Abstract
C. elegans RPM-1 is a member of a conserved protein family, the PHR proteins, which includes human Pam, mouse Phr1, zebrafish Esrom, and Drosophila Highwire. PHR proteins play important roles in the development of the nervous system. In particular, mutations in rpm-1 cause a disruption of synaptic architecture, affecting the distribution of synaptic vesicles and the number of presynaptic densities. Using antibodies against RPM-1, we determined the localization of the endogenous RPM-1 protein in wild type and in several mutants that affect synaptic development. Our analyses show that in mature neurons, RPM-1 resides in a distinct region that is close to, but does not overlap with, the synaptic exo- and endo- cytosis domains. The localization of RPM-1 occurs independently of several proteins that function in the transport or assembly of synapse components, and its abundance is partially dependent on its binding partner the F-box protein FSN-1. RPM-1 has been shown to target the MAPKKK DLK-1 for degradation. We show that activated DLK-1 may be preferentially targeted for degradation. Furthermore, using transgene analysis we identified a critical role of the conserved PHR domain of RPM-1 in its subcellular localization.
Keywords: RPM-1; presynaptic terminal; PHR domain; RING-Finger; E3; C. elegans, synapse targeting; DLK; synaptogenesis
Introduction
Chemical synapses are specialized intercellular junctions that mediate neurotransmitter release and signal transduction. The molecules required for synaptic function are numerous, but may be grouped into broad categories based on their roles in exocytosis, endocytosis, synapse assembly and adhesion (Sudhof, 2004; Ziv and Garner, 2004; Piechotta et al., 2006). Studies in the past decade have shown that the molecules within the pre and post-synaptic terminals are organized into sub-synaptic domains to conduct distinct signaling activities. Pre-synaptic terminals typically appear as a cluster of synaptic vesicles surrounding the electron-dense presynaptic density at the synaptic membrane. The synaptic vesicles are organized into distinct pools based on their competency and probability of fusion and release. Although transport of synaptic vesicles is known to be dependent on anterograde kinesin motors, much less is known about transport and biosynthesis of other presynaptic components.
C. elegans RPM-1 is a conserved protein that functions in the regulation of presynaptic assembly and axon patterning (Schaefer et al., 2000; Zhen et al., 2000). Homologs of RPM-1 include human PAM, Drosophila Highwire, mouse Phr1, and zebrafish Esrom, collectively named PHR (Pam Highwire RPM-1) proteins . All PHR proteins are extremely large (containing between 3766–5233 amino acids) and share several conserved domains, including an RCC1–like domain (RLD), two repeats only found in PHR proteins known as the PHR domains, and a RING-H2 domain. The RLD domain in PAM has been shown to bind and negatively regulate adenylyl cyclase (Scholich et al., 2001; Ehnert et al., 2004). The PHR domain has no known function. The RING-H2 domain has been shown to function as an E3 ubiquitin ligase (Liao et al., 2004; D'Souza et al., 2005; Nakata et al., 2005; Wu et al., 2005; Lewcock et al., 2007). Both C. elegans RPM-1 and Drosophila Hiw interact with F-box containing proteins, FSN-1 (F-box SyNaptic protein) (Liao et al., 2004) and DFsn (Wu et al., 2007). A major substrate of RPM-1 or Hiw is the conserved dual-leucine zipper-bearing kinase DLK (Nakata et al., 2005). The N-terminal of C. elegans RPM-1 binds a guanine nucleotide exchange factor GLO-4 (Grill et al., 2007). Drosophila Hiw also binds to the Smad protein Medea (McCabe et al., 2004). Both Hiw and Pam can interact with the tuberin-hamartin complex (Murthy et al., 2004).
Despite the significant roles that PHR family members play in presynaptic signaling and development, little is known about how they are targeted to the synapse. Nor is it known what kinds of molecular and cellular cues are important for guiding and maintaining their sub-synaptic positioning. In this study, we report the characterization of the endogenous RPM-1 localization. We show that RPM-1 is positioned independently of proper presynaptic density assembly and synaptic vesicle clustering. We also identify a critical role for the PHR domain in RPM-1 protein localization.
Materials and Methods
C. elegans genetics
Strains were maintained at 22.5°C as described (Brenner, 1974). The strains used in this study are the following; Wild type Bristol strain (N2), CZ1262 DYN-1::GFP(juIs83); lin-15(n765ts), CZ1252 rpm-1(ju44ts), CZ2474 syd-2(ju37); Pflp-13 SNB-1::GFP(juIs137), CZ1893 syd-1(ju82), CZ469 sad-1(ju53); Punc-25 SNB-1::GFP(juIs1), CZ2244 syd-1(ju82); syd-2(ju37); Punc-25 SNB-1::GFP(juIs1), CZ3307 syd-2(ju37) sad-1(ju53), CB1196 unc-26(e1196), ZM16 fsn-1(hp1); Punc-25 SNB-1::GFP(juIs1), CZ4387 DLK-1::GFP(juIs192), CZ5743 RPM-1::GFP(juIs58), CZ6259 RPM-1::GFP(juIs58);PF25B3.3 FLAG::DLK-1(K162A)(juIs201), CZ6845 PF25B3.3 FLAG::DLK-1(juEx1301), CZ4632 rpm-1(ju23); Punc-25 SNB-1::GFP(juIs1); juEx743(pCZ612), CZ5722 juEx1093(pCZ693), CZ5358 juEx988(pCZ376), CZ5356 juEx986(pCZ375), CZ3427 juEx488 (pCZ374), CZ7109 juEx1350(pCZGY143), CZ6056 juEx1166(pCZGY62), CZ6057 juEx1167(pCZGY62), CZ6060 juEx1170(pCZGY64), CZ6062 juEx1172(pCZGY65), CZ333 Punc-25 SNB-1::GFP(juIs1), CZ455 rpm-1(ju44); Punc-25 SNB-1::GFP(juIs1) CZ7111 juEx1350(pCZGY143); rpm-1(ju44); Punc-25 SNB-1::GFP (juIs1).
Immunocytochemistry and image analysis
Whole-mount staining was performed using a Finney-Ruvkun based protocol (Finney and Ruvkun, 1990), with the exception that the reduction step was done in Borate buffer (pH 9.3). Anti-RPM-1 antibodies were raised in rat against fusion proteins corresponding to amino acids 3232–3492. The antisera were affinity-purified and used at 1:300. Rabbit antibodies to UNC-10 and SNT-1 were provided by M. Nonet (Nonet et al., 1993; Koushika et al., 2001). Anti-GFP antibodies (Rabbit #A11122 and Mouse 3E6) and secondary antibodies (goat anti-rat, goat anti-mouse and goat anti-rabbit) conjugated with Alexa 488, Alexa 594 or Cy5 were from Molecular Probes (Eugene, OR). Mouse anti-FLAG antibodies from Sigma (M2) were used in co-staining experiments with rabbit anti-GFP. Images were acquired on a Zeiss LSM 5 Pascal confocal microscope. Detector gain was tuned to minimize pixel saturation and maximize detection range. Projections were made of z-stack images using LSM 5 Pascal software and then exported as TIFF files. Images were cropped and processed in parallel using Adobe Photoshop. Fluorescent intensity analysis was performed using Metamorph (Molecular Devices Corporation, Chicago, IL). A threshold was set for each channel to include the above background signal. Quantitation of the distance between RPM-1 and RIM puncta in Fig. 1 was performed on images of single z-sections as follows: a circle was drawn around RPM-1 puncta using Zeiss LSM software and then enlarged until the circle abutted the closest RIM puncta; the radius of the circle was taken as the distance between the center of the RPM-1 puncta to the edge of the RIM puncta. Similar analysis was done for DLK-1::GFP with respect to RIM. The quantitation in Fig. 2 was performed on 20µm lengths of dorsal cords. For Fig. 3, measurements from 6 images of dorsal cords for each genotype were compared using a Welch Two Sample t-test performed in “R” (see The Foundation for Statistical Computing, http://www.R-project.org). For Fig. 4, colocalization was calculated as the DLK-1 positive area within the RPM-1 positive area. 34 puncta were compared per genotype using a Wilcox Rank Sum test.
