Abstract
The p53 tumor suppressor continues to hold distinction as the most frequently mutated gene in human cancer. The ability of p53 to induce programmed cell death, or apoptosis, of cells exposed to environmental or oncogenic stress constitutes a major pathway whereby p53 exerts its tumor suppressor function. In the past decade we have discovered that p53 is not alone in its mission to destroy damaged or aberrantly proliferating cells: it has two homologues, p63 and p73, that in various cellular contexts and stresses contribute to this process. In this review, the mechanisms whereby p53, and in some cases p63 and p73, induce apoptosis are discussed. Whereas other reviews have focused more extensively on the contribution of individual p53-regulated genes to apoptosis induction by this protein, in this review we focus more on those factors that mediate the decision between growth arrest and apoptosis by p53, p63 and p73, and on the post-translational modifications and protein-protein interactions that influence this decision.
Keywords: p53, p63, p73, apoptosis, transcription, mitochondria
The p53 family
The tumor suppressor p53 is vital in maintaining cellular genomic integrity and controlled cell growth (Levine, 1997; Bargonetti and Manfredi, 2002; Fridman and Lowe, 2003). Loss or gain of p53 function results in the aberrant growth of cells. Hence, both the cellular expression and activity of p53 are tightly regulated. p53 protein has a very short half life and thus is usually present at extremely low levels within cells. In response to stress, DNA damaging agents, and chronic mitogenic stimulation, p53 is transiently stabilized and activated. Depending on cell type, cell environment and oncogenic alterations, p53 activation leads to inhibition of cell cycle progression, induction of senescence, differentiation, or apoptosis (Vousden and Lu, 2002). Over a decade after the identification of the tumor suppressor p53, two p53-related genes, p63 and p73, were identified (Kaghad et al., 1997; Schmale and Bamberger, 1997; Osada et al., 1998; Trink et al., 1998; Yang et al., 1998; Zeng et al., 2001). Like p53, both p63 and p73 possess an amino-terminal transactivation domain (TAD), a DNA binding domain (DBD), and a carboxyl-terminal oligomerization domain (OD) (Murray-Zmijewski et al., 2006). While p63 and p73 demonstrate relatively little homology with p53 in their TAD and OD, both share approximately 60% similarity with the p53 DBD, including conservation of essential DNA contact residues (DeYoung et al., 2007) (see Figure 1). This similarity allows p63 and p73 to regulate p53 target genes and, similar to p53, to induce cell cycle arrest and apoptosis. However, studies of knockout mice have demonstrated that, even though these proteins clearly share some activities with p53, each of these proteins also has functions that are very distinct. TP53 null mice are viable and develop normally (Donehower et al., 1992). In contrast, p63 knockout mice show severe developmental defects, including failure to develop limbs, skin and other epithelial tissue; these mice do not survive beyond a few days after birth (Mills et al., 1999). p73 knockout mice exhibit neuro-developmental (hippocampal dysgenesis and hydrocephalus) and inflammatory (chronic infections and excessive inflammation) defects (Yang et al., 2000).
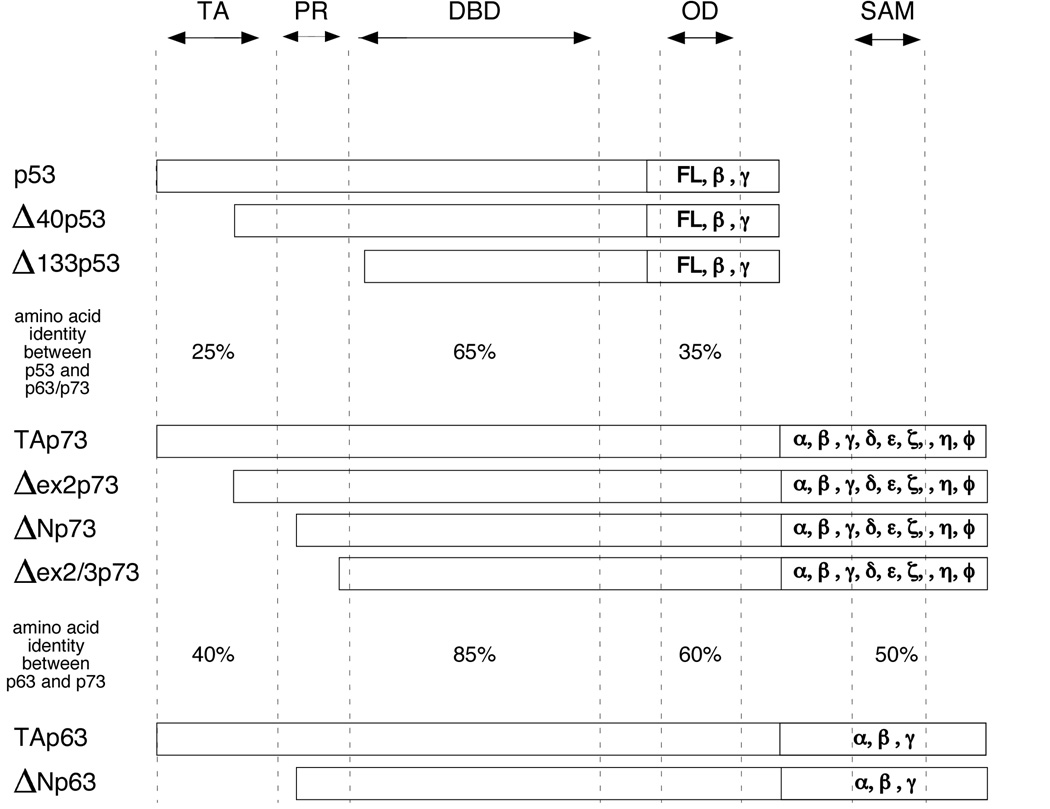
Figure 1. p53 family member isoforms.
Schematic presentation of p53, p63, and p73 isoforms. Approximate location of the transactivation (TA) domain, proline rich domain (PR), DNA binding domain (DBD), oligomerization domain (OD) and the sterile alpha motif (SAM) are indicated. The amino acid identity between the TA, DBD, and OD of p53 and p63/p73 as well as between p63 and p73 is denoted. Full length (FL) p53 or TAp63 and TAp73 protein are transcribed from a promoter located in the non-coding region of exon 1 (P1 promoter) of the p53, p63, and p73 gene. Δ133p53, ΔNp63, and ΔNp73 isoforms are generated by transcription from a promoter (P2 promoter) located in intron 3 of the p63 and p73 gene or intron 4 of the p53 gene. Further ΔN variants are generated by alternative splicing of the N-terminus (Δ40p53, Δex2p73, Δex2/3p73). Alternative splicing of the C-terminal region yields additional variants for p53 (FL, β, & γ), p63 (α, β, & γ), and p73(α, β, γ, δ, ε, ζ, η, & θ). p53 β and γ lack the oligomerization domain.
Isoforms of p53 family members
The p53, p63 and p73 genes are located on chromosomes 17 (17p13.1), 3 (3q27–29) and 1 (1q36), respectively, and all three genes are now known to express many differentially spliced isoforms (Muller et al., 2006; Murray-Zmijewski et al., 2006). All three genes encode two primary transcripts that are controlled by separate promoters (P1 and P2) (Figure 1). The P1 promoter of each gene is embedded in a non-coding region of exon 1. The P2 promoter of p63 and p73 is located in intron 3 while the P2 promoter of p53 is located in intron 4. Transcripts generated from the P1 promoter produce proteins that contain the TAD, the DBD, and the OD (TAp53, TAp63, and TAp73). In contrast, transcripts generated from the P2 promoter are missing the amino terminal TAD (Δ133p53, ΔNp63, and ΔNp73). These ΔN-terminal variants are generally regarded as dominant negative versions of p53 family members, as these can occupy promoter-binding sites but fail to transactivate gene expression. For both p53 and p73 additional ΔN variants can be generated by alternative splicing of the N-terminus or alternative initiation of translation (Δ40p53, Δex2p73, and Δex2/3p73) (Muller et al., 2006; Murray-Zmijewski et al., 2006). Additional complexity to p53 family member isoforms is added by virtue of the fact that P1 and P2 transcripts can be spliced at the C-terminus into several splice variants (α, β, γ, etc.). For p53, transcription from the P1 and P2 promoters and alternative splicing of intron 9 can generate at least six p53 isoforms, including three TA variants (p53, p53β & γ) and three ΔN variants (Δ133p53, Δ133p53β & γ). For p63 three TA variants (TAp63α, TAp63β, TAp63γ) and three ΔN variants (ΔNp63α, β & γ) have been identified. Finally, for p73, eight different isoforms from the p73 P1 promoter and P2 promoter elements have been described (TA and ΔN p73α, β, γ, δ, ε, ζ, η, & θ).
