helicases (for review, Bennett and Keck, 2004). The lack of a RQC domain in RECQ4 allowed us to determine the minimum domain requirement for the helicase activity of RECQ4. For this, we overexpressed and purified three RECQ4 fragments (Figure 3A and B): (1) RECQ41–830 containing the N terminus and the SFII helicase domain; (2) RECQ4450–1208 containing the helicase domain and the C terminus and (3) the helicase domain alone, RECQ4450–830. We found that RECQ41–830 exhibits ATP-dependent helicase activity similar to the full-length protein, whereas RECQ4450–1208 shows very little unwinding activity (Figure 3C). When compared with the helicase domain alone (RECQ4450–830), we found that the presence of the N-terminal sequence in RECQ41–830 drastically increased the efficiency of DNA unwinding. In addition, we found that the presence of the C-terminal domain had a mild inhibitory effect on the activity of the SFII helicase domain (Figure 3C, compare RECQ4450–830 and RECQ4450–1208). These results indicate that the N-terminal region is required for the efficient DNA unwinding activity of the RECQ4 protein.
Figure 3.
Domain analyses of the RECQ4 protein. (A) Schematic diagram of the RECQ4 fragments. The conserved SFII helicase domain is shown in dark grey, whereas the Sld2-like domain is shown in light grey. (B) Visualization of recombinant RECQ4 fragments purified from E. coli by SDS–PAGE and Commassie blue staining. (C) Helicase activities of the RECQ4 fragments (20 nM each) using duplex DNA in the presence of ATP or AMP-PNP. (D) Helicase activities of the RECQ4 fragments (20 nM each) using splayed arm (upper panel) or 12-nt bubble (lower panel). (E) Schematic diagram of the RECQ4 SFII helicase domain with the seven conserved helicase motifs. Walker A motif (GAGKS) and DEAH box belonging to the Walker B motif are shown. Walker A mutant K508R has Lys508 mutated to Arg508, whereas Walker B mutant D605A has Asp605 mutated to Ala605. (F) Helicase activities of the full-length RECQ4 WT, K508R and D605A proteins (20 nM each) using duplex DNA in the presence of ATP or AMP-PNP.
Surprisingly, when RECQ41–492, containing only the N-terminal sequence was analysed, we found that this fragment alone showed efficient ATP-dependent DNA unwinding activity (Figure 3C). The DNA unwinding activity observed with RECQ41–492 is significantly more efficient than RECQ4450–830. RECQ41–492 was found to possess strong DNA unwinding activity on duplex DNA, similar to the full-length protein, and also unwinds splayed arm (Figure 3D, upper panel) and 12-nt bubble (Figure 3D, lower panel) substrates. In all cases, RECQ4450–830 exhibited much weaker helicase activity than RECQ41–492. These data suggest that the majority of the ATP-dependent DNA unwinding activity observed with the full-length protein may be contributed by the N-terminal domain. If this is the case, mutations in the conserved Walker A and B motifs of the SFII domain should not abolish the helicase activity observed in the full-length protein. To test this, we generated two full-length RECQ4 point mutants: (1) K508R, in which the conserved lysine residue within the Walker A box of the SFII helicase domain important for ATP hydrolysis was converted to arginine, and (2) D605A, in which the conserved aspartate residue within the ‘DEAH' box important for metal ion binding was converted to alanine (Figure 3E). When compared with the WT protein, ATP-dependent DNA unwinding activity could still be detected in both K508R and D605A mutants (Figure 3F). Interestingly, the D605A mutant consistently showed at least a two-fold decrease in helicase activity compared with WT and K508R. These results indicate that this mutation may affect the stability or overall conformation of the full-length protein and indirectly influence the DNA unwinding efficiency of the N terminus domain.