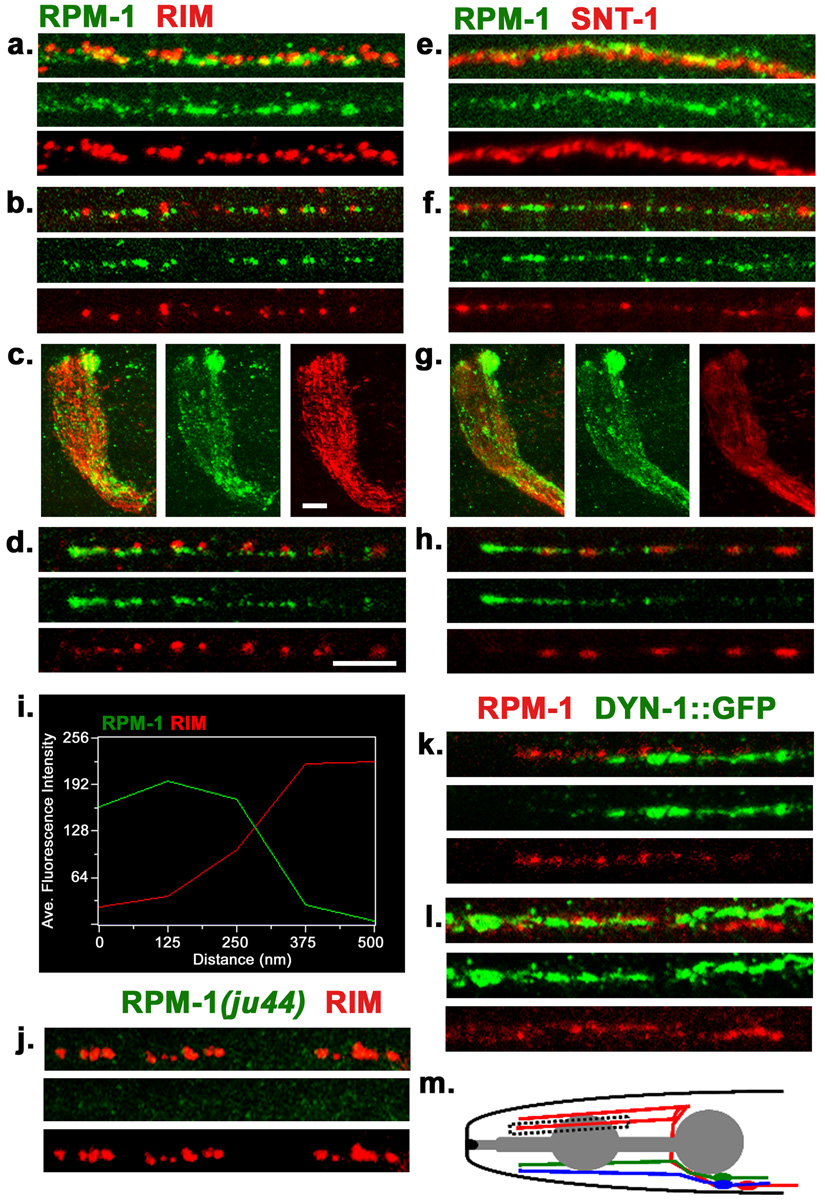
Figure 1. Endogenous localization of RPM-1.
(a–d) Confocal images of wild type (N2) worms co-stained with anti-RPM-1 (green) and anti-RIM (red) in (a) dorsal cord, (b) sub-lateral cord, (c) nerve ring, (d) SAB neuron. (e–h) Confocal images of N2 worms co-stained with RPM-1 (green) and synaptotagmin/SNT-1 (red) in (e) dorsal cord, (f) sub-lateral cord, (g) nerve ring, (h) SAB neuron. (i) Fluorescence intensity plot showing the typical distance between RPM-1 puncta and RIM in the dorsal cord. (j) The RPM-1 antibody is specific. Confocal images of the dorsal cord of rpm-1(ju44) mutants. RIM stain (red) is positive and shows gaps that are characteristics of the Rpm-1 mutant phenotype, whereas no RPM-1 staining is detected. (k–l) juIs83 (DYNAMIN-1::GFP) worms co-stained with RPM-1(red) and GFP (green) in (k) SAB neuron and (l) dorsal cord. (m) Schematic drawing of the SAB neurons. Blue, SABVL; green, SABVR; red, SABD. All SABs have their cell bodies in the retro-vesicular ganglion, project axons towards the anterior, and form en passant synapses onto head muscles. Box indicates region of SAB shown in d and h. All SAB images shown are in the same orientation with the anterior end on the left, individual SAB processes were chosen at random. Scale bars: 5µm, a, b, d, e, f, h, k, l, j . Scale bar, 10µm c, g,
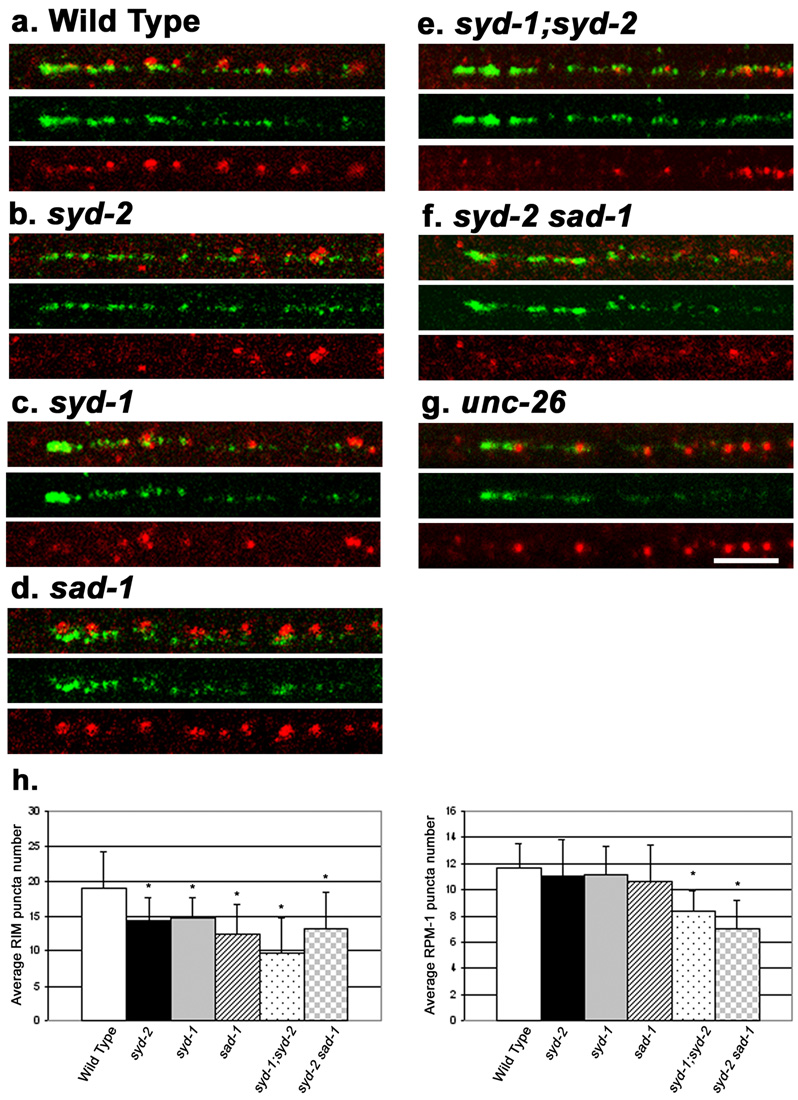
Figure 2. RPM-1 localization in mutants that have altered synaptic morphology.
Shown are confocal images of SAB neurons co-stained with RPM-1 (green) and RIM (red). SAB regions are same as indicated in figure 1m. (a) Wild type (N2). (b) syd-2(ju37) (c) syd-1(ju82) (d) sad-1(ju53) (e) syd-1(ju82);syd-2(ju37) (f) syd-2(ju37)sad-1(ju53) (g) unc-26(e1196). In syd-1 and syd-2 mutants, RIM expression is diffuse and reduced in intensity, but RPM-1 expression is normal. (h) Quantitation of RIM (left plot) or RPM-1 (right plot) puncta number per 20µm length of dorsal cord. t-tests were performed for RPM-1 and RIM puncta number relative to wild type, * p<0.05. Scale bar, 5µm, all panels are the same scale.
