Abstract
The ultraviolet-B (UV-B) portion of the solar radiation functions as an environmental signal for which plants have evolved specific and sensitive UV-B perception systems. The UV-B-specific UV RESPONSE LOCUS 8 (UVR8) and the multifunctional E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) are key regulators of the UV-B response. We show here that uvr8-null mutants are deficient in UV-B-induced photomorphogenesis and hypersensitive to UV-B stress, whereas overexpression of UVR8 results in enhanced UV-B photomorphogenesis, acclimation and tolerance to UV-B stress. By using sun simulators, we provide evidence at the physiological level that UV-B acclimation mediated by the UV-B-specific photoregulatory pathway is indeed required for survival in sunlight. At the molecular level, we demonstrate that the wild type but not the mutant UVR8 and COP1 proteins directly interact in a UV-B-dependent, rapid manner in planta. These data collectively suggest that UV-B-specific interaction of COP1 and UVR8 in the nucleus is a very early step in signalling and responsible for the plant's coordinated response to UV-B ensuring UV-B acclimation and protection in the natural environment.
Keywords: Arabidopsis, photomorphogenesis, plant– environment interaction, stress acclimation, UV-B
Introduction
Sunlight is of utmost importance to plants, both as the ultimate energy source and as an environmental signal regulating growth and development. For the latter, higher plants possess several classes of photoreceptors, including the molecularly known phytochromes for the red/far-red, and cryptochromes, phototropins and members of the Zeitlupe family for the UV-A/blue part of the spectrum (e.g. Chen et al, 2004). Ultraviolet-B (UV-B; 280–315 nm) radiation is an integral part of the sunlight reaching the surface of the Earth and induces a broad range of physiological responses. The UV-B-induced photomorphogenic responses, in contrast to damage responses, are thought to be mediated by a molecularly unidentified UV-B-specific photoreceptor different from the known receptors acting in the visible part of the light spectrum (Brosche and Strid, 2003; Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins and Brown, 2007). Key regulatory factors involved in the UV-B-induced photomorphogenic pathway, such as the bZIP transcription factor ELONGATED HYPOCOTYL 5 (HY5), the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and the seven-bladed propeller protein UV RESPONSE LOCUS 8 (UVR8), have been identified and plants harbouring hy5, cop1 and uvr8 loss of function mutations display reduced tolerance to UV-B stress (Kliebenstein et al, 2002; Ulm et al, 2004; Brown et al, 2005; Oravecz et al, 2006).
UVR8 was found to exclusively act in UV-B signalling, thus showing high functional specificity (Brown et al, 2005). In planta, UV-B stimulates rapid nuclear accumulation of the UVR8 protein, which seems to be required but is not sufficient for UV-B-responsive gene expression changes (Kaiserli and Jenkins, 2007). UVR8 associates constitutively with chromatin regions of several UV-B-activated genes, including the HY5 genomic locus (Brown et al, 2005; Cloix and Jenkins, 2008). Recently, it was suggested that HY5 and its homologue HYH are key effectors of the UVR8 pathway and act redundantly to control expression of most, if not all, UVR8 target genes (Brown and Jenkins, 2008).
COP1 is a known repressor of photomorphogenesis in darkness as well as in light, but is a promoter of UV-B-specific responses: cop1 mutants have a light-grown phenotype in darkness, show features of enhanced photomorphogenesis in light but are deficient in UV-B photomorphogenic responses (Yi and Deng, 2005; Oravecz et al, 2006). At the molecular level, COP1 targets different photomorphogenesis-promoting transcription factors for degradation in the dark, among them HY5 (Osterlund et al, 2000; Saijo et al, 2003). Upon activation of photoreceptors by visible light, COP1 is inactivated and physically separated from HY5 by nuclear exclusion, allowing HY5 stabilization and activation of light-responsive genes (von Arnim and Deng, 1994; Yi and Deng, 2005). Light-induced, early inactivation of this E3 ligase is most likely mediated by direct interaction with active phytochromes and cryptochromes, but the precise molecular mechanism underlying this process is still unknown (Yi and Deng, 2005). However, a number of characteristics clearly distinguish COP1 function under UV-B from that in visible light signalling, including (i) promotive versus repressive function, (ii) primarily nuclear versus cytoplasmic localization, (iii) structure–function differences displayed by different cop1 alleles and (iv) independence versus dependence on accessory SPA proteins (Oravecz et al, 2006). Altogether, this set of data indicated a distinct UV-B signalling function of the multifunctional COP1 protein.
Despite the ecological and economic impact of the UV-B response (e.g. Caldwell et al, 2007), very little is known about the underlying signalling mechanisms linking UV-B perception to specific photomorphogenic responses. Both UVR8 and COP1 impinge on the UV-B-mediated activation of HY5 gene expression; however, the relationship of COP1 and UVR8 UV-B-specific functions has remained unknown. Here, we show that COP1 and UVR8 proteins interact specifically in a UV-B-dependent manner in planta, suggesting that physical association between these two proteins contributes to their specific activities in UV-B signalling. This conclusion is supported by the findings that mutant alleles of COP1 or UVR8 displaying UV-B signalling deficiencies do not interact with their respective wild-type partner. Furthermore, we demonstrate the absence of UV-B-induced photomorphogenesis in uvr8 mutants at the phenotypic level and show that UVR8 overexpression, on the other hand, leads to UV-B hyperresponsiveness. As a result, uvr8 mutants are more, whereas UVR8 overexpressors are less affected than their corresponding wild type under UV-B regimens simulating natural conditions.
Results
A luciferase-based genetic screen identifies novel cop1 and uvr8 mutant alleles
To uncover players involved in early UV-B signalling, we screened for mutants altered in UV-B-induced expression of the HY5 gene. This was accomplished by generating an Arabidopsis line carrying a transgene consisting of the HY5 promoter fused to the firefly luciferase coding sequence (Ws/ProHY5:Luc) (Ulm et al, 2004). A number of mutants showing no UV-B induction were identified in the M2 generation after EMS mutagenesis. The identified mutants fell into two complementation groups, and we found that these constituted new cop1 and uvr8 alleles. In addition to the cop1-4 allele described before (Gln-283 to Stop) (Oravecz et al, 2006), we identified a novel allele carrying a point mutation in the region encoding the WD40 repeats of COP1, namely Gly-608 (GGA) changed to Arg (AGA). The corresponding mutant, designated as cop1-19, has a weak constitutively photomorphogenic (cop) phenotype in dark and enhanced photomorphogenesis in light, similar to cop1-4. This genetic screen also identified nine novel uvr8 alleles different from any of the previously described ones (uvr8-1 to uvr8-5) (Kliebenstein et al, 2002; Brown et al, 2005) (Supplementary Figure S1). In addition, we identified an uvr8 T-DNA insertion line from the SALK collection (uvr8-6, SALK_033468; see Supplementary Figure S1 for molecular characterization). Throughout the remainder of the work described, we used the uvr8-6 (Col) and uvr8-7 (Ws; Gln-124 to Stop)-null mutant alleles. The results were comparable for both alleles.