ATP binding and DNA unwinding of the RECQ4 N-terminal domain
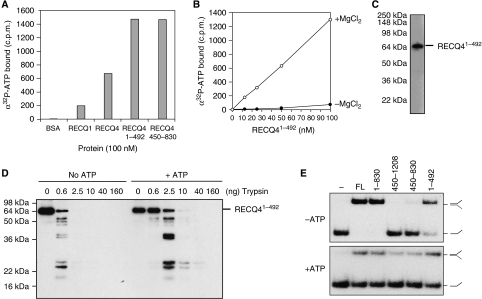
Given that the N-terminal ATP-dependent DNA unwinding activity of RECQ4 was unexpected, we next carried out a direct assessment of the ability of this fragment to bind α32P-labelled ATP using spin-column assays. First, ATP-binding assays were carried out using full-length RECQ1 and RECQ4 proteins, and showed that both proteins were capable of ATP binding (Figure 4A). The addition of 10-fold excess of unlabelled ATP competed away α32P-ATP binding by the RECQ proteins (data not shown). To control for nonspecific binding, we used BSA protein, and found that no radiolabel was retained. When RECQ41–492 and RECQ4450–830 were subjected to similar analyses, we found that RECQ41–492 bound α32P-ATP as efficiently as the helicase domain-containing fragment, RECQ4450–830. The amount of bound α32P-ATP was directly proportional to the amount of RECQ41–492, and ATP binding was found to be dependent on the presence of MgCl2 (Figure 4B). We further confirmed that only one band, corresponding to the RECQ41–492 fragment, became radioactively labelled after UV crosslinking with ATP analogue, α32P-8-azido-ATP (Figure 4C). These results demonstrate that the RECQ4 N-terminal fragment, RECQ41–492, is capable of binding ATP.
Figure 4.
Novel ATP-binding domain of the RECQ4 N terminus. (A) Spin-column analysis of the binding of α32P-ATP by the indicated protein. Following centrifugation, the α32P-ATP present in the flow through or void volume indicates the amount bound by the indicated protein. (B) Spin-column analysis of the binding of α32P-ATP by the indicated amounts of RECQ41–492 in the presence (open circle) or absence (close circle) of MgCl2. (C) Visualization of RECQ41–492 fragment binding to α32P-8-azido-ATP after UV crosslinking by autoradiography following SDS–PAGE. (D) Trypsin proteolysis mapping of RECQ41–492 in the presence or absence of ATP. Protein fragments were separated on SDS–PAGE followed by western blot analysis using anti-FLAG antibody. (E) DNA strand annealing activities of the RECQ4 full length (FL) and fragments (5 nM each). DNA annealing of 32P-labelled S1 and unlabelled complementary S2 oligos were carried out using the indicated proteins without ATP (upper panel) or in the presence of ATP (lower panel).
Cofactor binding to a protein often induces a conformational or oligomerization state change. We therefore tested whether the binding of ATP causes structural changes in RECQ41–492. Partial proteolysis mapping of RECQ41–492 was carried out in the presence or absence of ATP. When RECQ41–492 was digested with trypsin, we found that it was at least four-fold more resistant to proteolytic cleavage in the presence of ATP (Figure 4D). These results indicate that ATP binding induces a conformational change in RECQ41–492.
It has been shown that the presence of ATP reduces the DNA annealing efficiency of other RECQ helicases (Garcia et al, 2004; Cheok et al, 2005; Machwe et al, 2005; Muzzolini et al, 2007). We therefore analysed the DNA annealing activity of the RECQ4 fragments and found that fragments of RECQ4 containing the N-terminal region exhibited strong DNA annealing activity in the absence of ATP (Figure 4E, upper panel). As observed with other RECQ helicases, ATP reduced the amount of annealed products in all reactions, including those containing the RECQ41–492 fragment. This may be due to ATP-induced protein conformational change or activation of the DNA unwinding activity that counteracts strand annealing (Figure 4E, lower panel).