Tumor suppression by p53 family members
p53 can be found biallelically mutated or deleted in over 50% of human tumors, and there are p53 mutation databases containing a wealth of information in this regard (Olivier et al., 2002). Germline mutations in p53 are found in Li Fraumeni syndrome, a tumor-prone disorder predisposing affected individuals to tumors of the brain, breast, bone and adrenal cortex (Malkin et al., 1992). Finally, the p53-knockout mouse is predisposed to early onset tumors, chiefly thymic lymphoma, sarcoma, and testicular tumors (Donehower et al., 1992).
p63 and p73 map to two regions within the human genome that are often altered and deleted in cancers, respectively (Kaelin, 1999). This observation, combined with the observation that p63 and p73 mimic p53 action in tissue culture when over-expressed, suggests that p63 and p73 have tumor suppressive properties analogous to p53. However, characterization of these proteins as bona fide tumor suppressors has not been straightforward. Studies aimed at identifying mutations within the p63 and p73 gene in human cancers have demonstrated that, while p53 is mutated in 50% of all human cancers, p63 and p73 mutations occur rarely (Irwin and Kaelin, 2001; Melino et al., 2003; Deyoung et al., 2007). Rather, altered expression of p63 and p73 isoforms is more commonly observed (Muller et al., 2006). For example, p63 is upregulated in 80% of head and neck squamous cell carcinomas (HNSCCs), with ΔNp63 being the predominant isoform overexpressed (Weber et al., 2002; Sniezek et al., 2004; DeYoung et al., 2006; Rocco et al., 2006). ΔNp63 overexpression is observed in over 60 % of bladder carcinomas (Park et al., 2000), while loss of TAp63 expression occurs in bladder cancer and is associated with metastasis and poor prognosis (Urist et al., 2002; Koga et al., 2003). Similar to p63, altered expression of p73 occurs in a multitude of different cancers (Muller et al., 2006). Recent studies analyzing p73 isoform expression indicate that both TAp73 and ΔNp73 isoforms are upregulated in ovarian cancer and rhabdomyosarcomas (Concin et al., 2004; Cam et al., 2006), while exclusive upregulation of ΔNp73 isoforms can be observed in gliomas as well as carcinomas of the breast and colon (Dominguez et al., 2006; Wager et al., 2006). Down-regulation of TAp73 is found in lymphoblastic leukemias and Burkitt’s lymphoma as a result of p73 P1 promoter methylation (Corn et al., 1999; Kawano et al., 1999). The general, but by no means definitive, consensus appears to be that either methylation-mediated silencing of the entire gene, or upregulation of the dominant-negative ΔN variant, predominates in human tumors for p53 family members. How upregulation of specific ΔN variants occurs in human tumors is not presently understood.
Research on the predisposition of p63 and p73 knockout mice to tumor development has been conflicting and has added to the complexity of defining p63 and p73 as tumor suppressor proteins. p73 knockout mice do not develop spontaneous tumors (Yang et al., 2000) and heterozygous p63 mice are not tumor prone, nor do they develop tumors at an accelerated rate upon exposure to chemical carcinogens (Keyes et al., 2006). In fact, p63 deficiency was found to induce a cellular senescence program, increased aging, and shortened life span (Keyes et al., 2008). In contrast to these observations is a study examining tumor development in aging heterozygous p63 and p73 knockout mice. This study demonstrated that p63+/− and p73+/− mice develop spontaneous tumors, similar to p53+/− mice, with a median survival time of 10 months for p53+/−, 15 months for p63+/−, and 14 months for p73+/− (Flores et al., 2005). These latter data offer the firmest evidence that p63 and p73 have tumor suppressive properties analogous to p53.
Further evidence that p63 and p73 have tumor suppressor function comes from four independent observations: 1.) siRNAs specific for p63 and p73 enhance the transformation potential of p53 −/− mouse embryo fibroblasts (Lang et al., 2004); 2.) Both p63 and p73 can mediate chemo-sensitivity independent of p53 status by induction of apoptosis (Irwin et al., 2000; Bergamaschi et al., 2003a; Irwin et al., 2003; Lang et al., 2004; Gressner et al., 2005); 3.) Within tumor cells mutant p53 directly binds to both p63 and p73 via the DNA binding domain, rendering p63 and p73 impaired in their ability to induce growth suppression and apoptosis (Di Come et al., 1999; Marin et al., 2000; Gaiddon et al., 2001; Strano et al., 2002); and 4.) Silencing mutant p53 expression by siRNA-mediated knockdown sensitizes cells to anticancer agents (Bergamaschi et al., 2003a). These observations indicate that inactivation of p63 and p73 confers a growth advantage to tumor cells that have mutant forms of p53. Binding of mutant p53 to p73 is influenced by a polymorphism at codon 72, which encodes either arginine or proline. p53 mutants associated with the arginine polymorphic variant bind and inactivate p73 most efficiently (Marin et al., 2000; Bergamaschi et al., 2003a). Consistent with this observation, these p53 mutants demonstrate a less favorable response to chemotherapy (Bergamaschi et al., 2003a). Likewise, in osteosarcoma cells from p53R172H (equivalent to human hotspot mutant R175H) knock-in mice, mutant p53 co-immunoprecipitates and functionally inactivates p63 as well as p73, resulting in enhanced cellular transformation (Lang et al., 2004). Collectively, these observations suggest that p63 and p73 may complement, and in some situations may substitute for, the actions of p53. However, which effects p63 and p73 exert on cell proliferation and death may ultimately depend on the cellular context and genetic background, since p63 and p73 may not be required for the development and apoptosis of all cell types (Senoo et al., 2004).
Apoptosis by p53
Transcription-dependent apoptosis
The best understood activity of p53 is as a transcription factor that binds to the promoters and introns of target genes, and recruits the basal transcriptional machinery to activate expression of that gene. In 1992, a paper that searched for the consensus binding site for p53 using an unbiased affinity binding approach revealed that p53 bound best to two tandem repeats of the consensus 5’-Pu-Pu-Pu-C-(A/T)-(T/A)-G-Pyr-Pyr-Pyr-3’, where Pu is purine, and Pyr is pyrimidine, separated by 0–13 bases (el-Deiry et al., 1992). Since that time, literally hundreds of p53 target genes have been identified, and each contain one or more consensus sites in their 5’ promoter regions, introns, and in some cases, exons. These include p53AIP1, Apaf-1, BAX, Caspase-1, Caspase-6, cathepsin D, DINP1, DR4, DR5, Fas, IGF-BP3, NOXA, p85, PERP, PIDD, PIG3, PTEN, PUMA, Scotin, EI24/PIG8, as well as others (for review see Riley et al., 2008). Once bounds to the promoter regions of these genes, p53 recruits general transcription factors as well as histone acetyltransferases (HATs), such as CBP, p300 and PCAF. These HATs acetylate lysine residues of histones, thereby increasing the accessibility of chromatin to the transcriptional apparatus.
Two p53 target genes deserve specific mention as being particularly important in the apoptotic armamentarium of p53: these are PUMA and NOXA, BH-3 only members of the BCL-2 family. Mice lacking either PUMA or NOXA, or mice doubly-knocked out for both genes (PUMA/NOXA double knockout) develop normally and are not tumor-prone (Jeffers et al., 2003; Villunger et al., 2003; Michalak et al., 2008). The importance of PUMA to p53-dependent apoptosis is underscored by the fact that thymocytes and cells from the developing nervous system from the PUMA knockout mice are almost completely impaired for p53-dependent apoptosis, to levels equivalent to the p53 knock-out mouse (Jeffers et al., 2003). In contrast, apoptosis in other cell types of the PUMA knockout mouse is only partially impaired, and in these cells NOXA appears to contribute. For example, in mouse embryo fibroblasts, both PUMA and NOXA proteins play critical roles, as MEFs from the PUMA/NOXA double knock-out are most protected from cell death by etoposide, to levels similar to p53 knockout MEFs (Michalak et al., 2008). Finally, there are other cell types in the mouse where even the PUMA/NOXA double knockout mouse is not protected from apoptosis as well as the p53 knockout mouse; this includes mature T and B cells, which are partially but not completely protected by PUMA/NOXA loss (Michalak et al., 2008). These data firmly implicate PUMA and NOXA proteins as critical players in the p53 apoptotic program, at least in these cell types, but also suggest that the overall importance of individual p53 target genes in apoptosis is likely to be cell context and/or stress-specific.