UV-B-mediated inhibition of hypocotyl growth is absent in uvr8 mutants
To increase our understanding of UVR8 function in regulating UV-B-induced photomorphogenesis, we examined UV-B-responsive hypocotyl shortening. These experiments were performed under specific UV-B irradiation conditions using white light supplemented with narrowband UV-B. Under these conditions, 4-day-old wild-type Arabidopsis seedlings are grown without any sign of damage, but display about 50% inhibition of hypocotyl growth accompanied by anthocyanin and flavonoid accumulation (Oravecz et al, 2006). Figure 1A and B and Supplementary Figure S1 show that hypocotyl growth of the uvr8 mutant seedlings, in stark contrast to wild-type seedlings, was not inhibited by UV-B. Importantly, in contrast to cop1 (Oravecz et al, 2006), the hypocotyl growth of uvr8 under visible light is not different from wild type. Thus, we conclude that uvr8 mutants are non-responsive to UV-B as a photomorphogenic signal. Moreover, these data strongly indicate that the narrowband UV-B irradiation conditions used are ideal to specifically analyse UV-B-induced photomorphogenesis and distinguish it from UV-B damage/stress responses.
Figure 1.
Absence of UV-B-induced hypocotyl growth inhibition and gene expression changes in uvr8 and cop1 mutants. (A, B) Wild type (Ws) and uvr8-7 mutant were grown under white light with or without supplementary narrowband UV-B. Here, 4-day-old seedlings were photographed and their hypocotyl length was measured. Error bars represent s.d. (n=30). (C) Venn diagrams showing the number of genes classified as responding to narrowband UV-B (⩾2-fold) in uvr8-6, cop1-4 and wild type (Col) and their overlap. The corresponding gene lists can be found in Supplementary Tables S1, S2 and S3.
UV-B-mediated changes in gene expression are absent in uvr8 and cop1 mutants
Next to the hypocotyl phenotype, analysis of the uvr8 alleles showed that all of them are completely insensitive to UV-B concerning HY5 gene activation (data not shown). However, the same uvr8 mutants showed normal HY5 activation by red, far-red and blue light (Supplementary Figure S2). These data, together with the previous data from Brown et al (2005), indicate a UV-B-specific function of the UVR8 protein.
To have a more global view on gene expression changes underlying the UV-B photomorphogenic response, we carried out Affymetrix ATH1 Genechip analysis. We investigated, parallel to wild type, the impact of the loss of UVR8 and COP1 under these low-level, narrowband (∼312 nm) UV-B conditions using the uvr8-6-null and the cop1-4 mutants. We analysed gene expression changes in 4-day-old seedlings grown under continuous light with or without supplementary UV-B in the same light field (under WG305 and WG345 cutoff, respectively). In addition, to analyse the early UV-B response (see Oravecz et al, 2006 for experimental scheme), we grew seedlings for 4 days without UV-B under a WG345 cutoff filter and then exchanged it for a WG305 cutoff filter 1 or 6 h before harvesting. These different treatments are designated as 96 h −UV-B, 1 h +UV-B, 6 h +UV-B and 96 h +UV-B.
Data obtained demonstrate that in wild-type seedlings already after 1 h UV-B irradiation numerous transcripts are altered (e.g. 377 and 102 genes up- and downregulated, respectively), whereas these changes are virtually absent in the uvr8-6 and cop1-4 mutants (Figure 1C). These effects are similarly true for genes activated at 6 h +UV-B and 96 h +UV-B, as well as for genes downregulated at all time points (Figure 1C). These UV-B-activated classes include genes associated with UV-B tolerance such as photorepair of UV-B-induced DNA damage and phenylpropanoid biosynthesis to mount a sunscreen effect and their transcriptional regulators (see Supplementary Tables S1, S2 and S3). Most importantly, these data strongly indicate that almost all genes of the postulated UV-B photoreceptor-specific regulatory pathway(s) are dependent on functional UVR8 and COP1 proteins, supporting their major role.
Overexpression of UVR8 results in an enhanced UV-B photomorphogenic response
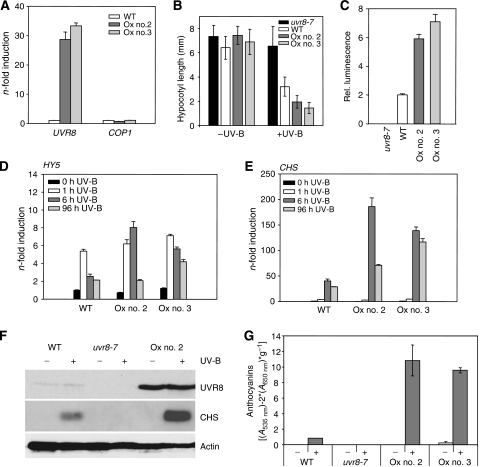
To determine whether UVR8 protein is a rate-limiting factor in the Arabidopsis UV-B response, we generated transgenic lines overexpressing UVR8 under the control of the constitutive strong CaMV35S promoter. Using western blot analysis, levels of UVR8 overexpression were estimated and two transgenic lines in which quantitative RT–PCR also detected an approximately 30-fold overexpression of UVR8 mRNA compared with wild type were used for detailed analysis (Figure 2A). In these lines, a marked UV-B photomorphogenic hypersensitivity was observed in all assays employed, including hypocotyl growth inhibition, HY5 and CHS gene activation, and anthocyanin accumulation (Figure 2B–G). Thus, we conclude that UVR8 has a rate-limiting function in the UV-B photomorphogenic pathway.
Figure 2.