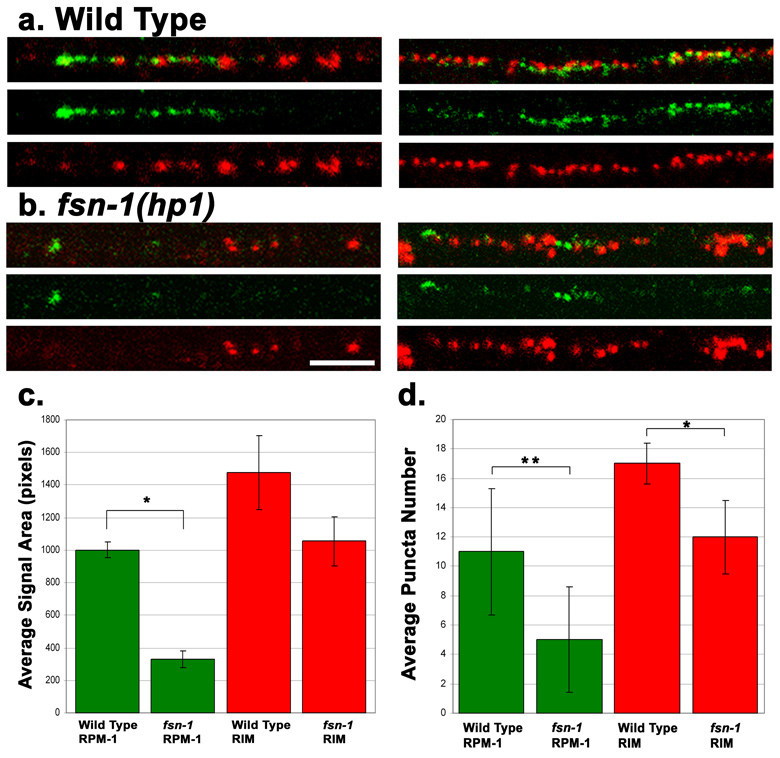
Figure 3. RPM-1 expression is reduced in fsn-1(hp1) mutants.
(a–b) Shown are confocal images of SAB neurons (left panels) or dorsal cord (right panels) co-stained with RPM-1 (green) and RIM (red). (a) N2 (wild type), a gradient of RPM-1 staining is observed in the SAB neurons that is brightest near the anterior tip of SAB axon, and tapers off in the posterior part of the axon. Dorsal cord regions do not show a gradient pattern. (b) In fsn-1(hp1) mutants, the abundance of RPM-1 is significantly decreased in the SAB and dorsal cord, though the RPM-1 staining is still brightest at the anterior end of the SAB. (c) Quantitation of RPM-1 staining abundance. Bar graph shows the average total number of pixels that contained above background signal for a section of dorsal cord, with error bars as std. dev. RPM-1 abundance is significantly decreased in fsn-1(hp1) animals, green bars *(p<10−6). RIM abundance is not significantly different from wild type, red bars (p=0.09). n = 6 dorsal cords for each data set. P-value is for a Welch Two Sample t-test. (d) Quantitation of puncta number shows a greater decrease in RPM-1 compared to RIM (for RPM-1, P<10−4; for RIM, p=0.02). Scale bar 5µm, same scale for all panels.
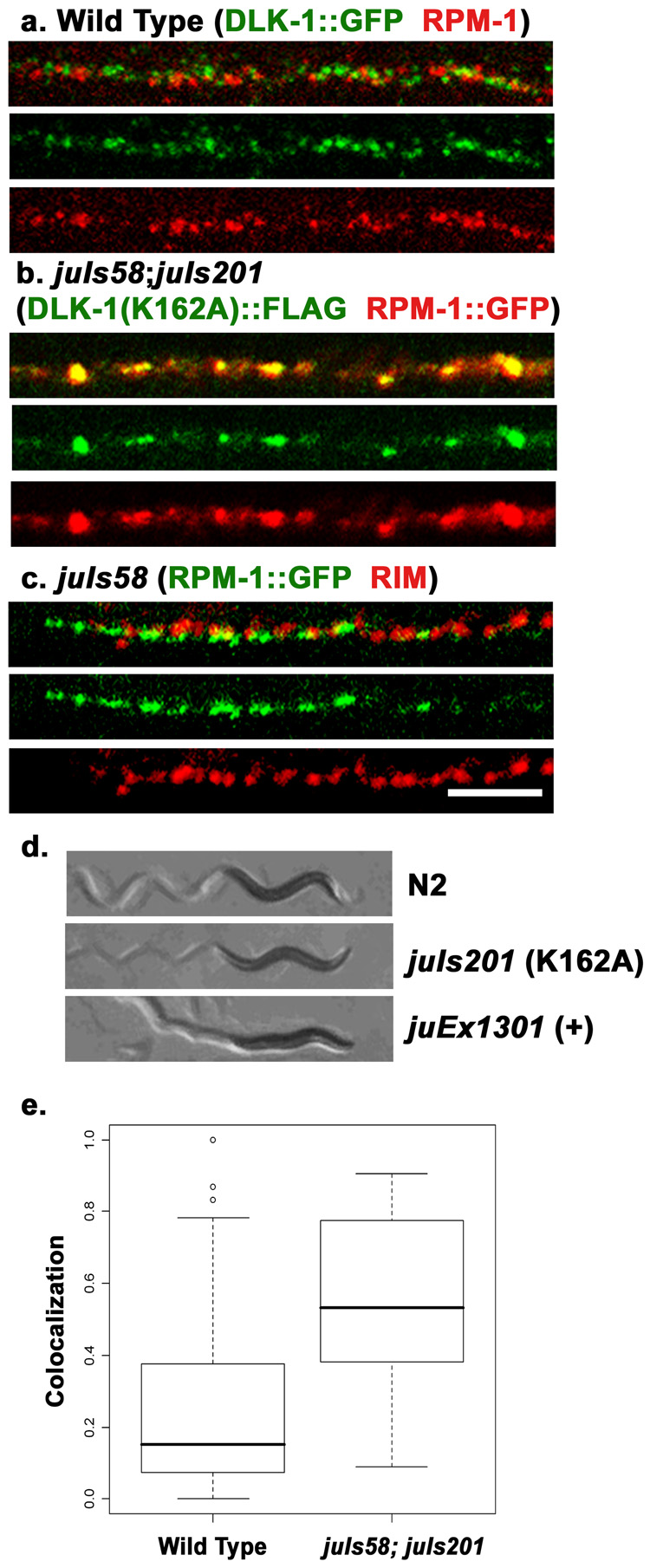
Figure 4. Colocalization studies of RPM-1 and DLK-1.
(a) Confocal image of the dorsal cord of juIs192[DLK-1::GFP] animals co-stained with GFP (green) for DLK-1::GFP and endogenous RPM-1 (red). (a). In wild type animals expressing DLK-1::GFP at a low level that does not cause detectable synaptic defects, RPM-1 and DLK-1::GFP are seen in close proximity and rarely overlap. (b) Confocal image of juIs58 (RPM-1::GFP); juIs201[ DLK-1(K162A)::FLAG] animals, co-stained with anti-GFP (red) and anti-FLAG (green). the kinase dead DLK-1 shows extensive colocalization with RPM-1. (c) In juIs58 animals, RPM-1::GFP (green) shows normal spatial relationship to RIM (red). (d) Shown are bright-field images of animals and their movement trails. Wild type animals move sinusoidally. Over expression of wild type DLK-1 (juEx1301) causes animals to be hyper-contracted and uncoordinated (bottom). Overexpression of the kinase dead DLK-1 (K162A)(juIs201) does not have any effects on locomotion (middle). (e) Quantitation of the amount of DLK-1 signal present per RPM-1 puncta. Colocalization is significantly increased in juIs58;juIs201 animals compared to wild type, but is not complete. Colocalization is the ratio of DLK-1 signal area within RPM-1 signal area/puncta (see methods). N = 34 RPM-1 puncta per genotype, p<0.0001. Scale bar 5 µm, same scale for a–c.