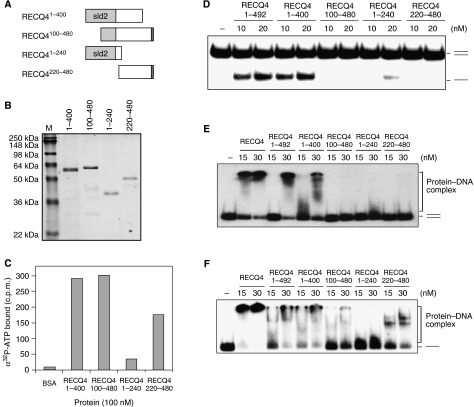
To date, diverged nucleotide-binding motifs, often glycine rich, have been described in proteins involved in a variety of cellular processes. The RECQ4 N-terminal domain contains several glycine-rich clusters potentially important for ATP binding. To further narrow down the potential ATP-binding site located at the N terminus of the RECQ4, we constructed and purified four smaller N-terminal fragments: (1) RECQ41–400, (2) RECQ4100–480, (3) RECQ41–240 and (4) RECQ4220–480 (Figure 5A and B). ATP-binding assays revealed that ATP-binding efficiency is significantly reduced in RECQ41–240 compared with other fragments. This result suggests that the ATP-binding site is located within the domain containing amino-acid residues 240–400 (Figure 5C). We next analysed these N-terminal fragments for their DNA unwinding activities. Interestingly, even though RECQ4100–480 binds ATP as efficiently as RECQ41–400, the lack of the first half of the Sld2-like domain (1–99 aa) in RECQ4100–480 abolished its DNA unwinding activity (Figure 5D). Therefore, even though the Sld2-like domain is not responsible for ATP binding, it is required for efficient DNA unwinding.
Figure 5.
Biochemical analysis of the RECQ4 N-terminal domain. (A) Schematic diagram of the N-terminal fragments. The amino-acid residues covered by each of the fragments are indicated, and the Sld2-like domain is shown in light grey. (B) Visualization of recombinant N fragments purified from E. coli by SDS–PAGE and Commassie blue staining. (C) Spin-column analysis of the binding of α32P-ATP by the indicated N-terminal fragments. (D) Helicase activities of different RECQ4 N-terminal fragments using duplex DNA. (E) Binding of 32P-end-labelled duplex oligos by RECQ4 full length and different RECQ4 N-terminal fragments. DNA–protein complex was analysed on 5% native PAGE. (F) Binding of 32P-end-labelled single-stranded oligos by RECQ4 full length and different RECQ4 N-terminal fragments. DNA–protein complex was analysed on 5% native PAGE.
We next determined whether the lack of DNA unwinding activity in RECQ4100–480 is due to a decrease of its affinity for DNA. For this, we analysed the DNA-binding activities of the various N-terminal fragments using electrophoresis mobility shift assays. Full-length RECQ4 binds efficiently to the duplex (Figure 5E) and single-stranded (Figure 5F) substrates. Similar results were obtained with either RECQ41–492 or RECQ41–400. The presence of RECQ41–240 results in a small mobility shift, as indicated by the smearing signal seen close to the free DNA. This less stable but detectable association of RECQ41–240 to both ssDNA and dsDNA was observed at the higher protein concentration (i.e. 30 nM), indicating that a ds/ssDNA-binding motif is located within the Sld2 domain. This dsDNA interaction by RECQ41–240 may also explain the low amount of ATP-independent DNA melting products observed (Figure 5D). On the other hand, RECQ4100–480 and RECQ4220–480 fails to bind to dsDNA. These results indicate that the affinity to dsDNA exhibited by the Sld2 domain is crucial for the DNA unwinding activity of the N terminus of RECQ4.
RECQ4 SFII helicase domain
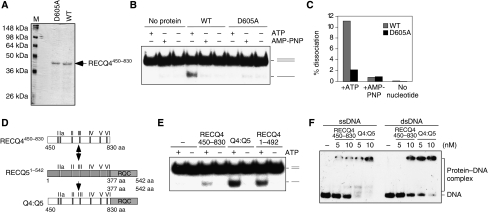
The RECQ4 SFII helicase domain contains the conserved seven amino-acid motifs, including the Walker A box located at motif I and Walker B box at motif II (Figure 3E). To further confirm an active DNA helicase activity associated with the RECQ4 conserved helicase domain, we compared DNA unwinding by RECQ4450–830 with a helicase-dead mutant fragment, RECQ4450–830 (D605A; Figure 6A). We found that the WT RECQ4450–830 was capable of unwinding 10% of duplex DNA, whereas, unlike the full-length D605A protein (Figure 3C), RECQ4450–830 (D605A) fragment failed to unwind any duplex DNA (Figure 6B and C).