UVR8 protein amount is rate limiting for UV-B-induced photomorphogenesis. (A) Quantitative RT–PCR data showing overexpression of UVR8 but no effect on COP1 expression in lines Ox nos. 2 and 3 compared with wild type. (B) Hypocotyl length measurements of 4-day-old seedlings grown with or without supplemental UV-B. Error bars represent s.d. (n=30). (C) Luciferase assays visualizing HY5 promoter activation in response to UV-B in UVR8 overexpression lines nos. 2 and 3 compared with wild type. Error bars represent s.e. (n=30). (D, E) Quantitative RT–PCR analysis of HY5 and CHS gene activation in response to UV-B in UVR8 overexpressor lines compared with wild type. (F) Immunoblot analysis of UVR8, CHS and actin (loading control) protein levels in 4-day-old seedlings grown with or without supplementary UV-B. (G) Anthocyanin accumulation of 4-day-old seedlings grown with or without supplemental UV-B. Error bars represent s.d. (n=3). (A–G) WT=Ws/ProHY5:Luc; Ox no. 2/no. 3=Pro35S:UVR8 in WT, lines 2 and 3.
Both COP1 and UVR8 are required for the UV-B photomorphogenic response
Using quantitative RT–PCR assays, we found no detectable UV-B-mediated early activation of the endogenous HY5 and CHS genes in cop1 and uvr8 mutants (Figure 3A and B). However, it is of note that uvr8 mutants do not show any constitutively photomorphogenic phenotype, indicating normal function of COP1. Reciprocally, to analyse the UVR8 protein levels in cop1 mutants, we have generated polyclonal antibodies against a specific C-terminal peptide of UVR8. The antibody detects a single band (about 47 kDa) in wild-type cell extracts that corresponds to the expected size of the UVR8 protein (440 amino acids with predicted mass 47 kDa) and this is absent in the uvr8-6-null mutant. Importantly, levels of UVR8 protein are comparable in cop1-4, hy5-215 mutant and wild-type seedlings (Figure 3C), thereby excluding an indirect cause of their previously described UV-B phenotypes (Ulm et al, 2004; Oravecz et al, 2006). In addition, we conclude that COP1 does not affect UVR8 protein levels under standard growth conditions. Moreover, chromatin immunoprecipitation showed that UVR8 associates with the HY5 promoter region independent of COP1 (Supplementary Figure S3A).
Figure 3.
UV-B-induced HY5 and CHS gene activation strictly requires UVR8 and COP1. (A, B) Quantitative RT–PCR of HY5 and CHS gene activation in response to UV-B in cop1-4 and uvr8-6 compared with wild type (Col). Error bars represent s.d. of triplicate. (C) Immunoblot analysis with anti-UVR8 and anti-actin (loading control) antibodies on protein extracts from 4-day-old mutant and wild-type seedlings.
The total absence of a UV-B regulatory response, for example, in HY5 and CHS gene activation, indicates that the COP1 and the UVR8 proteins function in the same genetic pathway. We thus hypothesized that COP1 and UVR8 might function together in the UV-B photomorphogenic signalling pathway.
UVR8 and COP1 colocalize and interact directly in a UV-B-dependent manner
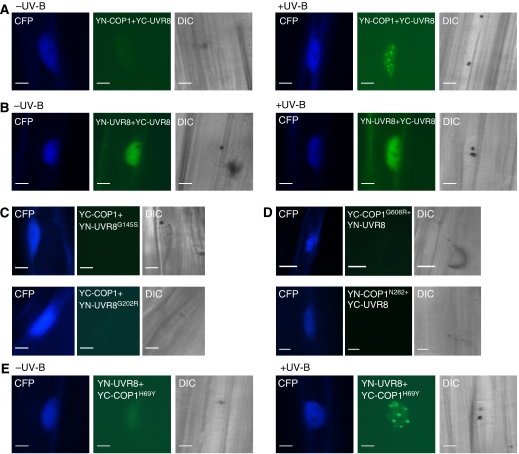
To investigate whether COP1 and UVR8 proteins interact, we made use of a transient expression system in mustard (Sinapis alba), a plant with a well-established photomorphogenic response (Stolpe et al, 2005, and references therein) (Supplementary Figure S4A). We generated expression constructs of YFP–COP1 and CFP–UVR8 and delivered the corresponding plasmids into mustard hypocotyls by biolistic gene transfer. Under standard conditions without UV-B, YFP–COP1 localized to nuclear bodies in mustard hypocotyl cells (Supplementary Figure S4B), as described before for onion epidermal cells (e.g. Ang et al, 1998). In contrast, CFP–UVR8 is detected as diffuse nuclear fluorescence in the same cells. However, when the co-bombarded plants were irradiated with UV-B, also CFP–UVR8 formed nuclear bodies that largely colocalized with YFP–COP1 (Supplementary Figure S4B). This indicates that CFP–UVR8 was recruited into YFP–COP1 nuclear bodies in a UV-B-dependent manner and that these two proteins might reside in the same protein complex under UV-B specifically.
To investigate whether UVR8 and COP1 are indeed directly interacting under UV-B, we used the bimolecular fluorescent complementation (BiFC) assay (Kerppola, 2006). By using this assay, we could clearly identify reconstitution of a functional YFP signal from the complementary ‘split YFP' parts attached to the UVR8 and COP1 proteins. However, similar to the colocalization, the direct interaction of UVR8 and COP1 was again UV-B dependent (Figure 4A; Supplementary Figure S4C, right), as there was barely any YFP signal detectable when the supplementary UV-B was removed (Figure 4A; Supplementary Figure S4C, left). Importantly, we could not detect any YFP signal when empty vector controls were used in combination with YN-/YC-UVR8 and YN-/YC-COP1 (Supplementary Figure S4D). It is also of note that in sharp contrast to the UV-B-dependent interaction of COP1 with UVR8, interaction of UVR8 with itself was readily detectable independent of supplementary UV-B (Figure 4B).
Figure 4.
Wild-type UVR8 and COP1 proteins interact directly in a UV-B-dependent manner, but not the mutant versions that are impaired in UV-B signalling. (A) Direct interaction of YN-COP1 with YC-UVR8 under UV-B. (B) BiFC visualization of UVR8 dimerization independent of UV-B. (C) No interaction of mutant UVR8 proteins with wild-type COP1 under UV-B detectable by BiFC. (D) No interaction of mutant COP1 proteins with wild-type UVR8 under UV-B. (C, D) No YFP signal was detected in at least 20 CFP positive cells and in two independent repetitions. (E) Direct interaction of YN-UVR8 with YC-COP1H69Y under UV-B. (A–E) A Pro35S:CFP control plasmid was always co-bombarded to identify transformed cells prior to the analysis of YFP fluorescence. Specific CFP and YFP filter sets were used for microscopic analysis. DIC (differential interference contrast images) are shown. Bars=10 μm.