Molecular Biology and transgene analysis
Plasmid clones pCZ374, pCZ375, pCZ376 and pCZ693 were constructed using restriction enzyme sites from a full length rpm-1 genomic clone pCZ160 (Zhen et al., 2000) and GFP vectors (from A. Fire), following standard procedures. pCZ612 was constructed by deleting a 5.3 kb BstEII DNA fragment from pCZ160. All pan-neural promoter driven PHR domain constructs (pCZGY62, pCZGY63, pCZGY64, pCZGY65) were made by PCR and using the Gateway cloning system (Invitrogen). Briefly, DNA corresponding to the region of the open reading frame was amplified, cloned into the PCR8 vector, and then recombined into a destination vector that contained the pan-neural promoter PF25B3.3 and GFP with the unc-54 3’ UTR (from pPD117.01). Sequences of DNA were confirmed, and primers used are available upon request. To generate the RPM-1ΔPHR construct (pCZGY143) the 5’ genomic sequences corresponding to amino acids M1-K1046 and the 3’ genomic sequences corresponding to amino acids S1747-STOP were amplified separately, cloned into PCR8, and then ligated into a Gateway vector containing the PF25B3.3. promoter. pCZ742 and pCZ702 constructs contained wild-type dlk-1 and dlk-1(K162A) cDNA driven by the F25B3.3 promoter with a 5’ in-frame Flag tag.
Transgenic animals expressing each rpm-1 construct were generated by injecting 10ng/µl or 1ng/µl of the DNA of interest, along with 40ng/µl pRF4 (rol-6(dm)) or 50 ng/µL Pttx-3::RFP as a co-injection marker, following standard procedures (Mello et al., 1991). At least two lines were analyzed in detail for each transgene.
Results
Endogenous RPM-1 Localizes to Perisynaptic Regions
To investigate the endogenous subcellular localization of RPM-1, we generated and affinity purified antibodies against amino acids 3232–3492 of RPM-1. In whole mount staining, RPM-1 expression was seen predominantly in the nervous system. In the nerve ring and nerve cords, endogenous expression of RPM-1 was mostly punctate, with varying degrees of intensity (Fig. 1 a–c, e–g). No signal was detected in the cell bodies. The specificity of the RPM-1 antibody was confirmed by staining rpm-1 mutant animals (Fig. 1 j, and supplemental Fig. 3).
To define the spatial relationship of RPM-1 with respect to other synaptic proteins, we performed co-immunostaining of anti-RPM-1 with either anti-RIM (UNC-10) or anti-Synaptotagmin (SNT-1) antibodies, respectively. RIM is a component of the presynaptic density and functions in vesicle priming (Koushika et al., 2001; Weimer et al., 2006). Synaptotagmin is an integral membrane protein of synaptic vesicles and functions in vesicle exocytosis and endocytosis (Nonet et al., 1993; Sudhof, 2004). Within the nerve ring, dorsal and ventral nerve cords anti-RPM-1 signal was found to be close to, but not overlap with, those of anti-RIM or anti-SNT-1 (Fig. 1a–c, e–g). Measurement of the average distance between RPM-1 and RIM puncta in the dorsal cords showed that 75% of RPM-1 puncta were within 250 nm of a RIM puncta (n=100 RPM-1 puncta, see materials and methods) (Fig. 1i). As the boundaries between individual synapses in the nerve cords and nerve ring are not resolvable, we examined the staining pattern of RPM-1 in the synapses of the SAB neurons. The SAB neurons are cholinergic neurons that form synapses en passant onto muscles in the head (Fig. 1m) (White et al., 1986; Zhao and Nonet, 2000). Anti-SNT-1 or anti-RIM staining showed a string of fluorescent puncta spaced evenly along the SAB axons (Fig. 1d, h), while anti-RPM-1 staining was seen adjacent to those of anti-RIM or anti-SNT-1. Moreover, anti-RPM-1 signal showed a graded pattern such that higher immunofluorescent intensity was consistently detected at the anterior ends of the SAB axons (Fig. 1d, h, m).
To evaluate RPM-1 localization with respect to proteins involved in the endocytosis of synaptic vesicles, we co-stained animals expressing a functional GFP-tagged dynamin (DYN-1::GFP) transgene (juIs83) with RPM-1. Dynamin is associated with synaptic vesicles and is required for endocytosis (Labrousse et al., 1998). We found that DYN-1::GFP and RPM-1 showed an adjacent, non-overlapping, distribution (Fig.1k, l). Thus, endogenous RPM-1 is localized in a region that is close to, but not overlapping with, synaptic vesicle release and recycling domains. This expression pattern also confirms the previously reported results using a functional RPM-1::GFP transgene (Zhen et al., 2000). We term the RPM-1 localization domain ‘perisynaptic’ to indicate that it is adjacent to but largely non-overlapping with known synaptic vesicle or presynaptic active zone markers.
RPM-1 Localization is Largely Normal in Mutants that Have Altered Presynaptic Architecture
Since loss of function in rpm-1 affects both synaptic vesicle accumulation and presynaptic density distribution (Schaefer et al., 2000; Zhen et al., 2000), we next asked how RPM-1 expression might be altered in mutants with defects in synapse morphology. We focused this analysis on mutants known to show genetic interactions with rpm-1.
The Liprin-α protein SYD-2 is associated with the presynaptic density, and mutations in syd-2 cause alterations in the size of the presynaptic density and the accumulation of synaptic vesicles (Zhen and Jin, 1999). Mutations in rpm-1 enhance locomotion defects caused by loss of function mutations in syd-2 (Liao et al., 2004). We found that the spatial position of RPM-1 relative to RIM in syd-2(ju37) mutant animals was unaltered (Fig. 2b, Supplemental Fig. 1b), although the number of RIM puncta was reduced and RIM localization became diffuse, as previously reported (Ackley et al., 2005) (Fig. 2h). The average numbers of RPM-1 puncta per 20µm of the dorsal cord in wild type and in syd-2 mutants were not significantly different (p>0.3, n=10)(Fig. 2h). The co-staining pattern of anti-RPM-1 with anti-SNT-1 in syd-2(ju37) animals was also comparable to that in wild type (data not shown).
The SYD-1 PDZ RhoGAP protein colocalizes with RIM, and has been shown to regulate the synaptic localization of several proteins, such as SYD-2 and SNB-1 (Hallam et al., 2002). rpm-1(ju44); syd-1(ju82) double mutants also display enhanced movement defects (Nakata et al., 2005). We found that the RPM-1 localization pattern relative to RIM and SNT-1 was grossly normal in syd-1(ju82) mutants (Fig. 2c, h, Supplemental Fig. 1c and data not shown for SNT-1). In syd-1(ju82); syd-2(ju37) double mutants, the average RPM-1 puncta number was significantly reduced, although punctate staining of RPM-1 was still observed (Fig. 2e, h) (p<0.001, n=10).
SAD-1 is a Ser/Thr kinase related to the PAR-1 and MARK kinases, and is an important regulator of synaptic vesicle clustering pattern and neuronal polarity (Crump et al., 2001; Kishi et al., 2005; Inoue et al., 2006). In sad-1 mutant animals, synaptic vesicles are broadly distributed, and vesicle markers are also localized improperly to dendrites (Crump et al., 2001). We found that in sad-1(ju53) animals, RPM-1 localization remained punctate and non-overlapping with RIM or SNT-1 (Fig. 2d, Supplemental Fig. 1d, and not shown for SNT-1 co-staining). The average RPM-1 puncta number in sad-1 mutants was also comparable to that in wild type animals (Fig. 2h), indicating that like syd-1 and syd-2, loss of sad-1 function alone does not affect RPM-1 localization. In syd-2(ju37) sad-1(ju53) double mutants RIM or SNT-1 expression was greatly reduced and showed more severe disruption, so was the average number of RPM-1 puncta (Fig. 2f,h Supplemental Fig. 1f, and not shown for SNT-1). However, the gross spatial pattern of RPM-1 was largely unaltered. Thus, these observations demonstrate that the genes functioning in the regulation of presynaptic assembly are not required for targeting RPM-1 to synaptic regions, but contribute to stable localization or maintenance of RPM-1 at synapses directly or indirectly.
