Abstract
The two living groups of flying vertebrates, birds and bats, both have constricted genome sizes compared with their close relatives. But nothing is known about the genomic characteristics of pterosaurs, which took to the air over 70 Myr before birds and were the first group of vertebrates to evolve powered flight. Here, we estimate genome size for four species of pterosaurs and seven species of basal archosauromorphs using a Bayesian comparative approach. Our results suggest that small genomes commonly associated with flight in bats and birds also evolved in pterosaurs, and that the rate of genome-size evolution is proportional to genome size within amniotes, with the fastest rates occurring in lineages with the largest genomes. We examine the role that drift may have played in the evolution of genome size within tetrapods by testing for correlated evolution between genome size and body size, but find no support for this hypothesis. By contrast, we find evidence suggesting that a combination of adaptation and phylogenetic inertia best explains the correlated evolution of flight and genome-size contraction. These results suggest that small genome/cell size evolved prior to or concurrently with flight in pterosaurs. We predict that, similar to the pattern seen in theropod dinosaurs, genome-size contraction preceded flight in pterosaurs and bats.
Keywords: genome size, palaeogenomics, pterosaurs
1. Introduction
Disciplines ranging from morphology and histology to biomechanics have supplied many insights into pterosaur biology (Padian et al. 2004). Pterosaurs were the first vertebrate group to evolve powered flight, over 70 Myr before birds and over 150 Myr before bats (Unwin 2003). However, studies into the genetic and genomic characteristics associated with flight and endothermy, among other traits, have focused necessarily on living species of bats and birds. For example, both birds and bats appear to have smaller genome sizes compared with their close relatives (Hughes & Hughes 1995). This pattern led to the hypothesis that genome size was under selective pressure to contract and remain small due to constraints that an elevated endothermic metabolism placed on cell size (Olmo 1983; Szarski 1983). We explore this problem by estimating genome size for four species of pterosaurs and analysing genome-size evolution in all three groups of flying vertebrates.
2. Material and methods
We combined data from several studies (see the electronic supplementary material) and added osteocyte lacunae size and genome-size data for nine additional living species (see table 1 in the electronic supplementary material). Osteocyte lacunae were also measured from palaeohistological samples of four pterosaur species and seven extinct basal archosauromorphs species.
Genome sizes, obtained from the Animal Genome Size Database (www.genomesize.com), were averaged for species with multiple entries. Genome-size and cell-size data were log transformed to accommodate the large range of values in the dataset. To test whether non-adaptive or neutral forces have played a large role in genome evolution within tetrapods we followed the protocol of Lynch (2007) and generated a regression model for genome size and body size for 87 extant species.
Phylogenetic trees were created in Mesquite v. 2.01 using the StratAdd package to date nodes according to the geological time scale. Data were analysed using a Markov chain Monte Carlo approach that normalizes trait data by their shared evolutionary history. To detect proportional evolution, the Pdap package for Mesquite was used to estimate node values and standardized contrasts (Oliver et al. 2007; see the electronic supplementary material for additional details).
3. Results
We found that bats evolved smaller sized genomes compared with other mammals when we controlled for phylogeny (phylogenetic t-test (PTT) of genome size in picograms, p=0.028, n=39). Although birds have the smallest amniote genomes, we find that contrary to previous reports (Hughes & Hughes 1995), when the data are normalized for phylogenetic relatedness, flightless birds have not secondarily evolved larger genomes than other birds (PTT, p=0.33, n=16). Previous work on birds has shown that small genome size evolved preceding flight in non-avian dinosaurian ancestors (Organ et al. 2007).