Single amino-acid changes in COP1 and UVR8 proteins impair UV-B signalling function and also abrogate direct interaction with their partner proteins
We further investigated whether mutant alleles of COP1 and UVR8 are still able to interact with their corresponding wild-type partner or whether COP1–UVR8 interaction correlates with a functional UV-B response. A number of uvr8 mutants express mutant UVR8 proteins at about wild-type level (Supplementary Figure S1B). This is of note as the mutants with single amino-acid changes in UVR8 displayed absence of the UV-B response, apparently identical to the null alleles (e.g. uvr8-1 and uvr8-6). Thus, we have tested interaction of UVR8G145S (corresponding to uvr8-15) and UVR8G202R (corresponding to uvr8-9) with wild-type COP1 and found that these non-functional UVR8 alleles were not capable of interacting with COP1 anymore (Figure 4C).
By using the COP1N282 (corresponding to cop1-4) truncation and COP1G608R (corresponding to cop1-19) protein, we found that it is the WD40 repeats of COP1 that are important for interaction with UVR8 (Figure 4D). In contrast to cop1-4 and cop1-19, the cop1eid6 mutant is still able to respond to UV-B (Oravecz et al, 2006), despite their comparable enhanced photomorphogenic phenotype in visible light (Dieterle et al, 2003). In agreement, we found that the corresponding COP1H69Y protein, mutated in a conserved histidine residue of the RING finger domain, still interacts with UVR8 under UV-B (Figure 4E). Thus, we conclude that functional UVR8 and COP1 are required for direct interaction with their wild-type partner protein.
YFP–COP1 and UVR8 are co-immunoprecipitated from UV-B-treated seedlings
To further investigate COP1–UVR8 interaction in planta, we performed co-immunoprecipitation experiments. To do this, we have generated transgenic lines constitutively expressing YFP-tagged COP1 in cop1-4 mutants, which led to complementation of the cop1-4 UV-B response (Oravecz et al, 2006). In agreement with the BiFC data, endogenous UVR8 protein was co-immunoprecipitated with YFP–COP1 from cop1-4/Pro35S:YFP-COP1 under UV-B specifically (Figure 5A). In contrast, no co-immunoprecipitation of UVR8 was found under conditions devoid of UV-B or from control plants not expressing YFP–COP1 (Figure 5A). Similarly, no protein cross-reacting with our anti-UVR8 antibodies was detected in the YFP control pull downs from plants expressing YFP–COP1 in a cop1 uvr8 double mutant background (cop1-4 uvr8-6/Pro35S:YFP-COP1) (Figure 5A). It should also be pointed out that YFP–COP1 protein levels are stabilized under UV-B and that this effect is dependent on the presence of UVR8 protein (Figure 5A). Notwithstanding this, we could detect co-immunoprecipitation of UVR8 with YFP–COP1 as early as 5 min after UV-B irradiation, when YFP–COP1 levels are not yet elevated (Figure 5B). Thus, we conclude that COP1 and UVR8 interact in vivo in a specific, rather rapid and UV-B-dependent manner.
Figure 5.
UV-B-dependent co-immunoprecipitation of UVR8 with YFP–COP1. (A) Co-immunoprecipitation of proteins using anti-YFP antibodies in extracts from wild-type (Col), cop1-4, cop1-4/Pro35S:YFP-COP1 and cop1-4 uvr8-6/Pro35S:YFP-COP1 transgenic seedlings. Here, 6-day-old seedlings were UV-B irradiated for 24 h (+UV-B) or mock treated under a cutoff filtering out UV-B (−UV-B). *A nonspecific cross-reacting band. (B) Early UV-B-dependent interaction detected by co-immunoprecipitation of UVR8 with YFP–COP1 from 5-day-old cop1-4/Pro35S:YFP-COP1 seedlings exposed to UV-B for the indicated times.
UV-B-induced photomorphogenesis is required for UV-B acclimation and survival in sunlight
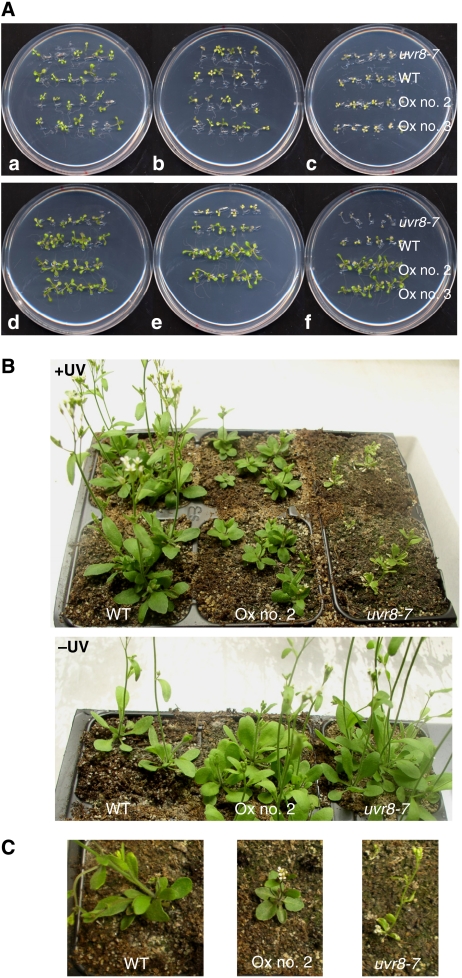
Altogether, our and published data predict an important role of the UVR8/COP1-mediated UV-B photomorphogenic pathway in UV-B acclimation and tolerance. To further support this notion and provide a physiological demonstration of UV-B acclimation, we combined weak narrowband UV-B exposure with subsequent broadband UV-B stress. Exposure of wild-type seedlings for 7 days to narrowband UV-B that activates photomorphogenic responses resulted in tolerance to a subsequent broadband UV-B stress treatment (Figure 6A). This acclimation effect was absent in uvr8 mutants and enhanced in UVR8 overexpressor lines (Figure 6A). Similarly, cop1-4 mutants were impaired in their acclimation response, whereas the cop1eid6 displayed higher UV-B stress tolerance after acclimation (Supplementary Figure S5). This is in good agreement with the previously demonstrated absence and presence of UV-B photomorphogenic response in cop1-4 and cop1eid6 alleles, respectively (Oravecz et al, 2006). Thus, weak photomorphogenic UV-B promotes plant survival under higher fluence rates of UV-B in a UVR8- and COP1-dependent manner.