We next asked if depleting synaptic vesicles could have an effect on RPM-1 localization. The majority of synaptic vesicles are delivered by anterograde kinesin motors and replenished through endocytosis. unc-26 encodes the C. elegans synaptojanin; in unc-26(e1196) mutants, synaptic vesicles are severely depleted and synapses show collapsed morphologies (Harris et al., 2000). We found that despite the absence of large pools of synaptic vesicles, RPM-1 localization, relative to RIM, was unaltered in these animals (Fig. 2g and Supplemental Fig. 1g). Previously, we showed that in unc-104 kinesin mutants the localization of a functional RPM-1::GFP transgene was unaffected (Zhen et al., 2000). Taken together, these results indicate that RPM-1 localization is not dependent on the machinery that maintains the synaptic vesicle pool at the synapses.
RPM-1 Synaptic Abundance is Partially Dependant on its Binding Partner FSN-1
RPM-1 forms an SCF-like ubiquitin ligase complex with the F-box protein FSN-1, a SKP homolog SKR-1, and a cullin protein CUL-1 (Liao et al., 2004). The formation of this complex is dependent on the presence of FSN-1. fsn-1 mutants display synaptic defects resembling those of rpm-1 mutants. In whole mount staining of fsn-1(hp1) mutants, RIM and RPM-1 showed non-overlapping patterns and the gross localization pattern of RPM-1 was similar to that in wild type animals (Fig. 3a, b). For example, RPM-1 localization appeared to form a gradient in SAB neurons with highest intensity detected at the anterior tip. In fsn-1 mutants RIM expression was mildly altered such that the average number of RIM puncta in 20µm dorsal nerve cord was 12, compared to 17 in wild type (Fig. 3c, p=0.02, n=10). However, the intensity of anti-RPM-1 signal was severely reduced in fsn-1 mutants than in wild type animals (Fig. 3c) (p<10−6). Fewer RPM-1 puncta were detected in fsn-1 mutants than in wild type animals (p<10−4, n=10). These observations suggest that formation of the SCF complex is not essential for the perisynaptic targeting of RPM-1, but that RPM-1 may be destabilized in the absence of FSN-1.
Cellular Interactions of RPM-1 with DLK-1, a Target for RPM-1 E3 Ligase Activity
The dual-leucine zipper MAP kinase DLK-1 is a target of RPM-1 ubiquitin ligase activity, and in rpm-1 mutants, the synaptic abundance of DLK-1::GFP is increased about two fold (Nakata et al., 2005). A functional DLK-1::GFP does not colocalize with anti-RIM or anti-SNT-1 staining (Nakata et al., 2005, supplemental Figure 4). The mean distance between DLK-1::GFP puncta with respect to RIM puncta in the sub-lateral cords was similar to that between RPM-1 and RIM, averaging 288nm and 346 nm for DLK-1::GFP and RPM-1, respectively (p=0.04, n=45). To further investigate the spatial relationship between RPM-1 and DLK-1, we used animals expressing DLK-1::GFP(juIs192) at a level that does not cause detectable synapse abnormalities. We found that anti-RPM-1 and DLK-1::GFP were consistently detected within the same confocal z-section, but the brightest staining signals for either protein were mostly non-overlapping (Fig. 4a, e). This non-overlapping pattern is unlikely to be due to improper staining, (e.g. as the result of the exclusion of antibodies), because the appearance of RPM-1 or DLK-1::GFP in the doubly-stained animals were indistinguishable from those singly stained with each antibody. Instead, we infer that this mutually exclusive pattern reflects the degradation of DLK-1 at synapses by RPM-1. To test this, we generated a strain that expressed a kinase-dead DLK-1 transgene (FLAG::DLK-1(K162A) (juIs201)). Overexpression of a wild type DLK-1 tagged with FLAG (FLAG::DLK-1(+) (juEx1301)) caused the worms to be uncoordinated, but high level expression of the kinase-dead DLK-1(K162A) did not (Fig. 4d), indicating that DLK-1(K162A) is inactive. We then introduced juIs201 into juIs58 (RPM-1::GFP), and co-stained the animals with anti-FLAG and anti-GFP. RPM-1::GFP(juIs58) showed normal localization relative to RIM, demonstrating that transgenic expression of RPM-1 did not lead to abnormal localization (Fig. 4c). To quantitate the degree of colocalization, we measured the presence of FLAG::DLK-1 per RPM-1 puncta. In wild type animals 15% of the RPM-1 puncta overlapped with DLK- 1::GFP signal whereas in juIs58; juIs201 animals 53% of the RPM-1::GFP puncta contained FLAG::DLK-1(K162A) signal, (p<0.0001). These observations suggest that kinase dead DLK-1 can colocalize with RPM-1 because it is not degraded and that active DLK-1 is preferentially degraded by RPM-1, leading to non-overlapping subsynaptic localization.
Domains Required for RPM-1 Perisynaptic Localization
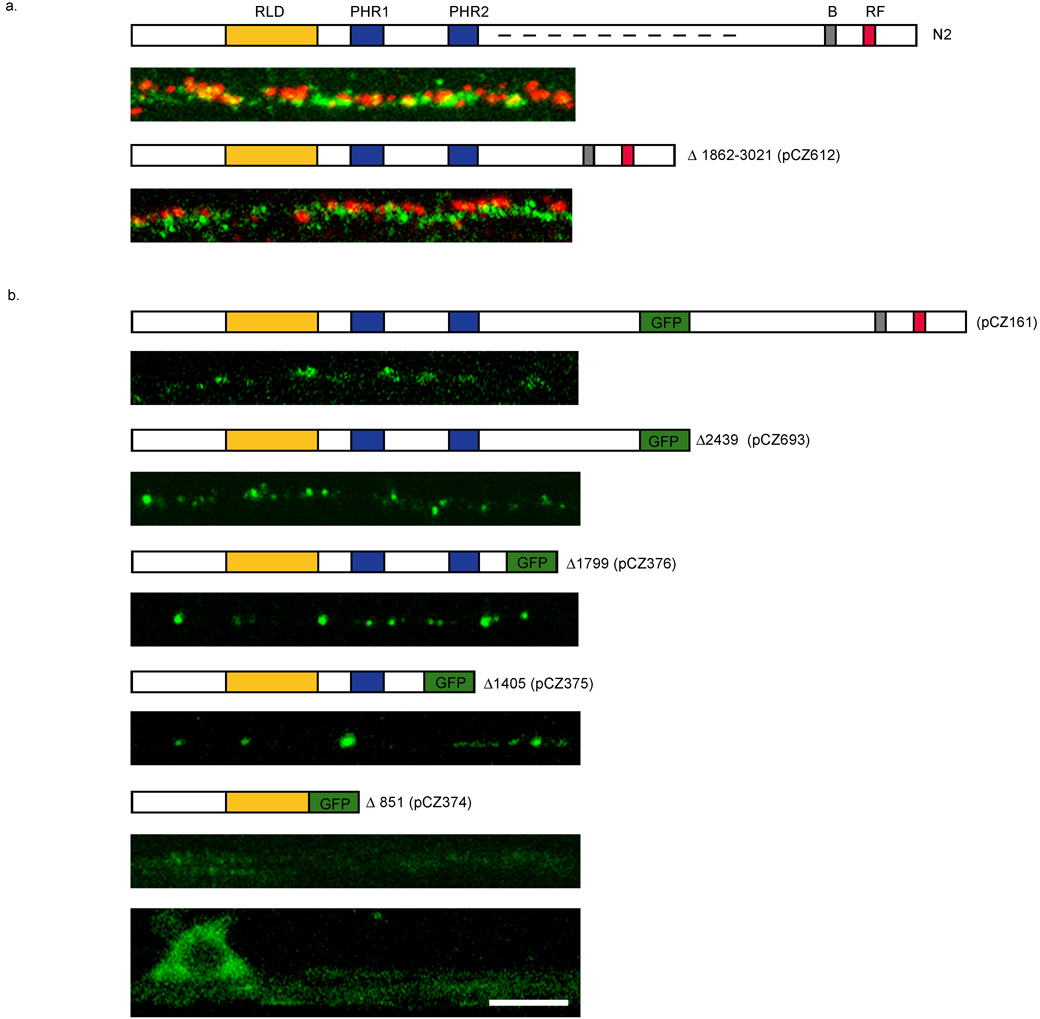
RPM-1, like other PHR proteins, is a very large protein with several domains: an RLD domain at the N-terminal, the PHR repeats in the middle, and the C-terminal E3 ligase domain. To define which region of RPM-1 mediates its perisynaptic targeting, we created transgenes expressing deletion derivatives of RPM-1 and analyzed their localization either using GFP as a reporter or by anti-RPM-1 immunostaining (Fig. 5, see Materials and Methods).