Figure 6.
DNA unwinding by the RECQ4 SFII domain. (A) Visualization of recombinant RECQ4450–830 fragment and the corresponding D605A mutant fragment purified from E. coli by SDS–PAGE and Commassie blue staining. (B) Helicase activities of the RECQ4 helicase domain fragment and D605A mutant (20 nM each) in the presence of ATP or AMP-PNP or without nucleotide cofactor. (C) Product formation in (C) was quantified by phosphorimaging. (D) Schematic diagram of the RECQ4 SFII helicase domain, RECQ51–542 fragment and the chimaera protein. The seven conserved helicase motifs were shown. (E) Helicase activities of the RECQ4450–830, RECQ4:RECQ5 chimaera and RECQ41–492 (20 nM each) in the presence or absence of ATP. (F) Binding of 32P-end-labelled single-stranded (left) and duplex oligos (right) by RECQ4450–830 fragment and RECQ4:RECQ5 chimaera protein. DNA–protein complex was analysed on 5% native PAGE.
The DNA unwinding activity observed with the RECQ4 conserved SFII helicase domain is significantly weaker than that of the N-terminal domain. As mentioned previously, the efficient helicase activities by the other RecQ family helicases require the conserved RQC domain (for review, Bennett and Keck, 2004). We therefore tested whether the RQC domain could also enhance the DNA unwinding activity of the RECQ4 helicase domain. To do this, we generated a chimaeric protein by replacing the SFII helicase domain of a RECQ51–542 fragment containing the SFII helicase and RQC domain, with the RECQ4 SFII helicase domain (Figure 6D). We found that the chimaeric protein, Q4:Q5 unwinds DNA more efficiently than RECQ4450–830, and the activity of the chimaeric protein is comparable to the activity of RECQ41–492 (Figure 6E). The enhancement of the helicase activity by the RQC domain may be due to the increased DNA affinity of the chimaeric protein (Figure 6F). These results indicate that the RECQ4 SFII helicase domain is functionally active, and behaves in a manner similar to the helicase domains found in other RECQ homologues. Taken together, these results show that the conserved SFII helicase domain of RECQ4 is proficient at ATP-dependent DNA unwinding.
Discussion
In previous studies, it was not possible to demonstrate in vitro DNA helicase activity associated with RECQ4, and as such RECQ4 appeared to be distinct from all other members of the RECQ helicase family. As a consequence, RECQ4 is the least biochemically characterized member of this protein family, despite the clinical impact of this protein with links to cancer development and premature ageing. Here, for the first time, we demonstrated an ATP-dependent RECQ4 helicase activity in vitro, which most likely has been masked by its strong intrinsic DNA re-annealing activity. This activity is similar to the previously described strand-exchange reaction catalysed by BLM and WRN through the concerted action between DNA unwinding and strand annealing (Machwe et al, 2006). However, unlike other RECQ helicases, the presence of the third strand (e.g. unlabelled S1 oligo) is required to demonstrate unwinding by RECQ4.
Importantly, we found that the ATP-dependent DNA unwinding activity is contributed by two distinct domains in RECQ4. Thus, in addition to the known helicase domain, we have uncovered an additional Mg-dependent ATP-binding pocket located at the N-terminal region between amino acids 240 and 400 of the protein. Moreover, we also demonstrated that the Sld2-like domain located at the N-terminal region of the RECQ4 protein, even though not crucial for ATP binding, is essential for efficient DNA unwinding activity possibly by promoting DNA–protein interactions. As RECQ4 is the only RecQ family helicase without a conserved RQC domain, it is possible that this Sld2-like domain may replace the RQC domain in facilitating DNA unwinding.