Figure 6.
UVR8-dependent acclimation to UV-B and its importance for survival under simulated sunlight. (A) Arabidopsis seedlings were grown for 7 days under white light (a, b, c; non-acclimated) or white light supplemented with narrowband UV-B (d, e, f; acclimated). Seedlings were then irradiated for 1 h (b, e) and 2 h (c, f) with broadband UV-B under a WG305 cutoff filter, or subjected to a 2 h mock treatment (a, d) under a WG345 filter (−UV-B). Treated seedlings were further grown for 7 days under standard conditions without UV-B before the picture was taken. (B) Here, 25-day-old plants grown in sunlight simulators under realistic conditions (+UV) or with the UV portion specifically filtered out (−UV). (C) Close up of 27-day-old single plants grown under +UV conditions. WT=Ws/ProHY5:Luc+, Ox no. 2/no. 3=Pro35S:UVR8 in WT, lines 2 and 3.
Moreover, to clarify the importance of the UV-B photomorphogenic pathway under natural conditions, we grew plants in sun simulators with a natural spectral balance throughout the ultraviolet to infrared spectrum (Thiel et al, 1996). Under these realistic conditions, uvr8 mutant plants were strongly affected by UV-B radiation. They displayed strong leaf curling and cell death, and were light green (Figure 6B). In contrast, UVR8 overexpressor lines were clearly tolerant to UV-B, but they were dwarf and dark green (Figure 6B and C). We conclude that a major role of the UV-B-induced photomorphogenic response is the acclimation of plants to finally establish UV-B tolerance, a role required for survival in sunlight.
COP1-mediated degradation of HY5 is inhibited under UV-B
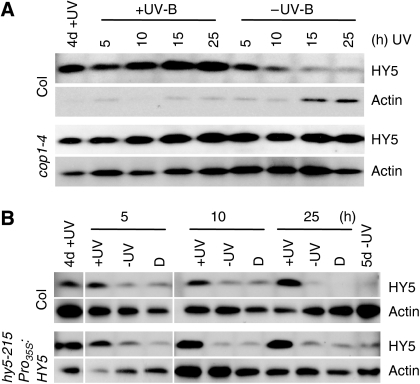
The dwarfed phenotype of UVR8 overexpression lines under UV-B closely resembles the cop1 mutant grown in light. This may be explained by a high cost of having an elevated UV-B photomorphogenic response or that UVR8 interaction results in COP1 inactivation. In agreement with the latter, we found that endogenous and constitutively expressed HY5 protein is stabilized under supplementary UV-B and is readily degraded under UV-B exclusion in a COP1-dependent manner (Figure 7). Thus, our data indicate that part of the UV-B signalling mechanism includes COP1 inactivation.
Figure 7.
COP1-mediated degradation of HY5 is inhibited under UV-B. (A) WT (Col) and cop1-4 mutant seedlings were grown for 4 days under white light supplemented with narrowband UV-B (4d+UV). Then the seedlings were either left under UV-B or the cutoff filter was exchanged to filter out UV-B (−UV-B) for 5, 10, 15 and 25 h before samples were analysed. (B) WT (Col) and a complemented hy5 transgenic line constitutively expressing HY5 (hy5-215/Pro35S:HY5) were grown for 4 days under white light supplemented with narrowband UV-B (4d+UV). Then the seedlings were either left under UV-B (+UV), UV-B was filtered out (−UV) or they were transferred to darkness (D) for the indicated times. (A, B) Protein gel blots were sequentially probed with anti-HY5 and anti-actin (loading control) antibodies.
Discussion
Plants are inevitably exposed to UV-B radiation in sunlight due to their sessile lifestyle and their need to capture light to fuel photosynthesis. Nonetheless, plants are well protected in nature and ‘sunburns' are seldom observed. This study demonstrates that (i) COP1 and UVR8 proteins are absolutely required for the UV-B photoregulatory pathway, (ii) UV-B induces direct interaction of UVR8 with COP1 in planta, (iii) UVR8 levels are rate limiting in this process, and that (iv) UV-B-induced photomorphogenesis is essential in establishing UV-B acclimation and tolerance under realistic climatic conditions. Our observations thus place COP1–UVR8 interaction as a very early event in the UV-B regulatory network responsible for conferring UV-B protection.
COP1 is a multifunctional protein that was initially identified as a repressor of photomorphogenesis (Yi and Deng, 2005). Recent work has indicated a promotive role in a response mediated by phytochrome B (Boccalandro et al, 2004) and has extended the functions of COP1 beyond seedling photomorphogenesis, including the regulation of flowering and regulation of stomatal opening (Mao et al, 2005; Jang et al, 2008; Liu et al, 2008). Our previous work also demonstrated a novel function of COP1 in UV-B photomorphogenesis that is mechanistically different from its repressor function in visible light (Oravecz et al, 2006). Moreover, recent data have provided more information about the function and regulation of COP1 in mammalian systems (Yi and Deng, 2005). Accordingly, it was shown in human cell lines that huCOP1 is a negative regulator of p53 and that DNA damage evoked by ionizing radiation induces an ATM-dependent phosphorylation of huCOP1 at Ser387, followed by its nuclear exclusion, rapid autoubiquitination and degradation (Dornan et al, 2006). In contrast, we showed that the Arabidopsis COP1 is stabilized and enriched in the nucleus under narrowband UV-B irradiation (Figure 5A and B; Oravecz et al, 2006) and existence of an Arabidopsis COP1 residue corresponding to Ser387 is not apparent by sequence analysis. Nonetheless, a cross-talk of the UV-B photomorphogenic pathway with DNA damage responses mediated by COP1 and ATM/ATR proteins might still exist in Arabidopsis. Interestingly, however, in human cell lines UV-B also induces dissociation of COP1 and thus stabilization of the bZIP transcription factor c-Jun (Yi et al, 2005; Savio et al, 2008). The inhibition of COP1-mediated degradation and thus stabilization of HY5 under UV-B in Arabidopsis seems related.