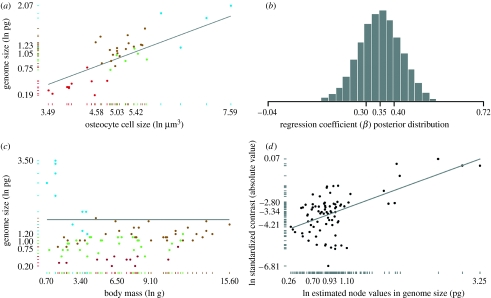
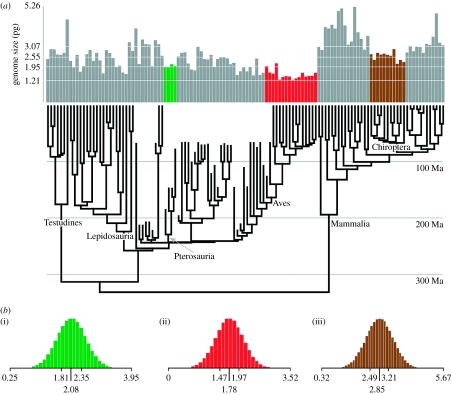
Consistent with previous research (Cavalier-Smith 1985), our analysis supports the hypothesized relationship between cell size and genome size (figure 1a,b). The inferred average haploid genome size for pterosaurs was estimated to be 2.0 pg (σ=0.08; see table 3 in the electronic supplementary material). The average haploid genome size for basal archosauromorphs was estimated to be 3.07 pg with a wider posterior distribution (σ=0.75). These estimates place pterosaur genome sizes within the upper quartile of the avian range (figure 2a) and the basal archosauromorphs (not shown) close to the average for living crocodylians. The analyses also suggest that pterosaurs evolved genomes smaller than their relatives (PTT, p=0.026, n=13). Adding the inferred genome sizes reported here to a larger dataset (130 tetrapod taxa in total), we find that the rate of genome-size evolution was proportional to genome size within tetrapods over long periods of geological time (figure 1d).
Figure 1.
Genome size regression models. (a) The phylogenetic generalized least-squares (PGLS) regression line relating genome size to bone cell size in 38 extant tetrapod species is drawn from the average of a Bayesian posterior distribution of regression models and takes the form (ln genome size)=−0.81+0.35(ln cell size), r2=0.43 (p<0.0001, H0: β=0). The axes denote the distributions of the x and y data, with labels marking the minimum, 25 per cent quartile, median, 75 per cent quartile, and the maximum. (b) The Bayesian posterior distribution of β, the regression coefficient relating genome size to cell size (mean=0.35, σ=0.07). (c) The maximum-likelihood PGLS line relating genome size to body mass in 87 extant tetrapods takes the form (ln genome size)=1.64+0.00003(ln body size), r2=0.0 (p=0.94, H0: β=0). (d) The regression line relating estimated node values to the amount of change in immediate descendants (standardized contrasts) in 130 tetrapods. It takes the form ln(standardized contrasts)=−4.93+1.53 ln(estimated node values in genome size), r2=0.32 (p<0.001, H0: β=0). Coloured markers in (a,c) denote amphibians (blue), mammals (brown), non-avian reptiles (green), and birds (red).
Figure 2.
Genome-size evolution in birds, bats and pterosaurs. (a) Phylogeny and bar graph of average genome size in picograms including the inferred genome size of non-avian dinosaurs, basal archosaurs and pterosaurs (green bars), as well as birds (red bars) and bats (brown bars). Estimates of genome size for extinct dinosaurs are re-estimates of Organ et al. (2007). (b) Posterior distributions of ancestral genome size in picograms for (i) pterosaurs (median=2.08, σ=0.40), (ii) birds (Aves; median=1.72, σ=0.38) and (iii) bats (Chiroptera; median=2.85, σ=0.54). The axes in both panels are labelled with the minimum, 25 per cent quartile, median, 75 per cent quartile, and the maximum of the distributions.
All three amniote groups to have evolved flight (birds, bats and pterosaurs) have also evolved smaller genomes compared with terrestrial amniotes (PTT, p=0.042, n=122). This test was highly significant under an ordinary t-test (p<0.0001, n=122). Ancestral state reconstructions of genome size (figure 2b) were also estimated. The results of this analysis suggest that the ancestral genome size in bats (average of the posterior distribution) was 2.85 pg (σ=0.54), in birds was 1.72 pg (σ=0.38) and in pterosaurs was 2.08 pg (σ=0.09) (figure 2b).
We found no significant relationship between body size and genome size in tetrapods (PGLS regression, r2=0.0, p=0.94, n=87; figure 1c). The relationship also remained insignificant when amphibians were removed from the dataset (PGLS regression, r2=0.005, p=0.52). These results suggest, assuming that body size is a rough inverse proxy for population size (following the protocol of Lynch 2007), that drift does not solely explain the patterns of genome diversity in tetrapods.