Previous studies using Xenopus extracts suggest that RECQ4 may be the functional homologue of yeast replication initiation factor Sld2 (Sangrithi et al, 2005; Matsuno et al, 2006). In these studies, depletion of the RECQ4 proteins from Xenopus extracts abolishes DNA replication initiation. Interestingly, although Matsuno et al showed that the replication defect can be partially complemented by the addition of a Xenopus RECQ4 protein fragment containing only the N terminus, Sangrithi et al were able to only partially restore DNA replication with full-length human RECQ4 but not the full-length D605A mutant. Our results may provide an explanation for both observations. First, if the catalytic activity found in the N terminus of the human RECQ4 protein is evolutionarily conserved, the biochemical activity exhibited by the N-terminal fragment of Xenopus RECQ4 alone may be sufficient to restore the replication defect. Second, even though human full-length RECQ4 protein carrying the D605A mutation still shows helicase activity, the DNA unwinding efficiency is reduced compared with the WT and K508R mutant (Figure 3F). These results suggest that the D605A mutation may affect protein folding or stability and indirectly influence the activity of RECQ4 N terminus. As a consequence, the human D605A mutant may be less efficient compared with the WT protein in complementing the replication defect in RECQ4 depleted Xenopus extracts.
Genetic studies in human RTS patients and mouse models reveal the importance of RECQ4 in mammalian systems. Disruptions at different regions of the RECQ4 proteins affect various aspects of normal development and genome stability. In particular, the N-terminal region of the RECQ4 protein is proven to be essential for cell survival and embryonic development (Ichikawa et al, 2002). In addition to the N-terminal domain, mutations within the conserved SFII helicase domain have often been linked to RTS (Siitonen et al, 2009). Though viable, cells lacking a functional RECQ4 SFII helicase domain showed slow cell growth and defects in sister chromatid cohesion and chromosome segregation (Hoki et al, 2003; Mann et al, 2005). Our unexpected finding of the RECQ4 dual DNA dissociation activities, together with the genetic studies revealing the different consequences in disrupting various RECQ4 domains, suggests that RECQ4 may have multiple roles in DNA metabolism in mammalian cells, and that each specific function may require different domains of RECQ4.
Materials and methods
Plasmids
To generate N-terminal His-tagged RECQ1, RECQ1 full-length cDNA was cloned into pET16b vector (Novagen). To generate N-terminal His-tagged and C-terminal FLAG-tagged RECQ4 full-length and fragments, the corresponding DNA fragments were PCR amplified and cloned into pET16b-FLAG vector containing 2 × FLAG inserted at XhoI site. To generate N-terminal His-tagged RECQ4450–830 fragment, the DNA fragment was PCR amplified and cloned into pET16b vector. Stop codon was provided by vector sequence downstream of the multi-cloning sites. RECQ4450–830 D605A mutant was made by mutating GAT codon encoding Asp to GCT codon encoding Ala using QuikChange Site-directed Mutagenesis Kit.
Proteins
All RECQ4 full-length and truncated proteins, except RECQ4450–830 WT and D605A mutant, containing N-terminal His tag and C-terminal FLAG tag were purified using the same method. Briefly, proteins were expressed in Rosetta (DE3) pLysS cells. Cells were grown at 37°C to a cell density OD600 ∼0.4, followed by overnight induction at 16°C with 0.1 mM isopropyl-β-D-thio-galactoside. Cells were harvested by centrifugation and lysed in lysis buffer (50 mM potassium phosphate, pH 8.0, 10% glycerol, 300 mM KCl, 0.5% Triton X-100, 5 mM β-mercaptoethanol, 1 × protease inhibitor cocktail (Roche), 1 mM PMSF and 0.2 mg/ml lysozyme), followed by sonication. The supernatant was clarified by centrifugation and applied to High-Q column, and the flow through was collected and brought to 0.1% (w/v) in polyethyleneimine (PEI). Precipitate was removed by centrifugation, and the supernatant was applied to Ni-NTA column, washed with buffer B (50 mM potassium phosphate, pH 6.0, 10% glycerol, 500 mM KCl, 0.5% Triton X-100 and 5 mM β-mercaptoethanol) containing 50 mM imidazole, and the His-tagged proteins were eluted with buffer B containing 1 M imidazole. The eluate was dialysed against buffer D (50 mM Tris–HCl, pH 7.6, 10% glycerol and 500 mM KCl) and incubated with 80 μl α-FLAG M2 beads (Sigma) overnight at 4°C. The M2-bound proteins were washed with buffer E (50 mM Tris–HCl, pH 7.6, 10% glycerol, 500 mM KCl, 0.2% Triton X-100 and 1 mM EDTA), eluted with buffer E containing 200 μg/ml FLAG peptide, and dialysed against buffer F (50 mM Tris–HCl, pH 8.0, 10% glycerol, 500 mM KCl, 1 mM EDTA and 1 mM DTT).