In contrast to the COP1 protein, the UVR8 protein is specifically involved in UV-B photomorphogenic responses, even though non-UV-B-related functions may still be discovered under conditions not tested so far. The UVR8 protein shows sequence similarity to the human regulator of chromatin condensation 1 (RCC1) (Kliebenstein et al, 2002), a GEF for the small GTP-binding protein Ran with important roles in nucleo-cytoplasmic transport, mitosis and nuclear envelope assembly (Hetzer et al, 2002). The three-dimensional structure of RCC1 revealed a seven-bladed β-propeller, of which the blades consist of seven homologous repeats of 51–68 amino-acid residues that are different from the WD40 β-propeller motif (Renault et al, 1998). This structure is very likely to be conserved in Arabidopsis UVR8 proteins, where a majority of the relevant residues are conserved (Kliebenstein et al, 2002). Similar to RCC1, nuclear localized UVR8 is associated with chromatin through histones (Cloix and Jenkins, 2008). Notwithstanding these features, several evidences indicate that UVR8 is not an RCC1 orthologue. For example, UVR8 is predominantly localized to cytoplasm (Kaiserli and Jenkins, 2007), and seems not to interact with Arabidopsis Ran proteins in directed yeast two-hybrid assays nor does it have substantial GEF activity (Brown et al, 2005). Moreover, in contrast to lethality or highly pleiotropic effects of RCC1 mutations in fungi and other species, the uvr8 mutants display no visible effect on standard growth and development, except in the presence of UV-B.
COP1 interaction with UVR8 seems crucial for the UV-B photomorphogenic pathway. We emphasize that this light response of COP1 occurs in the range of minutes, much faster than any of the presently known reactions of COP1, including nucleo-cytoplasmic trafficking in the range of 12–24 h (e.g. von Arnim and Deng, 1994; Oravecz et al, 2006). This is more in agreement with rapid effects of UV-B and other light qualities on gene expression and stabilization of COP1 target proteins (Duek et al, 2004; Yang et al, 2005). Our data suggest that UVR8 provides UV-B-specific signalling function to the multifunctional COP1 protein, which is necessary to relay the UV-B signal. In agreement, we provide evidence that mutations of UVR8 and COP1 residues hampered both protein–protein interaction with their wild-type partner and UV-B response, indicating that these processes are intimately connected.
Low levels of UV-B stimulate transcription of genes, among which many are involved in UV-B protection. A previous report using microarray analysis identified 639 genes induced by broadband UV-B in mature wild-type plants, with a majority of those normally induced in the uvr8-1 mutant (namely 567 genes; Brown et al, 2005). This initial analysis indicated that 72 (i.e. 11%) of the UV-B-induced genes depend on UVR8 protein. A subsequent report using several selected marker genes in RT–PCR experiments suggested that a large portion of these UVR8-independent genes are actually output of non-UV-B-specific signalling pathways, including those involved in UV-B stress (Brown and Jenkins, 2008). Similarly, our previous microarray analysis under broadband UV-B indicated that approximately 31 and 75% of the UV-B-induced genes depend on HY5 and COP1, respectively (Oravecz et al, 2006). Taken together, it is likely that in these assays, broadband UV-B may have activated to some extent both UV-B photomorphogenic and stress pathways. The gene profiling data presented here, using supplementary narrowband UV-B irradiation, clarify this issue. Under these conditions, specific for activation of UV-B photomorphogenesis, expression of the vast majority of the early UV-B-regulated genes depends on COP1 (namely 99.5% of the upregulated and 100% of the downregulated genes at 1 h; 99.8 and 99.4% at 6 h) and UVR8 (98.6 and 100% at 1 h; 99.6 and 99.4% at 6 h). These data strongly indicate that (i) both COP1 and UVR8 are of utmost importance to the UV-B photoregulatory response, and that (ii) we have established UV-B irradiation conditions that specifically activate the UV-B photoreceptor pathway. The uvr8 mutant is instrumental in differentiating the UV-B stress and non-stress pathways, best illustrated by the fact that uvr8 mutants are UV-B stress hypersensitive (most obvious under conditions involving UV-B acclimation) and, reciprocally, are hyposensitive to UV-B as an informational signal. Thus, uvr8 mutants can be used as a genetic tool for detailed analysis of the postulated UV-B photoreceptor pathway. We have already taken advantage of this feature and provide unequivocal evidence for a UV-B photomorphogenic transcriptome and the necessity of a functional UV-B photomorphogenic response for UV-B acclimation and survival in sunlight. It is of note here that UV-B acclimation has interesting parallels with other acclimation processes in plants, such as cold acclimation and freezing tolerance (e.g. Penfield, 2008). Notwithstanding this, we could also show that the UV-B-induced hypocotyl growth inhibition is a bona fide UV-B photomorphogenic response. It is absent in uvr8 mutants but not affected by phytochrome, cryptochrome, phototropin photoreceptor, and uvr2 and uvr3 photolyase mutants (Oravecz et al, 2006 and data not shown). Thus, we conclude that in uvr8, as well as cop1 mutants, the UV-B photoreceptor pathway is non-functional.
Downstream of COP1 and UVR8, the bZIP transcription factors HYH and particularly HY5 have a prominent role in UV-B signalling. Accordingly, it was shown that HY5-dependent genes are also dependent on UVR8 and COP1 under broadband UV-B (Brown et al, 2005; Oravecz et al, 2006; Brown and Jenkins, 2008). Moreover, the association of UVR8 with chromatin in the HY5 promoter region indicates a function of UVR8 and COP1 close to HY5 gene transcription, but the exact mechanism of action remains to be determined. However, we could not detect chromatin association of GFP–COP1 with the HY5 promoter region, nor was the interaction of UVR8 with chromatin disrupted in cop1-4 mutants (Supplementary Figure S3). These data indicate that, even though functional COP1 is required for UVR8 function in response to UV-B, it is not at the level of chromatin association and may not involve chromatin association of COP1 protein itself. Moreover, UVR8 protein levels in cop1 mutants are similar to wild-type levels, indicating that COP1 is not targeting UVR8 for proteasomal degradation. In agreement, UVR8 protein levels were found to be unaffected by different light qualities tested, including UV-B (Kaiserli and Jenkins, 2007).
Regarding our understanding of UV-B perception and signalling, the rather rapid UV-B-dependent COP1–UVR8 interaction provides an important mechanistic link between the two major players. We propose that the interaction with UVR8 specifies COP1 function under UV-B through adjusting its substrate specificity (Figure 8). Part of the interaction of UVR8 with COP1 under extended UV-B might include taking out COP1 from phytochrome/cryptochrome signalling, as indicated by the cop1-like phenotype of UVR8 overexpressor lines under UV-B. Notwithstanding this, absence of the UV-B photoregulatory response suggests a very early and crucial function of UVR8 and COP1 proteins closely linked to UV-B photoreceptor activity or signal transmission. The interaction of UVR8 with COP1, its rapid UV-B-dependent nuclear accumulation and requirement on UV-B radiation for function is reminiscent of properties of known photoreceptors. Thus, a function of UVR8 as UV-B photoreceptor cannot be excluded at present, a notion that deserves further investigation.