4. Discussion
Based on their phylogenetic position and estimated genome size, we would expect active and extinct transposable elements in the chicken repeat 1 family of long interspersed nuclear elements and mammalian-wide interspersed repeat short interspersed nuclear elements to comprise the largest repetitive fraction of pterosaur genomes (Shedlock et al. 2007). Furthermore, our finding that the rate of genome-size evolution is proportional to genome size within amniotes supports recent work (Oliver et al. 2007), where larger genomes were found to evolve at faster rates. The proportional model accounts for genome-size lability, suggesting that lineages with larger genomes may more readily respond to changes in selection pressures on genome size and is not inconsistent with selection, in part, shaping genome-size evolution (discussed later). The proportional model of genome-size evolution also suggests that genome contraction evolved independently in pterosaurs and theropods, because it would be unlikely for a common ancestor of dinosaurs and pterosaurs with small genome size to give rise to the larger genomes estimated for ornithischian dinosaurs.
Population-level dynamics may play a large role in shaping many aspects of genome architecture (Lynch 2007). Small population sizes are predicted to reduce the efficiency of selection, increasing the likelihood of drift to increase genome size by the accumulation of mildly deleterious mutations, such as interspersed repeats. Previous work on neutral genome evolution (Lynch 2007) has focused on differences among population sizes spanning 20 orders of magnitude (prokaryotes versus eukaryotes), and although such differences are absent within tetrapods the neutral hypothesis should still be evaluated. Our results do not support the neutral hypothesis to account for patterns of genome-size variation within extant tetrapods and by extension genome-size variation within extinct tetrapods, including pterosaurs.
Furthermore, genome size within theropod dinosaurs remained relatively static (Organ et al. 2007), despite large shifts in body size in non-avian dinosaurs. The non-adaptive theory of genome evolution predicts that, compared with those of ornithischian dinosaurs, genome sizes would be larger in the smaller populations of carnivorous non-avian theropods where genetic drift should dominate under conditions of reduced selection efficiency, yet the reverse appears to be the case (Organ et al. 2007). Although these results do not match predictions made by neutral models of genome evolution, they do not rule out the importance of non-adaptive forces, such as genetic drift, on the evolution of genome size in tetrapods or the critical value in generating null hypotheses.
Correlated evolution between small genome size and flight in amniotes suggests that adaptation links these two characters. Although the analyses presented here do not favour drift solely as an explanation for the evolution of genome size in amniotes, ‘phylogenetic inertia’ (defined as the combined influence of genetic, phylogenetic, and adaptive constraints) could explain the observed variation (Hansen & Orzack 2005). The parameter λ describes the amount of genome-size covariation among species predicted by the phylogeny. When λ is high (close to one) and the slope of the regression line (β) supports the hypothesis (that there is correlated evolution between small genome size and flight), both current adaptation and inertia have influenced their coevolution (Hansen & Orzack 2005). In our analysis, λ has an estimated value of 1 (p<0.0001; H0: λ=0), which suggests that both adaptation and inertia best explain the correlated evolution of genome size and flight.
Well-known mechanisms, such as illegitimate recombination, can account for substantial changes of genome size within populations on which selection might act (Biémont 2008). Reliant on such mechanisms, the nucleotypic theory (Bennett 1971) addresses the relationship between genome size (and hence nucleus size) and cell size, but also states that the genome influences the phenotype independently of its information content. For example, cell size affects housekeeping dynamics, cellular metabolism and the rate of cell division, among other cytological traits (Kozlowski et al. 2003). Metabolic intensity has recently been tightly linked with genome size through relative heart mass (Vinogradov & Anatskaya 2006) and wing shape in birds (Andrews et al. 2009), and we hypothesize that a metabolic intensity required for flight, not flight itself, explains the correlated evolution between genome size and flight in amniotes. This hypothesis also predicts that pterosaurs possessed some form of elevated metabolism, consistent with other hypotheses about pterosaur biology (Padian et al. 2004). Based on the pattern seen in theropod dinosaurs, where small genomes evolved before flight, we hypothesize that genome-size contraction also preceded flight in pterosaurs and bats.