E. coli lysates expressing RECQ1, RECQ4450–830 WT or D605A mutant proteins were first passed through High-Q column followed by PEI precipitation as described for RECQ4. The flow through from Q column was applied onto Ni-NTA column as described, except that 5 mM imidazole was included in the binding buffer. The Ni-NTA eluate was dialysed against buffer G (50 mM Tris–HCl, pH 7.2, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.2% Trion X-100 and 70 mM KCl) and then loaded onto SP column. The SP column was washed with buffer G containing additional 30 mM KCl and eluted with buffer F. All purified proteins were stored as aliquots at −80°C.
DNA substrates
All DNA substrates were prepared by annealing appropriate oligos together and purified on native PAGE. The splayed-arm substrate was generated by annealing 32P-labelled S1 oligo (5′-GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTT TGCCCACCTGCAGGTTCACCC-3′) and unlabelled S2 oligo (5′-ATCGATAGTCGGATCCTCTAGACAGCTCCATGTAGCAAGG CACTGGTAGAATTCGGCAGCGT-3′). Blunt-ended duplex was made by annealing 32P-labelled S1 oligo and unlabelled S1c oligo (5′-GGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAAGGCA CTGGTAGAATTCGGCAGCGTC-3′). DNA bubble (12 nt) was made by annealing 32P-labelled S1 oligo and unlabelled S1BU12 oligo (5′-GGGTGAACCTGCAGGTGGGCAAAGAGTCAGTCAGTCAGCA CTGGTAGAATTCGGCAGCGTC-3′). DNA bubble (4 nt) was made by annealing 32P-labelled S1 oligo and unlabelled S1BU4 oligo (5′-GGGTGAACCTGCAGGTGGGCAAAGATGTCAGCTCAAGGCA CTGGTAGAATTCGGCAGCGTC-3′). 3′ overhang was made by annealing 32P-labelled S2c5 oligo (5′-CATGGAGCTGTCTAGAGGATCCGACTATCGA-3′) and unlabelled S2 oligo. 5′ overhang was made by annealing32P-labelled S1c5 oligo (5′-TGGGTGAACCTGCAGGTGGGCAAAGATGTCC-3′) and unlabelled S1 oligo. The sequence of S4 used as control oligo in the helicase reaction is 5′-AAGGTTCGAATCCTTCCCCCCCCACCACCCCCTCCCCCTC GGCCGAAATTCGGTACC-3′.
Helicase, DNA annealing and DNA-binding assays
Helicase reactions (10 μl) contained 32P-end-labelled substrate (0.2 ng) in helicase buffer (30 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM ATP, 1 mM DTT, 100 μg/ml BSA and 10% glycerol). Unlabelled S1 (2.5 ng) was included in all the reactions except those experiments shown in Figure 1C and E. Where indicated, 5 mM ATP was replaced by 5 mM AMP-PNP. The reactions were initiated by the addition of purified RECQ proteins and incubated for 30 min at 37°C. The DNA products were deproteinized and analysed on neutral PAGE. Annealing reactions (10 μl) were carried out by incubating the corresponding purified proteins with 10 pg of 32P-labelled S1 and 25 pg of unlabelled S2 in annealing buffer (30 mM Tris, pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA and 5 mM ATP where indicated). The reactions were incubated at 37°C for 30 min, deproteinized and analysed on neutral PAGE. DNA-binding reactions (10 μl) were carried out by incubating the corresponding purified proteins with 4 pg of 32P-end labelled dsDNA or ssDNA in DNA-binding buffer (30 mM HEPES, pH 7.5, 1 mM DTT and 100 μg/ml BSA). The reactions were incubated on ice for 15 min and analysed on neutral PAGE.