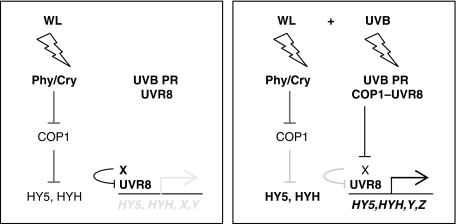
Figure 8.
Working model of COP1 and UVR8 function in the UV-B photoregulatory pathway. Left panel: under white light (WL), active photoreceptors partially inhibit COP1, which balances the response by repressing light signalling through degradation of HY5, HYH and other positive regulators of photomorphogenesis. A portion of UVR8 is constitutively associated with chromatin, for example, at the HY5 promoter region. A yet unidentified protein X represses HY5 transcription, possibly through keeping UVR8 inactive. Right panel: under supplementary UV-B (WL+UV-B), the specific perception by a UV-B photoreceptor (PR) results in rapid UVR8–COP1 interaction. This interaction is very closely linked to the UV-B PR function and confers UV-B-specific function to COP1, changing its substrate specificity away from HY5/HYH and functionally related proteins towards repressor protein X. Degradation of X then allows UVR8-mediated activation of genes, including HY5, that confers UV acclimation and protection.
Materials and methods
Plant material and growth conditions
cop1-4, hy5-215 and uvr8-6 (SALK_033468) are in the Columbia ecotype (Col) (McNellis et al, 1994; Oyama et al, 1997; Alonso et al, 2003), cop1eid6 in Landsberg erecta (Ler) (Dieterle et al, 2003), uvr8 and cop1 mutants derived from the ProHY5:Luc+ genetic screen are in the Wassilewskija background (Ws). The uvr8-7 mutant was backcrossed at least five times to wild type. Plants were grown exactly as described previously (Ulm et al, 2004).
Generation of transgenic Arabidopsis lines
The Ws/ProHY5:Luc+ reporter and the cop1-4/Pro35S:YFP-COP1 were described before (Ulm et al, 2004; Oravecz et al, 2006).
The wild-type and mutant COP1- and UVR8-coding regions were cloned into pDONR207 and sequenced to check integrity of the cloned fragment. Gateway-based cloning was then used to insert the ORF into the binary destination vectors pB2GW7, pB7WGC2 and pB7WGY2 (Karimi et al, 2002). The constructs were verified by sequencing and Arabidopsis plants were transformed by Agrobacterium using the floral dip method (Clough and Bent, 1998). The resulting transgenic lines described in this study were genetically determined to have the transgene integrated at a single locus.
Irradiation conditions
Conditions for narrowband UV-B irradiation were exactly as described before (Oravecz et al, 2006): plants were grown under continuous irradiation in a white-light field with Osram L18W/30 tubes (3.6 μmol m−2 s−1; measured with a LI-250 Light Meter; LI-COR Biosciences) supplemented with Philips TL20W/01RS narrowband UV-B tubes (1.5 μmol m−2 s−1; measured with a VLX-3W Ultraviolet Light Meter equipped with a CX-312 sensor; Vilber Lourmat). The UV-B range was modulated by the use of 3-mm transmission cutoff filters of the WG series with half-maximal transmission at the indicated wavelength (WG305 and WG345; Schott Glaswerke). In general, seedlings were grown for 4 days under continuous light supplemented with UV-B under a 345-nm cutoff filter (−UV-B) or 305-nm cutoff filter (+UV-B). The 345-nm cutoff filters were exchanged after 4 days for a 305-nm cutoff at 1 and 6 h before harvesting, as indicated.
For UV-B stress treatments, broadband UV-B lamps (Philips TL40W/12RS) were used exactly as described previously (Ulm et al, 2004).
A sun simulator was used to study the plants' response under natural light and UV radiation conditions (Thiel et al, 1996). The daylight period was 14 h with a mean PAR of 800 μmol m−2 s−1 and 12 h UV-B irradiance, which was weighted with the generalized plant action spectrum, normalized at 300 nm (Caldwell, 1971), giving the biologically effective (BE) quantity UVBE 400 mW m−2. Controls were grown under UV exclusion. The temperature was 23 and 18°C during the day and night, respectively, with relative humidity kept at 60%.
Anthocyanin and hypocotyl measurement
Anthocyanins were extracted and quantified according to Noh and Spalding (1998). Hypocotyl growth inhibition was analysed as described before (Oravecz et al, 2006). Experiments were carried out in at least three independent biological repetitions.
Microarray analysis
Arabidopsis RNA was isolated with the Plant RNeasy Kit (Qiagen), according to the manufacturer's instructions. RNA quality control, cRNA synthesis and labelling, and ATH1 array hybridizations were performed by the NASC's International Affymetrix Service. Expression values were estimated from the arrays using the GC-RMA function within Genedata's Refiner 4.5 package. Expression values were quantile normalized and genes with a Wilcoxon signed rank detection P-value ≤0.04 in at least 66% of the condition replicates were considered to be expressed.
Analysis was performed in Genedata's Analyst 4.5 application. Genes were required to pass a one-way ANOVA (P<0.05) and a Tukey post hoc test. The highest Storay–Tibshirani Q-value observed in the data with these settings was 0.025 (Storey and Tibshirani, 2003). The lists were further reduced to emphasize the largest changes by applying an ad hoc 1.5-fold difference in medians. These are the lists reported.
The microarray data are deposited under accession number E-MEXP-1957 in the ArrayExpress database (www.ebi.ac.uk/microarray-as/ae/).