Transcription-independent apoptosis by p53
Several studies, which have examined p53-dependent apoptosis in the presence of inhibitors of RNA and protein synthesis, indicate that p53 induces apoptosis not only by transcription-dependent, but also by transcription-independent mechanisms (Caelles et al., 1994; Wagner et al., 1994; Yan et al., 1997; Gao and Tsuchida, 1999). Moreover, deletion or mutation of the p53 transactivation domain, which is critical to the induction of p53 apoptotic target genes, does not eliminate the ability of p53 to induce apoptosis (Haupt et al., 1995). Likewise, a p53 deletion mutant containing only the first 214 N-terminal amino acid residues induces apoptosis and suppresses the transformation of rat embryo fibroblasts by several oncogenes, but it does not transactivate p53 target genes (Chen et al., 1996; Haupt et al., 1997).
Compelling evidence has accumulated over the last ten years indicating that p53 can directly activate components of the apoptotic machinery and that this involves translocation of p53 to the mitochondria. More specifically, addition of recombinant p53 to nuclear free cytosolic extract of irradiated cells leads to cytochrome C release, caspase activation, and apoptosis induction (Ding et al., 1998; Ding et al., 2000; Schuler et al., 2000). Therefore, activation of p53 in enucleated cytoplasts is sufficient to induce hallmarks of apoptosis. Immunofluorescence, electron microscopy and cell fractionation studies have demonstrated that in response to genotoxic stress, including DNA damage, hypoxia, and activated oncogenes, p53 rapidly translocates to the mitochondria (Marchenko et al., 2000; Mihara et al., 2003; Sansome et al., 2001; Nemajerova et al., 2005). Mitochondrial translocation of p53 precedes loss of mitochondrial membrane potential and caspase activation, suggesting that p53 may trigger mechanisms at the mitochondria that ultimately lead to induction of apoptosis (Marchenko et al., 2000). Indeed, mitochondrial translocation of p53 in irradiated mouse tissues triggers an early wave of caspase activation, which occurs long before the transcriptional program of p53 is activated (Erster et al., 2004). Notably, specific targeting of p53 to the mitochondria by fusion of the p53 N-terminus to the mitochondrial import leader peptide from ornithine transcarbamylase is sufficient to induce apoptosis, suppress colony formation, and to allow p53 to function effectively as a tumor suppressor in vivo (Marchenko et al., 2000; Dumont et al., 2003; Talos et al., 2005; Palacios et al., 2006).
The importance of p53 mitochondrial translocation in apoptosis is underscored by several observations. First, in cells or tissues that undergo cell cycle arrest (and not apoptosis), p53 mitochondrial translocation does not occur following exposure to genotoxic stress (Marchenko et al., 2000; Erster et al., 2004). Second, studies on the apoptotic potential of the proline and arginine p53 codon 72 polymorphic variants revealed that the proline codon 72 polymorphic variant, which is markedly impaired for apoptosis induction compared to the arginine codon 72 variant, does not translocate to the mitochondria as efficiently as the arginine variant (Dumont et al., 2003). In a mouse model for B cell lymphoma, p53 targeted exclusively to mitochondria is efficiently tumor suppressive in vivo (Talos et al., 2005). In contrast, an artificial p53 chimeric mutant that is capable of robustly transactivating pro-apoptotic p53 target genes is capable of inducing senescence, but incapable of inducing apoptosis in knock-in mouse embryo fibroblasts (Johnson et al., 2008). Moreover, the compound pifithrin μ, which selectively blocks p53’s mitochondrial function but leaves intact its transcriptional function, can completely block p53-mediated apoptosis of irradiated thymocytes (Strom et al., 2006). The apoptotic defect in thymocytes from the PUMA knockout mouse, once heralded as the single most important piece of evidence regarding p53’s transcriptional role in cell death, can now clearly be ascribed to a defect in both the transcriptional, and the direct mitochondrial, pathways of p53 (Chipuk et al., 2005). More recently the transcription-independent mitochondrial pathway of p53-mediated apoptosis has been shown to be particularly important for the apoptosis of embryonic stem cells. Specifically, reactive oxygen species, which efficiently induce apoptosis in embryonic stem cells, mediate apoptosis by inducing mitochondrial trafficking of p53, in a manner that is regulated by the histone deacetylase SIRT1 (Han et al., 2008).
Several pieces of evidence demonstrate that p53 regulates transcription-independent apoptosis at the mitochondria by regulating the activity of BCL-2 family members. Proteins of this family are key regulators of the intrinsic pathway of apoptosis (Cory and Adams, 2002). The main mechanism by which BCL-2 family members initiate or prevent apoptosis is by controlling release of cytochrome C and other pro-apoptotic factors from the mitochondria. The BCL-2 family of proteins is broadly divided into two groups that contain pro-apoptotic and anti-apoptotic members. Pro-apoptotic proteins within this family include BAK and BAX and the BH3-only proteins BID, BIM, BAD, NOXA, and PUMA. BCL-2, BCL-xl, and MCL-1 are anti-apoptotic family members, which bind to pro-apoptotic BCL-2 family members and thereby impair the apoptotic activity of these proteins. Among the pro-apoptotic family members, BH3-only proteins function as sensors of apoptotic signaling, and the key role of these proteins is to activate the main pro-apoptotic effector proteins BAK and BAX (Wei et al., 2001). BH3-only proteins are divided into “activator” and “de-repressor” proteins (Letai et al., 2002; Kuwana et al., 2005). In particular, a subset of BH3-only proteins directly activates BAK and BAX by inducing a conformational change in these proteins, allowing BAK and BAX to form homo-oligomers. These oligomers are thought to serve as channels within the outer mitochondrial membrane through which apoptotic factors are released. The second set of BH3-only proteins indirectly activates (derepresses) BAX and BAK by binding to BCL-2 and/or BCL-xl and thereby preventing these proteins from binding to and inhibiting the action of activator BH3-only proteins and the effecter proteins BAK and BAX.
Upon translocation to the mitochondria p53 seems to function analogous to BH3-only members. Specifically, p53 binds to pro- and anti-apoptotic BCL-2 family proteins and modulates their function. p53 physically interacts with the effector protein BAK, causes release of BAK from the inhibitory protein MCL-1 and induces a conformational change in BAK which results in oligomerization of this protein and subsequent cytochrome C release from mitochondria (Leu et al., 2004). Conserved residues within the DNA binding domain, located in the H2 α–helix and the L1 and L3 loop mediate BAK oligomerization (Pietsch et al., 2008). Significantly, tumor-derived mutations in the DNA binding domain of p53 render this protein unable to induce cytochrome C release and apoptosis (Pietsch et al., 2008). Although a physical interaction between BAX and p53 cannot be detected, p53 also induces BAX oligomerization and cytochrome C release (Chipuk et al., 2004). Specifically, p53 releases pro-apoptotic BAX and the BH3-only protein Bid from anti-apoptotic BCL-xl, and allows BAX to oligomerize (Chipuk et al., 2004).
In addition to interacting with pro-apoptotic BCL-2 family members, p53 also binds and inhibits the anti-apoptotic family members BCL-2 and BCL-xl (Mihara et al., 2003; Tomita et al., 2006). Molecular modeling studies predicted that the p53 DNA binding domain contacts BCL-xl via a domain encompassing the L3 loop (aa 239–248) and two flanking regions ranging from amino acid 135–141 and amino acid 173–187. NMR studies confirmed the involvement of these residues and pointed to additional residues within the H2 α–helix as important for p53-BCL-xl interaction (Petros et al., 2004). More recent NMR studies revealed that p53 utilizes very similar residues within the DNA binding domain to interact with BCL-2 (Tomita et al., 2006). Collectively, data from different laboratories indicate that p53 uses conserved residues within the DNA binding domain to contact BAK, BCL-2 and BCL-xl, although recent data suggest that slightly different contact regions of the p53 DNA binding domain are involved (Sot et al., 2007). One consistency in all of the combined studies, however, has been that tumor-derived mutants of p53 uniformly fail to induce transcription-independent cell death (Tomita et al., 2006; Pietsch et al 2008). This observation was unexpected since many of these mutations were thought to block only the DNA binding and transcriptional functions of p53.