ATP-binding assays
For ATP-binding assays using spin columns, ATP-binding reactions (20 μl) were carried out in the presence or absence of MgCl2 in a reaction buffer (30 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT and 10% glycerol) plus 1 μCi α32P-ATP (3000 Ci/mmol, NEN) for 30 min on ice. Reactions were then applied to Bio-Spin P30 Tris chromatography columns (Bio-Rad), which had been pre-equilibrated in a reaction buffer, and spun (4 min at 1000 g) to allow protein with bound α32P-ATP to pass through. The flow throughs were spotted and dried on Whatman paper and the radioactivity was quantified by phosphorimager. For crosslinking RECQ41–492 protein fragment to ATP, 0.4 μM of RECQ41–492 fragment was incubated with 10 μM α32P-8N3ATP (1 μCi; ALT BioScience) in a binding buffer (20 mM Tris, pH 7.5, 4 mM MgCl2 and 1 mM DTT). Incubations were performed for 20 min on ice in 10 μl of final volume. Samples were then exposed to 600 mJ/cm2 UV radiation and analysed on SDS–PAGE following autoradiography.
Proteolysis mapping
Digestion reactions (20 μl) contained 20 nM RECQ41–492 in ATP-binding reaction buffer with or without ATP were incubated at room temperature for 5 min before the addition of trypsin (Sigma). Reactions were stopped after 10 min incubation at 37°C by the addition of an equal volume of SDS–PAGE gel loading buffer, and ran on a 12% SDS–PAGE gel followed by western blotting.
Acknowledgments
This study was supported by grants from American Federation for Aging Research and Yale Rudolph J Anderson Endowed Fellowship. We thank Rebecca L Barnes for her contribution to the initial plasmid cloning.
References
- Bachrati CZ, Hickson ID (2008) RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117: 219–233 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL (2004) Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol 39: 79–97 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A (2008) The Mcm2–7 complex has in vitro helicase activity. Mol Cell 31: 287–293 [DOI] [PubMed] [Google Scholar]
- Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID (2005) The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res 33: 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Klima R, Ochem A, Arosio D, Falaschi A, Vindigni A (2003) Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem 278: 1424–1432 [DOI] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 23: 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA 94: 3860–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC (1999) RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell 3: 611–620 [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J Biol Chem 276: 232–243 [DOI] [PubMed] [Google Scholar]
- Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumua R, Nakamura M, Takahashi H, Noda Y, Kito S, Abe M (2003) Growth retardation & skin abnormalities of the Recql4-deficient mouse. Hum Mol Genet 12: 2293–2299 [DOI] [PubMed] [Google Scholar]
- Ichikawa K, Noda T, Furuichi Y (2002) Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund–Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi 119: 219–226 [DOI] [PubMed] [Google Scholar]
- Liu Y, West SC (2008) More complexity to the Bloom's syndrome complex. Genes Dev 22: 2737–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Matson SW, Orren DK (2005) RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem 280: 23397–23407 [DOI] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P (2006) Biochemical characterization of the RECQ4 protein, mutated in Rothmund–Thomson syndrome. DNA Repair (Amst) 5: 172–180 [DOI] [PubMed] [Google Scholar]
- Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Lou G (2005) Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund–Thomson syndrome. Hum Mol Genet 14: 813–825 [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H (2006) The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 26: 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Hickson ID (2002) Premature aging in RecQ helicase-deficient human syndromes. Int J Biochem Cell Biol 34: 1496–1501 [DOI] [PubMed] [Google Scholar]
- Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol 5: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund–Thomson syndrome. Cell 121: 887–898 [DOI] [PubMed] [Google Scholar]
- Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, Keymolen K, Lipsanen-Nyman M, Miny P, Plon SE, Riedl S, Sarkar A, Vargas FR, Verloes A, Wang LL, Kääriäinen H et al. (2009) The mutation spectrum in RECQL4 diseases. Eur J Hum Genet 17: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Nakayama K, Nakayama H (1990) Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA 87: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Kwon YT, Varshavsky A, Wang W (2004) RECQL4, mutated in the Rothmund–Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum Mol Genet 13: 2421–2430 [DOI] [PubMed] [Google Scholar]
- You Z, Ishimi Y, Mizuno T, Sugasawa K, Hanaoka F, Masai H (2003) Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J 22: 6148–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]