Figure 5. Identification of domains important for RPM-1 localization.
Constructs expressing RPM-1 or RPM-1::GFP are shown above the images. (a) Schematic of RPM-1 protein showing RCC1-like Domain (RLD), Pam, HIW ,RPM-1 (PHR1 and PHR2) domains, B-Box Zn finger domain (B) and RING-finger domain (RF). Dashed line in top panel of (a) indicates region removed in lower panel of (a). Confocal images in (a) show the dorsal cord in animals co-stained with RPM-1 (green) and RIM (red). The genotypes are wild type (top) and rpm-1(ju23);juEx743. Removal of residues 1862 to 3021 (dashed line) of RPM-1 abolishes RPM-1 activity but does not alter its spatial relationship to RIM. (b) Confocal images of GFP fluorescence in sub-lateral cords of transgenic animals expressing RPM-1::GFP. Number indicates the amino acid residue where RPM-1 is truncated and GFP is fused. Note that truncations all localize similarly to full length, until the first PHR domain is eliminated. Expression of RPM-1(1–851)::GFP is very diffuse and barely detectable in the sub-lateral cord (top), but is seen in the cell body in ventral cord (bottom). Scale bar 5 µm for all panels.
RPM-1 lacking amino acids 1862 through 3021 (pCZ612) showed a localization pattern similar to endogenous RPM-1, based on co-staining with anti-RIM (Fig. 5a). The deleted region of RPM-1 is not highly conserved. This construct did not rescue the synaptic defects of rpm-1 (Fig.5a). This analysis shows that although removal of this region does not affect the punctate expression pattern of RPM-1, it is required for RPM-1 function.
The expression of RPM-1 truncated at resides 2439 (pCZ693), or 1799 (pCZ376), or 1405 (pCZ375) and tagged with GFP revealed a localization pattern comparable to that of the full-length RPM-1::GFP (juIs58)(pCZ161) (Fig. 5b). However, expression of the N-terminal 851 amino acids of RPM-1 (pCZ374) showed abundant expression in the neuronal cell bodies and diffused localization in axons (Fig. 5b). This analysis suggests that the region between residues 851 and 1862 likely contains information for the proper synaptic targeting of RPM-1.
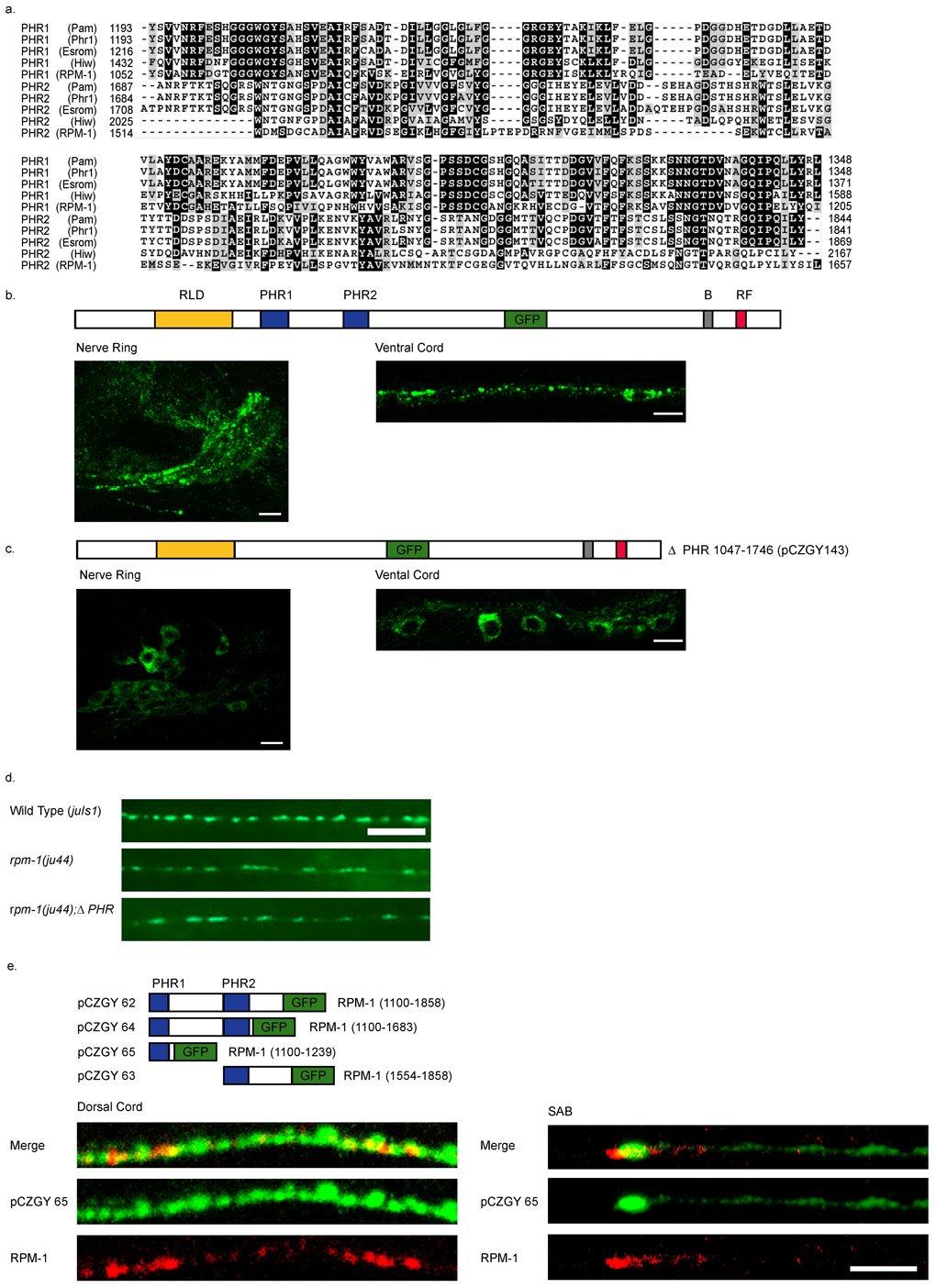
The PHR Domain of RPM-1 is Necessary and Sufficient for Perisynaptic Targeting
The region between residues 851 to 1862 of RPM-1 contains two domains found only in other PHR proteins, termed the PHR domain (Fig. 6a). The PHR domain is approximately 150 amino acids long. In RPM-1, the first PHR domain (PHR1) spans amino acids 1052–1205 and the second (PHR2) spans amino acids 1514–1657. The degree of conservation for PHR1 is higher than that for PHR2. To test whether these domains are important for the synaptic targeting of RPM-1, we deleted both domains from an RPM-1::GFP construct and assayed localization. Full length RPM-1::GFP(juIs58) localizes primarily to axonal regions (Fig. 6b). RPM-1ΔPHR::GFP (pCZGY143) expression was predominantly detected in the neuronal cell bodies in the head and ventral cord (Fig. 6c). Expression of this transgene in rpm-1 mutants failed to rescue rpm-1 phenotypes (Fig. 6d). These results show that the PHR domains are together required both for the localization of RPM-1 to synapses and for its synaptic function.
Figure 6. The PHR domain of RPM-1 is necessary and sufficient for synaptic localization.