Although most research with respect to p53 transcription-independent apoptosis has heavily focused on BCL-2 family members, it should be noted that p53 has transcription independent apoptotic functions unrelated to the BCL-2 family. For example, in the presence of inhibitors of RNA and protein synthesis, p53 causes the cell surface redistribution of the cytoplasmic death receptor Fas (CD95) by transport from the Golgi complex (Bennet et al., 1998). Notably, cells that contain inactivating mutations in Fas and Fas ligand have reduced p53-induced apoptosis compared to wild type cells.
Post-translational modifications that influence p53-mediated apoptosis
When cells encounter genotoxic stress, the levels of p53 protein rapidly increase. This correlates with a decrease in MDM2 catalyzed poly-ubiquitylation and an increase in a variety of other post-translational modifications (PTMs) including acetylation, phosphorylation, methylation, poly(ADP-ribosyl)ation, neddylation, sumoylation, and non-proteolytic monoubiquitylation (Bode and Dong, 2004; Toledo and Wahl, 2006; Olsson et al., 2007). These post-translational modifications are catalyzed by a large number of enzymes and they contribute to the activation of p53 through a variety of mechanisms. For example, p53 is phosphorylated on at least 18 different residues by over a dozen different kinases (Olsson et al., 2007). Most modifications are correlated with a generic increase in the ability of p53 to induce both cell cycle arrest and apoptosis. However, a small subset of these modifications are specifically linked to the apoptotic function of p53. Exploiting pathways that specifically enhance p53-mediated apoptosis has long been a goal of cancer therapeutics. Some of the better-understood examples from this subset will be discussed below.
Acetylation of Lysines 320 and 373 by PCAF and p300/CBP, respectively
The HATs p300, CBP and PCAF not only acetylate lysine residues in histones, they also acetylate p53 (Gu & Roeder, 1997; Liu et al., 1999; Sakaguchi et al., 1998). Interestingly, there is support for the premise that the specific HAT that gets recruited to p53 influences the outcome of p53 induction (growth arrest or apoptosis). More specifically, PCAF is the HAT that acetylates lysine 320 of p53 (Sakaguchi et al., 1998). Acetylation of this residue is often associated with cell cycle arrest, as mutating this residue to an unacetylatable arginine disrupts p53-mediated cell growth arrest but enhances p53-directed apoptosis (Barlev et al., 2001; Chao et al., 2006; Liu et al., 1999; Sakaguchi et al., 1998). Furthermore, acetylation at this residue allows for p53 to bind only at certain “high affinity” p53 binding sites, such as p21/waf1 (Di Stefano et al., 2005; Knights et al., 2006). In contrast, acetylation of lysine 373 of p53, catalyzed by p300 and/or CBP, leads to increased phosphorylation of N-terminal residues of p53, and markedly increased ability of this protein to transactivate lower affinity binding sites, such as those found at the pro-apoptotic target genes PIG3, Bax, and p53AIP1 (Knights et al., 2006). These results are most consistent with a model whereby acetylation of lysines 320 and 373 acts as a sensor system that enables p53 to choose between growth arrest and cell death, and to coordinate gene expression patterns appropriately following DNA damage.
Serine 46 phosphorylation
Among the PTMs specifically linked to apoptosis is phosphorylation of p53 at serine 46 (Ser46). Enhanced apoptosis occurs as a result of this phosphorylated form of p53 selectively activating pro-apoptotic target gene transcription (Oda et al., 2000; Rinaldo et al., 2007; Taira et al., 2007). For example, phosphorylation of p53-Ser46 is necessary for p53 to activate the transcription of the pro-apoptotic target gene p53AIP (Oda et al., 2000). Hints about the biochemical mechanism by which Ser46 phosphorylation might enhance the transcription of pro-apoptotic targets have come from studies of the prolyl isomerase PIN1. PIN1 binds proteins containing a motif that consists of a phosphorylated-serine or -threonine residue that is followed by a proline residue (Shaw, 2007). Ser46 within p53 is followed by a proline, and recently it was shown that binding of PIN1 to p53 following Ser46 phosphorylation promotes the dissociation of the apoptosis inhibitor iASPP from p53 (Mantovani et al., 2007). (The role of ASPP family proteins in p53-mediated apoptosis will be discussed below.) The isomerization of p53 by PIN1 has also been shown to facilitate the binding and acetylation of p53 by the acetyltransferase p300 (Mantovani et al., 2007).
There are two caveats regarding the importance of Ser46 phosphorylation in apoptosis. First, Ser46 is not conserved in mice. That being said, a large portion of the mouse p53 locus (corresponding to amino acids residues 33–332) can be replaced with the corresponding human p53 DNA sequence by homologous recombination (Hupki, Humanized p53 knock-in mouse) (Luo et al., 2001). This mouse/human chimera of p53 completely recapitulates the function of mouse p53, and Serine 46 is phosphorylated in mouse cells containing this protein (Feng et al., 2006). Further, mutation of Serine 46 to an alanine in the Hupki mouse (HupkiS46A) can inhibit p53-mediated apoptosis, though this inhibition is somewhat more modest than that seen in human cells (Feng et al., 2006). Second, the interaction with PIN1 cannot explain the totality of the effect of Ser46 phosphorylation on p53 function, because loss of PIN1 also reduces the ability of p53 to bind to the promoters of the non-apoptotic target genes p21 and hMDM2 (Zheng et al., 2002). Additional studies are clearly required before we have a better understanding of how Ser46 phosphorylation selectively regulates the ability of p53 to selectively transactivate pro-apoptotic genes and induce apoptosis.
Lysine 120 acetylation
Recent studies showing that the MYST family members TIP60 and hMOF acetylate p53 in the DNA binding domain at lysine 120 (Lys120) have provided additional insight regarding how p53 makes the decision between cell cycle arrest and apoptosis. These studies showed that mutation of Lys120 to a non-acetylatable arginine residue (K120R) diminishes the ability of p53 to induce apoptosis but does not affect p53-mediated cell cycle arrest (Sykes et al., 2006; Tang et al., 2006). Unlike most other amino acids in p53 that are substrates for PTMs, K120 is mutated in human cancer. These mutations, while rare, occasionally convert K120 to an arginine, thus conserving most of the biophysical properties at this site but eliminating acetylation. Significantly, loss of Lys120 acetylation prevents p53 from up-regulating the expression of target genes important for apoptosis, but not growth arrest. Additionally, chromatin immunoprecipitation studies indicate that the acetylated form at Lys120 appears to be selectively recruited to the promoters of pro-apoptotic p53 target genes, as opposed to those involved in cell cycle arrest (Sykes et al., 2006; Tang et al., 2006).
While S46 phosphorylation and K120 acetylation appear to enhance apoptosis by allowing p53 to selectively enhance the transcription of pro-apoptotic targets, other PTMs that modulate the apoptotic potential of p53 influence the transcription-independent apoptosis pathway.
Mono- versus poly-ubiquitylation
The addition of poly-ubiquitin chains to a protein commonly results in proteasome-mediated destruction. On the contrary, mono-ubiquitylation often acts as a protein modification that affects protein function without altering protein stability (Welchman et al., 2005). Consistent with this premise, while p53 poly-ubiquitylation acts as a protein destruction signal, mono-ubiquitylation of p53 promotes nuclear export and mitochondrial localization (Li et al., 2003; Marchenko et al., 2007). Interestingly, once targeted to the mitochondria by monoubiquitylation, the ubiquitin moiety must be removed before p53 can trigger transcription-independent apoptosis (Marchenko et al., 2007). At present it is unclear which residues of p53 must be ubiquitylated in order to target this protein to mitochondria. One group has found that the E3-ligase E4F1 catalyzes ubiquitylation of p53 at Lys319, 320 & 321 (Le Cam et al., 2006), and that ubiquitylation of these residues is associated with chromatin-bound p53, and specifically directs a p53-transcriptional program that promotes cell-cycle arrest and not apoptosis. Therefore, while it remains unclear what site(s) are mono-ubiquitylated in order to target p53 to the mitochondria, those residues targeted by E4F1 appear unlikely to be candidates.