Quantitative real-time PCR
Arabidopsis total RNA was treated with DNaseI according to the manufacturer's specifications (Qiagen). Per PCR reaction, cDNA was synthesized from 50 ng RNA with random hexamers using the TaqMan Reverse Transcription Reagents Kit (Applied Biosystems). Quantitative RT–PCR was carried out in 96-well format using a 7300 Real-Time PCR System and TaqMan probes (Applied Biosystems). PCR reactions were performed using the ABsolute QPCR Rox Mix Kit following the manufacturer's instructions (ABgene). The gene-specific probes and primers were as follows: CHS (At5G13930) probe 6-FAM-TCGAGCGCGTGCGTTCTCTTCA-TAMRA with CHS_for (5′-CGTGTTGAGCGAGTATGGAAAC-3′) and CHS-rev (5′-TGACTTCCTCCTCATCTCGTCTAGT-3′); HY5 (At5g11260) probe 6-FAM-CTCTGCTCCACATTTG-MGB with HY5_for (5′-CAAGCAGCGAGAGGTCATCA-3′) and HY5_rev (5′-CATCGCTTTCAATTCCTTCTTTG-3′). cDNA concentrations were normalized to the 18S rRNA transcript levels as standard using the Eukaryotic 18S rRNA Kit (Applied Biosystems). Expression was determined in triplicate measurements.
Microscopy and bimolecular fluorescence complementation
The COP1 and UVR8 gene fragments were transferred into BiFC binary vectors (Walter et al, 2004), pE-SPYNE-GW and pE-SPYCE-GW, that were made Gateway compatible and kindly provided by Caroline Carsjens and Wolfgang Dröge-Laser (University of Göttingen). Transient transformation of mustard seedlings using the biolistic PDS-1000/He system (Bio-Rad) and BiFC assays were carried out according to Stolpe et al (2005). Microscopical analysis was performed as described before (Oravecz et al, 2006). Microscopy data were confirmed in at least three independent experiments.
Generation of antibodies, immunoprecipitation assays and protein gel blot analysis
Rabbit polyclonal antibodies were generated against a synthetic peptide derived from the UVR8 protein sequence (amino acids C+426–440: CGDISVPQTDVKRVRI) and were affinity purified against the peptide (Eurogentec).
For YFP–COP1 immunoprecipitation, protein extracts were incubated with monoclonal anti-GFP antibodies (Invitrogen) and protein A-agarose (Roche Applied Science) in extraction buffer EB (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 0.1% Igepal, 2 mM benzamidine, 10 mM β-mercaptoethanol, 1 mM PMSF, 1 mM TPCK, 10 μM leupeptine, 10 μM dichloroisocumarin, 1% (v/v) protease inhibitor cocktail for plant extracts (Sigma), 10 μM MG132) for 1 h at 4°C, and beads were washed three times in buffer EB.
For protein gel blot analysis, total cellular proteins (10 μg) or immunoprecipitates were separated by electrophoresis in 10% SDS–polyacrylamide gel and electrophoretically transferred to PVDF membrane according to the manufacturer's instructions (Bio-Rad). We used polyclonal anti-UVR8, anti-HY5 (Oravecz et al, 2006), anti-actin (Sigma), anti-CHS (Santa Cruz Biotechnology) and monoclonal anti-GFP (BAbCO) as primary antibodies, with horseradish peroxidase-conjugated protein A (Pierce) or anti-rabbit, anti-goat and anti-mouse immunoglobulins (Dako A/S) as secondary antibodies, as required. Signal detection was performed using the ECL Plus Western detection kit (GE Healthcare).
Supplementary Material
Supplementary Figures S1–S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Acknowledgments
We are grateful to Klaus Harter and Wolfgang Dröge-Laser for providing the BiFC vectors. The Nottingham Arabidopsis Stock Centre is acknowledged for providing SALK mutant lines and carrying out Affymetrix microarray hybridizations. We are thankful to Werner Heller, Christian Osseforth, Stefan Kircher and Eberhard Schäfer for their help and support. CC was supported by a UK Biotechnology and Biological Sciences Research Council grant to GIJ. FN was supported by the Scottish Universities Life Sciences Alliance, an HHMI International Scholarship and OTKA (60106). This study was funded by the Excellence Initiative of the German Federal and State Governments (EXC 294), the SFB 746 and the Emmy Noether Programme of the Deutsche Forschungsgemeinschaft (UL341/1-1) to RU.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Boccalandro HE, Rossi MC, Saijo Y, Deng XW, Casal JJ (2004) Promotion of photomorphogenesis by COP1. Plant Mol Biol 56: 905–915 [DOI] [PubMed] [Google Scholar]
- Brosche M, Strid A (2003) Molecular events following perception of ultraviolet-B radiation by plants. Physiol Plant 117: 1–10 [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM (1971) Solar UV irradiation and the growth and development of higher plants. In Photophysiology, Giese AC (ed) Vol. 6, pp 131–177. New York: Academic Press [Google Scholar]
- Caldwell MM, Bornman JF, Ballare CL, Flint SD, Kulandaivelu G (2007) Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors. Photochem Photobiol Sci 6: 252–266 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cloix C, Jenkins GI (2008) Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dieterle M, Büche C, Schäfer E, Kretsch T (2003) Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol 133: 1557–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D, Shimizu H, Mah A, Dudhela T, Eby M, O'Rourke K, Seshagiri S, Dixit VM (2006) ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D (2003) Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Gruss OJ, Mattaj IW (2002) The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol 4: E177–E184 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI, Brown BA (2007) UV-B perception and signal transduction. In Light and Plant Development, Annual Plant Reviews, Whitelam GC, Halliday KJ (eds) Vol. 30, pp 155–182. Oxford: Blackwell Publishing Ltd [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kerppola TK (2006) Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol 7: 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Misera S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Spalding EP (1998) Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiol 116: 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A (1998) The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392: 97–101 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio MG, Rotondo G, Maglie S, Rossetti G, Bender JR, Pardi R (2008) COP1D, an alternatively spliced constitutive photomorphogenic-1 (COP1) product, stabilizes UV stress-induced c-Jun through inhibition of full-length COP1. Oncogene 27: 2401–2411 [DOI] [PubMed] [Google Scholar]
- Stolpe T, Susslin C, Marrocco K, Nick P, Kretsch T, Kircher S (2005) In planta analysis of protein–protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel S, Döhring T, Köfferlein M, Kosak A, Martin P, Seidlitz HK (1996) A phytotron for plant stress research: how far can artificial lighting compare to natural sunlight? J Plant Physiol 148: 456–463 [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Nagy F (2005) Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8: 477–482 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yi C, Li S, Chen X, Wiemer EA, Wang J, Wei N, Deng XW (2005) Major vault protein, in concert with constitutively photomorphogenic 1, negatively regulates c-Jun-mediated activator protein 1 transcription in mammalian cells. Cancer Res 65: 5835–5840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3