Acknowledgments
We are grateful to Rebecca Homan, Michael Reed, Fuzz Crompton and Jack Horner for access to histological slides, and Charles Marshall, Nicole Hobbs, Dave Blackburn, Michel Laurin, David Marjanovic, Steven Orzack, Christopher Bennett and an anonymous referee for comments. This research was supported by an NIH NSRA Fellowship granted to C.L.O.
Supplementary Material
data matrices, phylogenetic trees, and additional details of analysis
References
- Andrews C.B., Mackenzie S.A., Gregory T.R. Genome size and wing parameters in passerine birds. Proc. R. Soc. B. 2009;276:55–61. doi: 10.1098/rspb.2008.1012. doi:10.1098/rspb.2008.1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.D. The duration of meiosis. Proc. R. Soc. B. 1971;178:277–299. doi:10.1098/rspb.1971.0066 [Google Scholar]
- Biémont C. Within-species variation in genome size. Heredity. 2008;101:297–298. doi: 10.1038/hdy.2008.80. doi:10.1038/hdy.2008.80 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell volume and the evolution of eukaryotic genome size. In: Cavalier-Smith T., editor. The evolution of genome size. Wiley; Chichester, UK: 1985. pp. 104–184. [Google Scholar]
- Hansen T.F., Orzack S.H. Assessing current adaptation and phylogenetic inertia as explanations of trait evolution: the need for controlled comparisons. Evolution. 2005;59:2063–2072. doi:10.1111/j.0014-3820.2005.tb00917.x [PubMed] [Google Scholar]
- Hughes A.L., Hughes M.K. Small genomes for better flyers. Nature. 1995;377:391. doi: 10.1038/377391a0. doi:10.1038/377391a0 [DOI] [PubMed] [Google Scholar]
- Kozlowski J., Konarzewski M., Gawelczyk A.T. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc. Natl Acad. Sci. USA. 2003;100:14 080–14 085. doi: 10.1073/pnas.2334605100. doi:10.1073/pnas.2334605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. (ed.) 2007 The origins of genome architecture Sunderland, MA: Sinauer Associates.
- Oliver M.J., Petrov D., Ackerly D., Falkowski P., Schofield O.M. The mode and tempo of genome size evolution in eukaryotes. Genome Res. 2007;17:594–601. doi: 10.1101/gr.6096207. doi:10.1101/gr.6096207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E. Nucleotype and cell size in vertebrates: a review. Basic Appl. Histochem. 1983;27:227–256. [PubMed] [Google Scholar]
- Organ C.L., Shedlock A.M., Meade A., Pagel M., Edwards S.V. Origin of avian genome size and structure in nonavian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. doi:10.1038/nature05621 [DOI] [PubMed] [Google Scholar]
- Padian K., Horner J.R., de Ricqlès A.J. Growth in small dinosaurs and pterosaurs: the evolution of archosaurian growth strategies. J. Vert. Paleontol. 2004;24:555–571. doi:10.1671/0272-4634(2004)024[0555:GISDAP]2.0.CO;2 [Google Scholar]
- Shedlock A.M., Botka C.W., Zhao S., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V. Phylogenomics of non-avian reptiles and the structure of the ancestral amniote genome. Proc. Natl Acad. Sci. USA. 2007;104:2767–2772. doi: 10.1073/pnas.0606204104. doi:10.1073/pnas.0606204104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H. Cell size and the concept of wasteful and frugal evolutionary strategies. J. Theor. Biol. 1983;105:201–209. doi: 10.1016/s0022-5193(83)80002-2. doi:10.1016/S0022-5193(83)80002-2 [DOI] [PubMed] [Google Scholar]
- Unwin, D. M. 2003 On the phylogeny and evolutionary history of pterosaurs. In Evolution and palaeobiology of pterosaurs (eds E. Buffetaut & J.-M. Mazin). Geological Society of London Special Publications 217, 139–190.
- Vinogradov A.E., Anatskaya O.V. Genome size and metabolic intensity in tetrapods: a tale of two lines. Proc. R. Soc. B. 2006;273:27–32. doi: 10.1098/rspb.2005.3266. doi:10.1098/rspb.2005.3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
data matrices, phylogenetic trees, and additional details of analysis