While attractive, models that evoke selective PTM patterns to explain the ability of p53 to induce cell cycle arrest versus apoptosis tend to be plagued by considerable complexity. For example, lysine 320 of p53 (K320) is not only subject to acetylation, but can also be ubiquitylated or neddylated. Interestingly, for the Lys320 equivalent in mice, Lys317, mutation to an arginine leads to enhanced p53-mediated transcription of pro-apoptotic target genes, such as PIDD, PUMA, and NOXA (Chao et al., 2006). Correlating with enhanced target gene transcription, an increase in p53-mediated apoptosis in thymocytes, transformed MEFs, and intestinal crypt cells exposed to DNA damaging agents was also observed. These observations are consistent with a loss of E4F1-catalyzed ubiquitylation of human p53 at Lys320, which promotes p53 cell cycle arrest (Le Cam et al., 2006). Furthermore, FBXO11-mediated neddylation of human p53 at Lys320 represses p53 activity and therefore the observations made in p53K317R mice are also representative of a loss of neddylation (Abida et al., 2007). As discussed above, p53 transcriptional activity and growth arrest function is enhanced by acetylation at Lys320 by the acetyltransferase PCAF (Liu et al., 1999; Barlev et al., 2001). Thus, the increased apoptotic activity of mouse p53K317R may be explained by impaired acetylation, neddylation, or ubiquitylation at this residue. In addition to these caveats, it should be noted that PTMs on p53 may affect p53 function in a context-dependent manner, depending on tissue specificity and type of cellular stress.
Protein-protein interactions that influence p53-mediated apoptosis
Recent years have witnessed the identification of a number of p53-interacting proteins that direct either cell cycle arrest or apoptosis. These findings occur on a backdrop in which dozens and perhaps hundreds of partners for p53 have been identified. Here we will focus on the examples of Hzf, MUC1, and the Brn-3, ASPP and p63/p73 families.
Hematopoietic Zinc Finger protein
Hzf (Hematopoietic Zinc Finger) is a gene that is up-regulated in hematopoietic progenitor cells and is also a direct transcriptional target of p53 (Das et al., 2007). In mouse embryonic fibroblasts lacking Hzf (Hzf−/− MEFs), p53 cannot bind to the promoters and activate transcription of some growth arrest specific genes, such as p21 and 14-3-3 σ (Das et al., 2007). However, p53 displays enhanced promoter binding and transcription of the pro-apoptotic p53 target genes BAX, PUMA, NOXA and PERP in Hzf−/− MEFs. Furthermore, ectopic expression of Hzf enhances p21 and 14-3-3σ transcription in a p53-dependent manner but diminishes the ability of p53 to induce expression of BAX, PUMA, NOXA and PERP (Das et al., 2007). A model has thus been developed in which interaction between p53 and Hzf blocks apoptosis by inhibiting the transcription of the pro-apoptotic targets of p53.
MUC1
The trans-membrane protein, MUC1 (Mucin1) is another co-factor that selectively promotes p53-mediated cell cycle arrest, thereby inhibiting apoptosis (Wei et al., 2005). Upon proteolytic cleavage from the plasma membrane, the cytoplasmic domain of MUC1 (MUC1-CD) is targeted to the nucleus and mitochondria. In the nucleus, MUC1-CD binds and co-localizes with p53 at the p21/waf1 and BAX promoters. Depletion of MUC1 from human cells leads to an increase in p53-dependent BAX transcription and a decrease in p21/waf1 transcription (Wei et al., 2005). Furthermore, MUC1 attenuates BAX transcription by preventing loading of the pre21 initiation complex. Conversely, MUC1 cooperates with p53 to recruit transcriptional co-activators to the p21/waf1 promoter.
Brn3 family
The transcription factors Brn-3a and Brn-3b are important for proper neuronal development (Gan et al., 1996; Xiang et al., 1996). Although Brn-3a and Brn-3b share a domain of homology (POU domain) they have opposite effects on cell survival. Brn-3a supports cell survival while Brn-3b promotes cell death. Brn-3a can promote cell survival by inhibiting p53-mediated transcription of the pro-apoptotic genes BAX and NOXA (Hudson et al., 2005). Consistent with this, neurons derived from Brn-3a knockout mice display enhanced apoptosis. In contrast, Brn-3b −/− neurons are defective for BAX expression and p53-induced apoptosis, indicating that Brn-3b is important for p53 apoptotic activities in neuronal cells (Budhram-Mahadeo et al., 2006).
ASPP family
Some co-factors for p53 can specifically promote p53-mediated apoptosis. For example, three members of the ASPP (ankyrin repeats, SH3 domain, polyproline-rich domains) family of proteins ASPP1, ASPP2, and iASPP also influence the pro-apoptotic activity of p53. iASPP binds to the proline-rich and DNA-binding domains of p53 and inhibits the ability of p53 to induce transcription of BAX, as well as p53-dependent apoptosis (Bergamaschi et al., 2003b). Alternatively, ectopic expression of either ASPP1 or ASPP2 enhances p53-dependent apoptosis, and depletion of ASPP1 and/or ASPP2 suppresses the ability of p53 to induce apoptosis (Samuels-Lev et al., 2001). Mechanistically, ASPP1 and ASPP2 disrupt iASPP binding to p53, which results in increased p53 binding and transcription from the BAX promoter (Samuels-Lev et al., 2001; Bergamaschi et al., 2003b).
It is worth noting that the activity of p53 partners that specifically inhibit or enhance apoptosis may also be influenced by the selective PTMs discussed above. It is attractive to speculate that a given PTM might facilitate the binding of a specific partner whose activity is coupled to inhibiting or enhancing apoptosis.
p63 and apoptosis
Similar to p53, p63 protein levels increase upon treatment of cells with DNA damaging agents (Katoh et al., 2000; Okada et al., 2002; Bergamaschi et al., 2004; Petitjean et al., 2008). Increased p63 protein levels are not the result of transcriptional activation of the p63 gene, but most likely are due to post-translational modifications of this protein (Katoh et al., 2000; Okada et al., 2002). In addition, p63α, which contains an internal inhibitory domain in its C-terminal region, is cleaved by caspases at a non-classical caspase cleavage consensus sequence centered on amino acid 458, which enhances the apoptotic activity of this protein (Sayan et al., 2007).
p63-dependent apoptosis appears to be primarily regulated by transcriptional mechanisms and no transcription independent mechanisms have been identified to date. All p63 isoforms transcriptionally activate exogenously transfected promoters and endogenous mRNA or protein of pro-apoptotic p53 target genes (Katoh et al., 2000; Helton et al., 2008). p63 target genes include the death receptors CD-95, TNF-R and TRAIL-R, the pro-apoptotic Bc-2 family member BAX, the oxidoreductase PIG3, the tetraspan membrane protein PERP, the nuclear and mitochondrial localized apoptosis inducer RAD9, the apoptotic protease APAF1, and caspase-3, -8, and -9 (Gressner et al., 2005; Ihrie et al., 2005; Helton et al., 2008). When ectopically expressed in cells p63 isoforms induce apoptosis, although not as robustly as p53 or p73 (Dohn et al., 2001; Dietz et al., 2002). Evidence suggests that the transactivation domain (aa 1–59) and the proline rich domain (aa 67–127) of p63 are required for transactivation activity. Notably, deletion of the proline rich domain does not affect the ability of p63 to suppress growth, but does abolish apoptosis induction by p63 (Helton et al., 2008).
The regulatory mechanisms that drive p63-dependent apoptosis are not as well understood. A few post-translational modifications and protein interaction partners have been identified, and while there is some evidence that these affect p63 transactivation activity, it is not clear that these specifically influence the apoptosis function of this protein. Phosphorylation of p63 by as yet unknown proteins may have a role in regulating intracellular p63 protein levels. For example, treatment of cells with okadaic acid, a serine threonine phosphatase inhibitor, results in elevation of p63 protein levels, suggesting that phosphorylation by serine and threonine specific kinases may stabilize p63 protein (Okada et al., 2002). Additionally, upon treatment of cells with DNA damaging agents, p63α, p63β, and p63γ are phosphorylated on serine residues located between amino acids 160–162 (Petitjean et al., 2008). Thus far ASPP1 and 2 are the only p63 interacting proteins that specifically direct p63 activity towards apoptotic functions versus cell cycle regulatory functions (Bergamaschi et al., 2004). Both ASPP1 and 2 interact with p63, enhance transactivation of the BAX and PIG3 promoters, and increase the apoptotic activity of p63, while transactivation of p21/waf1 or MDM2 by p63 are not affected when ASPP1 and 2 are overexpressed.
p73 and apoptosis
p73 is activated in response to a variety of genotoxic agents, including DNA damaging agents and the oncogenes E1A and myc (Zaika et al., 2001). Once activated p73, like p53, can regulate induction of apoptosis and cell cycle arrest. p73-dependent apoptosis seems to be primarily regulated by its ability to transcriptionally activate pro-apoptotic p53 target genes. These include the BCL2 family members BAX, PUMA, NOXA, BAD and BIK, the oxidoreductase PIG3, the tetraspan membrane protein PERP, the death receptors CD95, TNFR1, TRAIL-R1 and TRAIL-R2, the mitochondrial membrane protein p53AIP1, as well as the caspases-3, -6 and -8 (Melino et al., 2002; Muller et al., 2005).