(a) A CLUSTAL alignment of the PHR regions. (b–c) Confocal images of GFP fluorescence in transgenic animals. (b) Expression of the functional full-length RPM-1::GFP(juIs58) is primarily in the nerve ring and nerve cords, and barely detectable in the cell bodies. (c) Deletion of the PHR domains juEx143 [pCZGY143] causes GFP to be confined to the cell bodies and is barely seen in nerve processes. (d) Expression of RPM-1ΔPHR::GFP (pCZGY143) does not rescue rpm-1(ju44). Wild type animals expressing juIs1[P unc-25 SNB-1::GFP] show an array of evenly sized, evenly spaced fluorescent puncta (top). rpm-1(ju44) animals display enlarged puncta with gaps in SNB-1GFP expression (middle), and are not rescued by the expression of RPM-1ΔPHR::GFP (pCZGY143) (bottom). (e) Schematics of the constructs expressing regions of PHR fused to GFP. The images are juEx1172 [pCZGY65] (pan neural PHR1::GFP) co-stained with endogenous RPM-1 and GFP in the dorsal cord (left panels) and in the SAB neurons (right panels). Scale bars 5µm.
To test if either PHR domain is sufficient for RPM-1 localization we tagged either or both domains with GFP and expressed them from a pan-neural promoter (Fig. 6e, see Materials and Methods). All PHR constructs gave GFP localization patterns indistinguishable from each other and similar to that of the full length RPM-1::GFP(juIs58). PHR1::GFP (pCZGY65) showed localization to the tip of the SAB neurons. We found that PHR1::GFP colocalized with endogenous RPM-1 at synapses in the SAB neurons and many places including the dorsal cord (Fig. 6e). We conclude that the PHR domains are both necessary and sufficient for RPM-1 transport and localization to perisynaptic regions, and that this localization is essential for RPM-1 function.
Discussion
Increasing evidence has indicated that the conserved PHR protein family plays important roles in neuronal development, from topographic mapping to axon patterning, and from synapse organization to synapse growth (Schaefer et al., 2000; Wan et al., 2000; Zhen et al., 2000; Burgess et al., 2004; D'Souza et al., 2005; Bloom et al., 2007; Lewcock et al., 2007). Studies of Drosophila Highwire reported that it is localized to the neuromuscular junctions, in a region surrounding the active zone, termed the periactive zone (Sone et al., 2000; Wan et al., 2000). However, subsequent studies show that the anti-Hiw antibodies used by Sone et al. are not specific for Hiw (Wu et al., 2005). The subcellular location of Hiw therefore remains unclear. Recent analysis of mouse Phr1 in the developing motor axons shows that Phr1 is associated with stable microtubules in the axon shaft (Lewcock et al., 2007). It remains unknown where Phr1 is localized in mature neurons.
Our immunocytochemical data here reveal that in mature neurons endogenous RPM-1 is localized in a distinct region that is adjacent to the synaptic vesicle exo- and endo-cytosis domain. The observations that loss of rpm-1 function causes changes in synaptic architecture, but that disruptions in different aspects of synaptic assembly and maintenance do not dramatically alter RPM-1 localization suggest that RPM-1 may be recruited to, or define, the sites that facilitate the development of the nascent synapses. RPM-1 colocalizes with two of its binding partners, FSN-1 and GLO-4 (Liao et al., 2004; Grill et al., 2007). RPM-1 localization is unaltered in glo-4 mutants (Grill et al., 2007). Biochemical experiments have shown that in the absence of FSN-1 RPM-1 does not bind CUL-1 and SKR-1 (Liao et al., 2004). The finding that RPM-1 staining is reduced in fsn-1 mutants suggests that RPM-1 could be destabilized or that its expression level is reduced. It is possible that the assembly of this protein complex is instructed by signals that initiate synaptogenesis, which in turn contributes to the stabilization of RPM-1 at correct locations. The graded pattern of RPM-1 localization in the SAB axons is intriguing. In rpm-1 mutants, the SAB axons frequently fail to terminate at their normal positions and over-extend for various lengths (Schaefer et al., 2000). Such a graded pattern may bare similarities to that of mouse Phr1 in the developing axons. The higher level of RPM-1 towards the anterior ends of the axons suggests that RPM-1 may sense and/or process the axon termination or extension signals. Conceivably, axon termination may facilitate stable synapse formation. The precise cellular complements in the subsynaptic domain where RPM-1 resides at different neuronal differentiation stages await for further analysis.
Down-regulation of the DLK-1 kinase is important for normal development of synapses. Our colocalization studies suggest that some DLK-1 likely resides in the same sub-synaptic region as RPM-1. However, DLK-1 is maintained at low levels where RPM-1 levels are high and appears more abundant where RPM-1 levels are low. This pattern is similar to that described for mouse Phr1 and DLK in the developing motor axons (Lewcock et al., 2007). It is likely that the activation of DLK-1 may trigger its degradation since a kinase dead DLK-1 shows colocalization with RPM-1. Our observation is consistent with the recent report that in COS7 cells the mammalian DLK kinase can be degraded by the E3 ligase CHIP upon activation induced by stress (Daviau et al., 2006). Such transient interactions between DLK-1 and RPM-1 are important for neuronal decisions in axonal and synapse development.
All PHR family members are large multi-domain proteins. With the exception of the C-terminal RING domain, little is known about the functions of other domains in these proteins. We find that RPM-1(Δ1802–3021) is localized normally, but fails to rescue rpm-1 mutant defects, even though the E3 ligase and other conserved domains are properly expressed. This observation suggests that the size of RPM-1 may be important for its function. The large size of RPM-1 may allow it to scaffold other synaptic molecules or to bridge and integrate multiple signaling events.
An important finding from our analyses of the deletion constructs of RPM-1 is the identification of a critical role for the conserved PHR domains. RPM-1 lacking the PHR domains is retained in cell bodies, whereas either PHR domain is sufficient to target GFP to regions close to where endogenous RPM-1 resides. This is the first function to be attributed to the PHR domain in any PHR family member. It is possible that the PHR domain may mediate the sorting of RPM-1 into proper carriers that are destined to synapses. Alternatively, the PHR domain may mediate RPM-1 interaction with other synaptic components to stabilize the localization of RPM-1 at synapses. The PHR domain is only found among the PHR proteins and is moderately conserved in another family of proteins known as BTBD proteins, which also contain BTB domains (Supplemental Fig. 2). Most of the BTBD proteins are uncharacterized, although the PHR domain in BTBD1 and BTBD2 mediates their binding to other proteins (Xu et al., 2003). It has been hypothesized that PHR domain may represent a novel type of β-propeller structure (Stogios et al., 2005). We did not detect homo-dimerization for the PHR domain in a yeast two-hybrid assay (B.A. and Y.J., unpublished). The isolation of PHR binding partners will provide clues into how the PHR domain mediates RPM-1 localization at the synapse, and what the identity of other players in synapse formation might be.
Supplementary Material
RPM-1 localization is normal in mutants that have altered synaptic morphology. Shown are confocal images of dorsal cord co-stained with RPM-1 (green) and RIM (red). (a) Wild type (N2). (b) syd-2(ju37) (c) syd-1(ju82) (d) sad-1(ju53) (e) syd-1(ju82);syd-2(ju37) (f) syd-2(ju37)sad-1(ju53) (g) unc-26 (e1196). In syd-1 and syd-2 mutants, RIM expression is diffuse and reduced in intensity (Hallam et al., 2002; Ackley et al., 2005), but RPM-1 expression is normal. Scale bars, 5 µm, same for all panels.
The PHR domain shows moderate conservation in a family of BTB domain containing proteins. 7.3% of amino acid residues are identical and 19.7% of amino acid residues are similar. (a) A CLUSTAL alignment of the PHR region of human PAM (AAC39928), mouse Phr1(NP_997098), zebrafish Esrom (NP_001012247), Drosophila Hiw (NP_511159), and C. elegans RPM-1 (NP_505267) and BTB domain containing proteins from rat (NP_001011932), mouse (NP_666305), human (NP_079514), and fish (CAI21055). (b) Phylogram showing grouping of PHR1 domains, PHR2 domains and BTB domain containing proteins.
The RPM-1 antibody is specific. Confocal images show RPM-1 and RIM co-staining in rpm-1(ju44) animals. (a–b) are positive for RIM staining, but negative for RPM-1 in the nerve ring (a), and SAB neuron (b). Animals were raised at the restrictive temperature (25° C). Regions shown are equivalent to those in figure 1 c, d. Scale bars: 10 µm, a. 5 µm b.
