Abstract
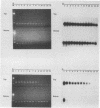
Specific diagnostic test results generated by polymerase chain reaction (PCR) depend upon control of amplicon contamination in the clinical laboratory. We compared photochemical (isopsoralen [IP]) and enzymatic (uracil N-glycosylase [UNG]) methods for their ability to prevent carryover of amplicons generated from genomic targets of five viruses. PCR products (amplicons) (herpes simplex virus, 342 bp; cytomegalovirus, 250 bp; Epstein-Barr virus, 240 bp) exposed to UV light in the presence of various concentrations of IP compound 10 (IP-10) resulted in apparent increased molecular sizes of the products, as indicated by migration patterns after gel electrophoresis, and were predictive of inactivation by the agent. For amplicons of < or = 100 bp, IP-10-induced electrophoretic shifts were related to the guanidine-cytidine (G + C) content of the PCR product; no apparent shift and no inactivation were observed for a 92-bp herpes simplex virus amplicon (G + C content, 65%), whereas the 100-bp human papillomavirus product (G + C content, 42%) showed a concentration-dependent shift (25 to 100 micrograms/ml) in electrophoretic migration and was partially inactivated. UNG effectively controlled amplicon carryover for target DNA of > or = 240 bp; however, this treatment did not inactivate the two amplicons of < or = 100 bp, regardless of the G + C content of the product. Larger products were inactivated efficiently by both methods, regardless of their G + C contents. We concluded that both IP and UNG effectively inactivated PCR amplicons but not short amplicons of < or = 100 bp. We recommend that with the adoption of PCR technology in clinical laboratories, primers should be designed to produce amplicons of at least 240 to 350 bp (depending on G + C content) and that at least one effective method of controlling carryover contamination should be incorporated into each PCR protocol.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aslanzadeh J., Helm K. F., Espy M. J., Muller S. A., Smith T. F. Detection of HSV-specific DNA in biopsy tissue of patients with erythema multiforme by polymerase chain reaction. Br J Dermatol. 1992 Jan;126(1):19–23. doi: 10.1111/j.1365-2133.1992.tb08397.x. [DOI] [PubMed] [Google Scholar]
- Aslanzadeh J., Osmon D. R., Wilhelm M. P., Espy M. J., Smith T. F. A prospective study of the polymerase chain reaction for detection of herpes simplex virus in cerebrospinal fluid submitted to the clinical virology laboratory. Mol Cell Probes. 1992 Oct;6(5):367–373. doi: 10.1016/0890-8508(92)90029-w. [DOI] [PubMed] [Google Scholar]
- Cimino G. D., Gamper H. B., Isaacs S. T., Hearst J. E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Cimino G. D., Metchette K. C., Tessman J. W., Hearst J. E., Isaacs S. T. Post-PCR sterilization: a method to control carryover contamination for the polymerase chain reaction. Nucleic Acids Res. 1991 Jan 11;19(1):99–107. doi: 10.1093/nar/19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. C., Ait-Khaled M., Webster A., Emery V. C. Eliminating PCR contamination: is UV irradiation the answer? J Virol Methods. 1991 Aug;33(3):375–382. doi: 10.1016/0166-0934(91)90037-z. [DOI] [PubMed] [Google Scholar]
- Isaacs S. T., Tessman J. W., Metchette K. C., Hearst J. E., Cimino G. D. Post-PCR sterilization: development and application to an HIV-1 diagnostic assay. Nucleic Acids Res. 1991 Jan 11;19(1):109–116. doi: 10.1093/nar/19.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S., Higuchi R. Avoiding false positives with PCR. Nature. 1989 May 18;339(6221):237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- Longo M. C., Berninger M. S., Hartley J. L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990 Sep 1;93(1):125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- Persing D. H. Polymerase chain reaction: trenches to benches. J Clin Microbiol. 1991 Jul;29(7):1281–1285. doi: 10.1128/jcm.29.7.1281-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rys P. N., Persing D. H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J Clin Microbiol. 1993 Sep;31(9):2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Shedding light on PCR contamination. Nature. 1990 Jan 4;343(6253):27–27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- Syvänen A. C. From one to millions--the polymerase chain reaction in diagnosis. Ann Med. 1992 Jun;24(3):179–182. [PubMed] [Google Scholar]
- Tompkins L. S. The use of molecular methods in infectious diseases. N Engl J Med. 1992 Oct 29;327(18):1290–1297. doi: 10.1056/NEJM199210293271808. [DOI] [PubMed] [Google Scholar]