Evidence for a transcription independent role of p73 in apoptosis induction has been controversial. One report has described cleavage of p73 by caspase 3 and caspase 8 in response to DNA damaging agents and TRAIL (TNF-related apoptosis inducing factor), along with localization of both full length and caspase-cleaved p73 to the mitochondria. Importantly, a transactivation deficient p73 mutant was found to enhance TRAIL-induced apoptosis, and addition of recombinant p73 to purified mitochondria resulted in cytochrome C release (Sayan et al., 2008). These findings contrast with an earlier publication that reported that, although p73 induces BAX translocation from the cytosol to the mitochondria, p73 does not itself localize to the mitochondria, and translocation of BAX is not a direct effect of interaction with p73 (Melino et al., 2004). More research is clearly needed in this area to clarify this issue.
p73 post-translational modifications and protein-protein interactions
Induction of apoptosis by p73 in response to DNA damaging agents is intimately linked to its transcriptional activation by p53, along with post-translational modifications and interaction with transcriptional co-activators. In response to DNA damaging agents and oxidative stress p73 is transcriptionally activated by p53 (Chen et al., 2001; Wang et al., 2007). Additionally, DNA damage activates E2F1, which in turn transactivates p73 (Pediconi et al., 2003) and induces p73 to upregulate its own expression (Chen et al., 2001). Several post-translational modifications, including ubiquitylation, acetylation, phosphorylation, in addition to transcriptional co-activator recruitment, have been identified that regulate the apoptotic activity of p73. Similar to p53, HATs, Pin1, and ASPP family members all modify p73’s apoptotic function, and these are discussed briefly below. Unique to p73 are the apoptotic regulators c-Abl and YAP (Yes-associated protein).
c-Abl
The first kinase identified to phosphorylate p73 and enhance the transcriptional and apoptotic activity of this protein is the tyrosine kinase c-Abl (Gong et al., 1999; Agami et al., 1999; Yuan et al., 1999). c-Abl overexpression stabilizes p73 (Gong et al., 1999; Sanchez-Prieto et al., 2002), and p73 and c-Abl physically interact with each other via the SH3 domain of c-Abl and a PXXP motif of p73 located in the linker region between the DNA binding domain and the oligomerization domain (Agami et al., 1999; Yuan et al., 1999). DNA damage induces phosphorylation of p73 at tyrosine 99 (Agami et al., 1999; Yuan et al., 1999) in a c-Abl dependent manner, and p73 proteins mutated at tyrosine 99 are impaired in mediating an apoptotic response upon ionizing radiation treatment. In addition to c-Abl, p38MAPK phosphorylates p73 on threonine residues, and the enhanced transcriptional function of p73 by c-Abl is further enhanced by p38MAPK (Sanchez-Prieto et al., 2002).
YAP
Interaction with the Yes-associated protein (YAP) appears to be uniquely important for p73 function, as it is critical to p73 transactivation of apoptotic target genes and induction of apoptotic cell death (Strano et al., 2001). Physical interaction between YAP and p73 is mediated by the WW domain of YAP and a PPXY motif within p73 that serves as a target sequence for WW domains (Strano et al., 2001). Mutation of tyrosine 487 within the PPXY motif of p73 abolishes binding of p73 to YAP (Strano et al., 2001). YAP-p73 interaction prolongs p73 half-life and prevents proteasomal degradation of p73, by blocking binding of the E3 ubiquitin-like protein ligase Itch to p73 (Levy et al., 2007). Interestingly, YAP appears to function in integrating converging signaling modules to potentiate or diminish p73 transcriptional and apoptotic activity; ultimately, association of p73 with YAP appears to determine whether 73 exerts apoptotic or cell cycle regulatory properties. Ectopic expression of YAP together with p73 increases BAX protein levels within cells and augments p73-mediated transactivation of a BAX and p53AIP1 promoter luciferase reporter construct, but not a p21/waf1 promoter-driven luciferase reporter (Strano et al., 2001; Strano et al., 2005). Concomitantly, p73 and YAP co-expression potentiate p73-mediated apoptosis (Strano et al., 2005), while siRNA-mediated silencing of YAP attenuates DNA damage induced p73 dependent apoptosis (Strano et al., 2005; Basu et al., 2003; Levy et al., 2007). YAP also enhances acetylation of p73 by p300 and association between YAP and p73 is required for p300 recruitment to the p73 pro-apoptotic target gene p53AIP1 (Strano et al., 2005).
Intracellular YAP protein levels are regulated by the c-Abl tyrosine kinase (Levy et al., 2008). YAP and c-Abl physically interact and phosphorylation of YAP on tyrosine 357 by c-Abl occurs in a DNA damage dependent manner resulting in stabilization of the YAP protein. Notably, phosphorylation of YAP by c-Abl promotes interaction with p73 and association with the promoter region of pro-apoptotic proteins, but not cell cycle regulatory proteins (Strano et al., 2005; Levy et al., 2008). In contrast to the ability of c-Abl to promote stabilization and activation of YAP, Akt phosphorylates YAP on serine 127 and promotes cytoplasmic localization of YAP due to enhanced interaction of YAP with the cytosolic 14-3-3 scaffold proteins (Basu et al., 2003). Hence, active Akt impairs YAP from associating with p73, leading to attenuation of p73’s transactivation and apoptotic activity (Basu et al., 2003; Strano et al., 2005).
p300 and CBP
The CH1 domain (amino acid 350 to 450) of the transcriptional coactivators p300 and CREB-binding protein (CBP) interacts with the N-terminus of p73 (Zeng et al., 2000). p300 acetylates p73 on lysines 321, 327, and 331 and transactivation or induction of apoptosis by p73 is impaired in p300 or CBP deficient cells, suggesting an important role of both of these proteins in p73-dependent apoptosis (Zeng et al., 2000; Costanzo et al., 2002). The importance of acetylation in controlling the apoptotic activity of p73 is underscored by the fact that a p73 protein mutated at lysine residues 321, 327, and 331 is able to transactivate p21 and induce cell cycle arrest, but demonstrates a reduced apoptotic response and an inability to suppress colony formation (Costanzo et al., 2002). These data suggest that, as in the case for p53, acetylation of p73 fine-tunes its apoptotic function. Consistent with this premise, whereas wild type p73 is recruited to the promoter of the pro-apoptotic gene, p53AIP1, a non-acetylatable mutant of p73 does not bind the p53AIP1 promoter and is impaired in transactivating a p53AIP1 promoter driven luciferase reporter gene construct (Costanzo et al., 2002).
Pin1
Pin1, a peptidyl-prolyl cis/trans isomerase, stabilizes the p73 protein and enhances the transactivation and apoptotic activity of p73 (Mantovani et al., 2004). Pin1 and p73 interact in a c-Abl dependent manner and interaction requires phosphorylation of p73 on tyrosine 99. Additional phosphorylation by the p38MAPK seems to further enhance p73-Pin1 interaction. Pin1 augments binding of p73 to p300 and stimulates subsequent acetylation of p73 by p300. Similarly to YAP, Pin1 may be an essential factor in determining transactivation of genes encoding pro-apoptotic proteins by p73. Specifically, siRNA mediated downregulation of PIN1 abrogates the increases in the protein levels of PIG3 and BAX by p73 upon DNA damage while transactivation of the p21/waf1 gene is unaffected.
ASPP family
Apoptosis stimulating proteins of p53 1 and 2 (ASPP1 and ASPP2) affect p73 dependent apoptosis very similarly to p53 dependent apoptosis. ASPP1 and -2 both bind to p73 and augment p73 transactivation of BAX and PIG3 promoter driven luciferase reporter gene constructs, but fail to stimulate p73 transactivation of a p21/waf1 and MDM2 promoter driven luciferase reporter gene construct. Consistent with these findings, ASPP1 and -2 enhance the apoptotic activity of p73 (Bergamaschi et al., 2004).