Staining of DLK-1::GFP and anti-RIM in sub-lateral cords. RPM-1 and DLK-1 puncta have similar distributions relative to RIM in the sub-lateral cord. Shown are confocal images of sub-lateral cord co-stained with RPM-1 (green) or DLK-1 (green) and RIM (red). (a) Wild type (N2). (b) DLK-1::GFP(juIs192). Scale bars, 5 µm, same for all panels.
Acknowledgments
We would like to thank Mike Nonet for kindly providing anti-RIM and anti-SNT-1 antibodies, Jason McEwen for juIs83, Brian Ackley for confocal training, Heather Van Epps for help with the manuscript. This work was supported by an NIH grant (NS035546) to Y.J.. Y.J. is an investigator of Howard Hughes Medical Institute.
References
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The Two Isoforms of the Caenorhabditis elegans Leukocyte-Common Antigen Related Receptor Tyrosine Phosphatase PTP-3 Function Independently in Axon Guidance and Synapse Formation. J. Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RW, Peterson KA, Johnson MJ, Roix JJ, Welsh IC, O'Brien TP. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–1105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Crump JG, Zhen M, Jin Y, Bargmann CI. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 2001;29:115–129. doi: 10.1016/s0896-6273(01)00184-2. [DOI] [PubMed] [Google Scholar]
- Daviau A, Proulx R, Robitaille K, Di Fruscio M, Tanguay RM, Landry J, Patterson C, Durocher Y, Blouin R. Down-regulation of the Mixed-lineage Dual Leucine Zipper-bearing Kinase by Heat Shock Protein 70 and Its Co-chaperone CHIP. J. Biol. Chem. 2006;281:31467–31477. doi: 10.1074/jbc.M607612200. [DOI] [PubMed] [Google Scholar]
- D'Souza J, Hendricks M, Le Guyader S, Subburaju S, Grunewald B, Scholich K, Jesuthasan S. Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc) Development. 2005;132:247–256. doi: 10.1242/dev.01578. [DOI] [PubMed] [Google Scholar]
- Ehnert C, Tegeder I, Pierre S, Birod K, Nguyen H-V, Schmidtko A, Geisslinger G, Scholich K. Protein associated with Myc (PAM) is involved in spinal nociceptive processing. J. Neurochem. 2004;88:948–957. doi: 10.1046/j.1471-4159.2003.02229.x. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Goncharov A, McEwen J, Baran R, Jin Y. SYD-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains, specifies axon identity in C. elegans. Nat Neurosci. 2002;5:1137–1146. doi: 10.1038/nn959. [DOI] [PubMed] [Google Scholar]
- Harris TW, Hartwieg E, Horvitz HR, Jorgensen EM. Mutations in synaptojanin disrupt synaptic vesicle recycling. J Cell Biol. 2000;150:589–600. doi: 10.1083/jcb.150.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue E, Mochida S, Takagi H, Higa S, Deguchi-Tawarada M, Takao-Rikitsu E, Inoue M, Yao I, Takeuchi K, Kitajima I. SAD: A Presynaptic Kinase Associated with Synaptic Vesicles and the Active Zone Cytomatrix that Regulates Neurotransmitter Release. Neuron. 2006;50:261–275. doi: 10.1016/j.neuron.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science. 2005;307:929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Shurland DL, van der Bliek AM. Contribution of the GTPase domain to the subcellular localization of dynamin in the nematode Caenorhabditis elegans. Mol Biol Cell. 1998;9:3227–3239. doi: 10.1091/mbc.9.11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader S, Maier J, Jesuthasan S. Esrom, an ortholog of PAM (protein associated with c-myc), regulates pteridine synthesis in the zebrafish. Dev Biol. 2005;277:378–386. doi: 10.1016/j.ydbio.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The Ubiquitin Ligase Phr1 Regulates Axon Outgrowth through Modulation of Microtubule Dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- McCabe BD, Hom S, Aberle H, Fetter RD, Marques G, Haerry TE, Wan H, O'Connor MB, Goodman CS, Haghighi AP. Highwire Regulates Presynaptic BMP Signaling Essential for Synaptic Growth. Neuron. 2004;41:891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V, Han S, Beauchamp RL, Smith N, Haddad LA, Ito N, Ramesh V. Pam and Its Ortholog Highwire Interact with and May Negatively Regulate the TSC1-TSC2 Complex. J. Biol. Chem. 2004;279:1351–1358. doi: 10.1074/jbc.M310208200. [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Piechotta K, Dudanova I, Missler M. The resilient synapse: insights from genetic interference of synaptic cell adhesion molecules. Cell Tissue Res. 2006;326:617–642. doi: 10.1007/s00441-006-0267-4. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Scholich K, Pierre S, Patel TB. Protein associated with Myc (PAM) is a potent inhibitor of adenylyl cyclases. J Biol Chem. 2001;276:47583–47589. doi: 10.1074/jbc.M107816200. [DOI] [PubMed] [Google Scholar]
- Sone M, Suzuki E, Hoshino M, Hou D, Kuromi H, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C. Synaptic development is controlled in the periactive zones of Drosophila synapses. Development. 2000;127:4157–4168. doi: 10.1242/dev.127.19.4157. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:15. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The Synaptic Vesicle Cycle. Annual Review of Neuroscience. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Weimer RM, Gracheva EO, Meyrignac O, Miller KG, Richmond JE, Bessereau J-L. UNC-13 and UNC-10/Rim Localize Synaptic Vesicles to Specific Membrane Domains. J. Neurosci. 2006;26:8040–8047. doi: 10.1523/JNEUROSCI.2350-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White YG, Southgate E, Thomson JN, Brenner S. The Structure of the Nervous System of the Nematode Caenorhabditis elegans. Phil. Trans Royal Soc. London. Series B, Biol Scien. 1986;314 doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wu C, Daniels RW, Diantonio A. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Develop. 2007;2:16. doi: 10.1186/1749-8104-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Wairkar YP, Collins CA, DiAntonio A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J Neurosci. 2005;25:9557–9566. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, Kaul S, Meroni G, Jensen JP, Weissman AM, D'Arpa P. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- Zhao H, Nonet ML. A retrograde signal is involved in activity-dependent remodeling at a C. elegans neuromuscular junction. Development. 2000;127:1253–1266. doi: 10.1242/dev.127.6.1253. [DOI] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Zhen M, Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci. 2004;5:385–399. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RPM-1 localization is normal in mutants that have altered synaptic morphology. Shown are confocal images of dorsal cord co-stained with RPM-1 (green) and RIM (red). (a) Wild type (N2). (b) syd-2(ju37) (c) syd-1(ju82) (d) sad-1(ju53) (e) syd-1(ju82);syd-2(ju37) (f) syd-2(ju37)sad-1(ju53) (g) unc-26 (e1196). In syd-1 and syd-2 mutants, RIM expression is diffuse and reduced in intensity (Hallam et al., 2002; Ackley et al., 2005), but RPM-1 expression is normal. Scale bars, 5 µm, same for all panels.
The PHR domain shows moderate conservation in a family of BTB domain containing proteins. 7.3% of amino acid residues are identical and 19.7% of amino acid residues are similar. (a) A CLUSTAL alignment of the PHR region of human PAM (AAC39928), mouse Phr1(NP_997098), zebrafish Esrom (NP_001012247), Drosophila Hiw (NP_511159), and C. elegans RPM-1 (NP_505267) and BTB domain containing proteins from rat (NP_001011932), mouse (NP_666305), human (NP_079514), and fish (CAI21055). (b) Phylogram showing grouping of PHR1 domains, PHR2 domains and BTB domain containing proteins.
The RPM-1 antibody is specific. Confocal images show RPM-1 and RIM co-staining in rpm-1(ju44) animals. (a–b) are positive for RIM staining, but negative for RPM-1 in the nerve ring (a), and SAB neuron (b). Animals were raised at the restrictive temperature (25° C). Regions shown are equivalent to those in figure 1 c, d. Scale bars: 10 µm, a. 5 µm b.
Staining of DLK-1::GFP and anti-RIM in sub-lateral cords. RPM-1 and DLK-1 puncta have similar distributions relative to RIM in the sub-lateral cord. Shown are confocal images of sub-lateral cord co-stained with RPM-1 (green) or DLK-1 (green) and RIM (red). (a) Wild type (N2). (b) DLK-1::GFP(juIs192). Scale bars, 5 µm, same for all panels.