Cooperation and antagonism between p53 family members in apoptosis induction and tumorigenesis
The majority of research with respect to p53 family members has focused on defining the individual activities of p53, p63, and p73 in apoptosis induction and have demonstrated that p73 and to a lesser extent p63 share similar activities with p53. A few research studies that examined the interplay between p53 family members in apoptosis induction and tumorigenesis have unveiled very interesting cooperative as well as antagonistic interactions between these family members. For example, both MEFs and neuronal cells derived from mice lacking p63 and p73 genes (p63−/−; p73−/−) are resistant to p53-mediated apoptosis (Flores et al. 2002). This defect results from the inability of p53 to bind to the promoters and activate transcription of the p53 pro-apoptotic target genes BAX, NOXA, and PERP in the absence of p63 and p73 expression. Notably, although p63−/−; p73−/− MEFs are resistant to apoptosis they are still competent for p53-induced growth arrest (Flores et al. 2002). Moreover, mice heterozygous for p53 and p63, or p53 and p73, display a more aggressive cancer phenotype (higher tumor burden and metastasis) compared to heterozygous p53 mice (Flores et al., 2005) again pointing to an important cooperative activity of p63 and p73 with p53. Further support for this notion comes from the observation that p73 is transcriptionally regulated by p53 under oxidative stress conditions and functions as an irreplaceable downstream target of p53 in controlling the cellular defense against stress (Wang et al., 2007).
While p53 family members cooperate with each other in apoptosis induction, members within this family can also negatively regulate each other’s functions. The transactivation domain-truncated isoforms of p53, p63, and p73 act in antagonizing the activity of full-length p53 and TA forms of p63 and p73. For example, ectopic expression of Δ133p53 impairs p53 apoptotic activity (Bourdon et al., 2005). Similar to p53 and p63, ΔNp73 overexpression down-regulates the transactivation activity of p53 and p73 and attenuates the apoptotic activity of p53 and p73 (Grob et al., 2001; Kartasheva et al., 2002), likely by direct competition for promoter binding (Stiewe et al., 2002). Interestingly, although ΔNp63 has been found to exert dominant negative effects over p53 and p63 (Yang et al., 1998; Westfall et al., 2003) two independent reports have demonstrated that ΔNp63 isoforms can also transactivate gene expression (Dohn et al., 2001; Wu et al., 2003).
Future Directions
Several of the most informative advances in the area of p53 family members and apoptosis have been made very recently. These include the identification of a role for p53 at the mitochondria and in transcription-independent apoptosis, the identification of apoptosis-specific partner proteins such as the ASPP family, and the characterization of PTMs that may help direct p53 toward apoptosis rather than induction of cell cycle arrest, such as Lysine 373 acetylation, Lysine 120 acetylation, and Serine 46 phosphorylation. How these PTMs and interacting proteins cooperate or compete with each other remains to be determined.
Given the prevalence of p53 pathway mutations in human cancer, strategies for rescuing the apoptotic activity of p53 and its family members have been long sought after. Recent studies in mice have now formally proven that restoration of p53 function in tumor cells lacking p53 can restore tumor suppression (Martins et al., 2006). A complete understanding of the role played by p53 in apoptosis in response to genotoxic stress is also of clinical importance because many traditional chemotherapeutic strategies rely on the induction of DNA damage and the subsequent activation of apoptosis. It is perhaps most remarkable that with over 45,000 publications related to p53, we are only now beginning to understand how this critical protein makes the decision between growth arrest and apoptosis. Among the central unresolved questions related to p53-mediated apoptosis that we envision are the following: How do different combinatorial patterns of PTMs influence p53 binding partner interactions, and how do these dictate the ability of p53 to induce apoptosis? At present, the relevance of many of these PTMs has been assessed individually, or perhaps in combination with one or a few others. Related to this, how do the p53 family members p63 and p73 contribute to apoptosis and what role do they play in human cancer? What is the link between p53-mediated apoptosis and its role in regulating autophagy and DNA repair? How are recent links between p53 and miRNA important in apoptosis?
Finally, the relative contributions of transcription-dependent and –independent pathways to apoptosis induction by p53 need to be resolved. There are now over sixty peer-reviewed manuscripts in the literature that report detection of p53 at mitochondria in apoptotic cells. Despite the breadth and depth of this literature, there remains skepticism in the field regarding this pathway. Certainly, the importance of the direct mitochondrial pathway of p53 will not be solidified until a mutant of p53 that can transactivate gene expression normally, but fails to traffic to mitochondria (or vice versa) is identified and characterized for its apoptotic function. However, this requirement exists on the backdrop of a protein for which there are few (or no) mutations that completely abrogate only one function: even the well-described 22/23 QS mutant of p53, once believed to be transcriptionally inactive, is now known to possess some transcriptional function, along with apoptotic ability (Johnson et al. 2005). Perhaps further analysis of the ancestral p53 family members can be a guide. For example, while it is known that C. elegans p53 can bind and oligomerize BAK (Pietsch et al., 2008), it is not clear if C. elegans p53 (or even Drosophila p53, for that matter) can localize to mitochondria, or interact with the BCL-2 family member CED-9. Such studies may help clarify the importance of transcription-independent apoptosis by p53. The very recent identification of the transcription-independent pathway makes it clear that we as a field need to be open minded and perhaps think more broadly about potential functions for p53.
Figure 2. Upstream mediators of p53 and p73-dependent cell death.
References
- Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003a;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, O'Neil NJ, Trigiante G, Crook T, Hsieh JK, et al. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet. 2003b;33:162–167. doi: 10.1038/ng1070. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol. 2004;24:1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhram-Mahadeo VS, Bowen S, Lee S, Perez-Sanchez C, Ensor E, Morris PJ, Latchman DS. Nucleic Acids Res. 2006;34:6640–6652. doi: 10.1093/nar/gkl878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10:281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y. Mol Cell Biol. 2006;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Chen X, Zheng Y, Zhu J, Jiang J, Wang J. p73 is transcriptionally regulated by DNA damage, p53, and p73. Oncogene. 2001;20:769–774. doi: 10.1038/sj.onc.1204149. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- Corn PG, Kuerbitz SJ, van Noesel MM, Esteller M, Compitello N, Baylin SB, et al. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt's lymphoma is associated with 5' CpG island methylation. Cancer Res. 1999;59:3352–3356. [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deissler H, Kafka A, Schuster E, Sauer G, Kreienberg R, Zeillinger R. Oncol Rep. 2004;11:1281–1286. [PubMed] [Google Scholar]
- DeYoung MP, Johannessen CM, Leong CO, Faquin W, Rocco JW, Ellisen LW. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz S, Rother K, Bamberger C, Schmale H, Mössner J, Engeland K. Differential regulation of transcription and induction of programmed cell death by human p53-family members p63 and p73. FEBS Lett. 2002;525:93–99. doi: 10.1016/s0014-5793(02)03093-4. [DOI] [PubMed] [Google Scholar]
- Ding HF, McGill G, Rowan S, Schmaltz C, Shimamura A, Fisher DE. Oncogene-dependent regulation of caspase activation by p53 protein in a cell-free system. J Biol Chem. 1998;273:28378–28383. doi: 10.1074/jbc.273.43.28378. [DOI] [PubMed] [Google Scholar]
- Ding HF, Lin YL, McGill G, Juo P, Zhu H, Blenis J, et al. Essential role for caspase-8 in transcription-independent apoptosis triggered by p53. J Biol Chem. 2000;275:38905–38911. doi: 10.1074/jbc.M004714200. [DOI] [PubMed] [Google Scholar]
- Di Stefano V, Soddu S, Sacchi A, D'Orazi G. Oncogene. 2005;24:5431–5442. doi: 10.1038/sj.onc.1208717. [DOI] [PubMed] [Google Scholar]
- Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–3205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- Domínguez G, García JM, Peña C, Silva J, García V, Martínez L, et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2006;24:805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hollstein M, Xu Y. Cell Cycle. 2006;5:2812–2819. doi: 10.4161/cc.5.23.3526. [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Tsuchida N. Activation of caspases in p53-induced transactivation-independent apoptosis. Jpn J Cancer Res. 1999;90:180–187. doi: 10.1111/j.1349-7006.1999.tb00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob TJ, Novak U, Maisse C, Barcaroli D, Lüthi AU, Pirnia F, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Tokuchi Y, Hayashi M, Kobayashi Y, Nishida K, Hayashi S, Ishikawa Y, Tsuchiya S, Nakagawa K, Hayashi J, Tsuchiya E. Cancer Res. 1999;59:5572–5577. [PubMed] [Google Scholar]
- Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Rowan S, Shaulian E, Kazaz A, Vousden K, Oren M. p53 mediated apoptosis in HeLa cells: transcription dependent and independent mechanisms. Leukemia. 1997;11:337–339. [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hayes VM, Dirven CM, Dam A, Verlind E, Molenaar WM, Mooij JJ, Hofstra RM, Buys CH. Brain Pathol. 1999;9:463–467. doi: 10.1111/j.1750-3639.1999.tb00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton ES, Zhang J, Chen X. The proline-rich domain in p63 is necessary for the transcriptional and apoptosis-inducing activities of TAp63. Oncogene. 2008;27:2843–2850. doi: 10.1038/sj.onc.1210948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS. J Biol Chem. 2005;280:11851–11858. doi: 10.1074/jbc.M408679200. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Irwin MS, Kaelin WG. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 2001;12:337–349. [PubMed] [Google Scholar]
- Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Meade K, Pathak N, Marques MR, Attardi LD. Knockin mice expressing a chimeric p53 protein reveal mechanistic differences in how p53 triggers apoptosis and senescence. Proc Natl Acad Sci U S A. 2008;105:1215–1220. doi: 10.1073/pnas.0706764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr The p53 gene family. Oncogene. 1999;18:7701–7705. doi: 10.1038/sj.onc.1202955. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Kartasheva NN, Contente A, Lenz-Stöppler C, Roth J, Dobbelstein M. p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncogene. 2002;21:4715–4727. doi: 10.1038/sj.onc.1205584. [DOI] [PubMed] [Google Scholar]
- Katoh I, Aisaki KI, Kurata SI, Ikawa S, Ikawa Y. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene. 2000;19:3126–3130. doi: 10.1038/sj.onc.1203644. [DOI] [PubMed] [Google Scholar]
- Kawano S, Miller CW, Gombart AF, Bartram CR, Matsuo Y, Asou H, et al. Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood. 1999;94:1113–1120. [PubMed] [Google Scholar]
- Keyes WM, Vogel H, Koster MI, Guo X, Qi Y, Petherbridge KM, et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc Natl Acad Sci U S A. 2006;103:8435–8440. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2008;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga F, Kawakami S, Fujii Y, Saito K, Ohtsuka Y, Iwai A, et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003;9:5501–5507. [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Leitao MM, Soslow RA, Baergen RN, Olvera N, Arroyo C, Boyd J. Gynecol Oncol. 2004;93:301–306. doi: 10.1016/j.ygyno.2004.01.043. [DOI] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007;14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- Malkin D, Jolly KW, Barbier N, Look AT, Friend SH, Gebhardt MC, et al. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. N Engl J Med. 1992;326:1309–1315. doi: 10.1056/NEJM199205143262002. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell. 2004;14:625–636. doi: 10.1016/j.molcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Sal GD. Nat Struct Mol Biol. 2007;14:912–920. doi: 10.1038/nsmb1306. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci. 2003;28:663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- Meyers FJ, Chi SG, Fishman JR, deVere White RW, Gumerlock PH. J Natl Cancer Inst. 1993;85:1856–1858. doi: 10.1093/jnci/85.22.1856. [DOI] [PubMed] [Google Scholar]
- Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Müller M, Schilling T, Sayan AE, Kairat A, Lorenz K, Schulze-Bergkamen H, et al. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–1577. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- Müller M, Schleithoff ES, Stremmel W, Melino G, Krammer PH, Schilling T. One, two, three--p53, p63, p73 and chemosensitivity. Drug Resist Updat. 2006;9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Nemajerova A, Wolff S, Petrenko O, Moll UM. Viral and cellular oncogenes induce rapid mitochondrial translocation of p53 in primary epithelial and endothelial cells early in apoptosis. FEBS Lett. 2005;579:6079–6083. doi: 10.1016/j.febslet.2005.09.074. [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Okada Y, Osada M, Kurata S, Sato S, Aisaki K, Kageyama Y, et al. p53 gene family p51(p63)-encoded, secondary transactivator p51B(TAp63alpha) occurs without forming an immunoprecipitable complex with MDM2, but responds to genotoxic stress by accumulation. Exp Cell Res. 2002;276:194–200. doi: 10.1006/excr.2002.5535. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Olsson A, Manzl C, Strasser A, Villunger A. Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- Palacios G, Moll UM. Mitochondrially targeted wild-type p53 suppresses growth of mutant p53 lymphomas in vivo. Oncogene. 2006;25:6133–6139. doi: 10.1038/sj.onc.1209641. [DOI] [PubMed] [Google Scholar]
- Park BJ, Lee SJ, Kim JI, Lee SJ, Lee CH, Chang SG, et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60:3370–3374. [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–281. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW. Defining the p53 DNA-binding domain/Bcl-x(L)-binding interface using NMR. FEBS Lett. 2004;559:171–174. doi: 10.1016/S0014-5793(04)00059-6. [DOI] [PubMed] [Google Scholar]
- Pietsch EC, Perchiniak E, Canutescu AA, Wang G, Dunbrack RL, Murphy ME. Oligomerization of bak by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem. 2008 doi: 10.1074/jbc.M710539200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S. Mol Cell. 2007;25:739–750. doi: 10.1016/j.molcel.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Sanchez-Arevalo VJ, Servitja JM, Gutkind JS. Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncogene. 2002;21:974–979. doi: 10.1038/sj.onc.1205134. [DOI] [PubMed] [Google Scholar]
- Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- Sayan BS, Sayan AE, Yang AL, Aqeilan RI, Candi E, Cohen GM, et al. Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc Natl Acad Sci. 2007;104:10871–10876. doi: 10.1073/pnas.0700761104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan AE, Sayan BS, Gogvadze V, Dinsdale D, Nyman U, Hansen TM, et al. p73 and caspase-cleaved p73 fragments localize to mitochondria and augment TRAIL-induced apoptosis. Oncogene. 2008 doi: 10.1038/onc.2008.64. in press. [DOI] [PubMed] [Google Scholar]
- Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- Senoo M, Manis JP, Alt FW, McKeon F. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell. 2004;6:85–89. doi: 10.1016/j.ccr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Shaw PE. EMBO Rep. 2007;8:40–45. doi: 10.1038/sj.embor.7400873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114:2063–2072. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- Sot B, Freund SM, Fersht AR. Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xl. J Biol Chem. 2007;282:29193–29200. doi: 10.1074/jbc.M705544200. [DOI] [PubMed] [Google Scholar]
- Stiewe T, Theseling CC, Pützer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. Mol Cell. 2007;25:725–738. doi: 10.1016/j.molcel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Talos F, Petrenko O, Mena P, Moll UM. Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res. 2005;65:9971–9981. doi: 10.1158/0008-5472.CAN-05-1084. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281:8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- Urist MJ, Di Como CJ, Lu ML, Charytonowicz E, Verbel D, Crum CP, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161:1199–1206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wager M, Guilhot J, Blanc JL, Ferrand S, Milin S, Bataille B, et al. Prognostic value of increase in transcript levels of Tp73 DeltaEx2-3 isoforms in low-grade glioma patients. Br J Cancer. 2006;95:1062–1069. doi: 10.1038/sj.bjc.6603410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AJ, Kokontis JM, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu YX, Hande MP, Wong AC, Jin YJ, Yin Y. TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J Biol Chem. 2007;282:29152–29162. doi: 10.1074/jbc.M703408200. [DOI] [PubMed] [Google Scholar]
- Weber A, Bellmann U, Bootz F, Wittekind C, Tannapfel A. Expression of p53 and its homologues in primary and recurrent squamous cell carcinomas of the head and neck. Int J Cancer. 2002;99:22–28. doi: 10.1002/ijc.10296. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, et al. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- Yan Y, Shay JW, Wright WE, Mumby MC. Inhibition of protein phosphatase activity induces p53-dependent apoptosis in the absence of p53 transactivation. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Proc Natl Acad Sci U S A. 1996;93:11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika A, Irwin M, Sansome C, Moll UM. Oncogenes induce and activate endogenous p73 protein. J Biol Chem. 2001;276:11310–11316. doi: 10.1074/jbc.M005737200. [DOI] [PubMed] [Google Scholar]
- Zeng X, Li X, Miller A, Yuan Z, Yuan W, Kwok RP, et al. The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol Cell Biol. 2000;20 doi: 10.1128/mcb.20.4.1299-1310.2000. 1299-12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Zhu Y, Lu H. NBP is the p53 homolog p63. Carcinogenesis. 2001;22:215–219. doi: 10.1093/carcin/22.2.215. [DOI] [PubMed] [Google Scholar]
- Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX. Nature. 2002;419:849–853. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]