Abstract
Most biomedical neuroscientists realize the importance of the study of brain evolution to help them understand the differences and similarities between their animal model of choice and the human brains in which they are ultimately interested. Many think of evolution as a linear process, going from simpler brains, as those of rats, to more complex ones, as those of humans. However, in reality, every extant species' brain has undergone as long a period of evolution as has the human brain, and each brain has its own species-specific adaptations. By understanding the variety of existing brain types, we can more accurately reconstruct the brains of common ancestors, and understand which brain traits (of humans as well as other species) are derived and which are ancestral. This understanding also allows us to identify convergently evolved traits, which are crucial in formulating hypotheses about structure–function relationships in the brain. A thorough understanding of the processes and patterns of brain evolution is essential to generalizing findings from ‘model species’ to humans, which is the backbone of modern biomedical science.
Keywords: scala naturae, brain evolution, neurocladistics, biomedical research, animal models, comparative neuroscience
1. Introduction
[Understanding brain evolution does not affect my research…] …[b]ecause [my research] is very much concerned about the way that neuronal systems operate NOW in normal subjects and those that suffer with affective disorders. How the system evolved to what it is, is irrelevant. (Anonymous, survey respondent working with rats)
To those who work on brain evolution and comparative neuroscience, the above quote represents how most biomedical researchers are thought to look at the study of brain evolution: ‘It may all be very interesting, but it does not really affect my research’. However, in a small anonymous survey of 47 neuroscientists in the UK, which I recently conducted through the British Neuroscience Association, this opinion was voiced by only a small minority. Thirty-nine respondents, among whom only three did not work in typical biomedical models, claimed that understanding brain evolution did indeed affect their own research. Of course, the sample of people who will take the time to respond to a survey about ‘brain evolution’ will be biased towards those who have opinions about brain evolution. Nevertheless, this suggests that evolutionary neuroscientists may have to rethink the biases that other neuroscientists just do not care about brain evolution: they clearly do.
When one gives it a bit more thought, it is not completely surprising that biomedical researchers care about brain evolution. After all, the whole concept of model animal research is based on the assumption that we share common ancestry with other animals (Preuss 2000). Therefore, there should be enough similarities in the ways our bodies (and nervous systems) work, to make research on these animals informative about humans. Indeed, Preuss (2004) has argued that most biomedical researchers stress similarities to the point of forgetting that there is another side to the evolutionary coin: the differences that make each species unique. However, contrary to Preuss's (2004) claim, in my small survey, I did not find this overwhelming emphasis on similarities alone. Across the respondents, arguments about similarities were approximately equally frequent as arguments about differences. Indeed, I believe that most biomedical researchers who use animals for their experiments are very well aware that these are not just smaller, simpler human beings, but that any findings need to be verified in humans.
It is clear, therefore, that neuroscientists care about evolution, because it helps us understand both similarities and differences between humans and typical laboratory animals. This focus on humans, however, creates the problem that most neuroscientists see brain evolution fairly narrowly as a process leading (up) to human brains. This leads to thinking of evolution as a linear process, from simpler to more complex forms, with humans at the pinnacle (Shimizu 2004). Of the 29 respondents who made relevant comments, 23 implied they thought of evolution as a mostly linear process leading up to humans. It is this general misconception that I will be addressing in the rest of this essay. For similar, often more in-depth arguments, see also Preuss (2000), Shimizu (2004) and Striedter (2004).
2. Understanding ancestral brains
It is not strictly speaking incorrect to speak about an evolutionary process leading to human brains. After all, there is a direct line from our ancestors' brains to our current brains. However, it is incorrect to assume a priori that this process was unidirectional. For example, there is no evidence to suggest that the brains of our ancestors (all our ancestors, not just the most recent ones in the hominid line) have always increased in size over time. It is very possible that during some stage of our evolution from the earliest mammals, brain sizes decreased. The same arguments can be made about complexity or any other trait one might choose to measure brain organization. Another problem with many people's views of brain evolution is that they assume that we can use other extant species as proxies for our ancestors (Shimizu 2004). However, any species alive today has undergone evolution for the same amount of time as have humans. To quote another respondent in the survey: ‘It may be worth remembering that a hedgehog's brain has taken just as many million years to evolve as ours!! Hedgehogs didn't stop and watch the evolution of primates.’ Therefore the same reasoning about a direct line leading from ancestral to modern human brains can be used for every other extant animal species. Each species has undergone its own brain evolution and will have its own species-specific brain adaptations, which are potentially as different from those of our shared common ancestor as our brains are.
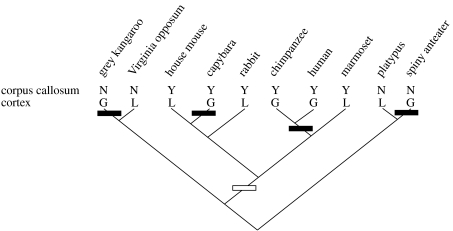
Therefore, if we want to understand the appearance of our ancestors' brains, we need to somehow reconstruct them. Fossils can contribute to this endeavour a little bit, through the use of endocasts from skulls (Wu et al. 2007). But most of the structural and functional elements of brains are lost in the fossil record. The best-established method for reconstructing ancestral form and function from available evidence is cladistics (Hennig 1966; Northcutt 1984; reviewed in Striedter 2004). Using an established phylogenetic tree or cladogram, based on non-neural characters, cladistic analysis can reconstruct what the traits of putative ancestors were by mapping traits of extant species onto this cladogram (figure 1). To do this, it makes the assumption that these traits should have undergone a minimum number of possible changes over evolutionary time (the principle of parsimony).
Figure 1.
A small selection of extant mammal species arranged on a cladogram to indicate their evolutionary relationships. The line labelled ‘corpus callosum’ indicates whether each species has a corpus callosum (Y) or not (N), while the line labelled ‘cortex’ indicates either gyrencephalic (G: folded cortex) or lissencephalic (L: smooth cortex) brains. The points on the cladogram where each of the two traits evolved are indicated by rectangles (black: gyrencephaly; white: corpus callosum). Information based on Striedter (2004) and http://www.brainmuseum.org.
It should be immediately obvious then that one needs more than two extant species in order to be able to deduce whether a trait is ancestral or derived. When you only have information on two species (e.g. a mouse and a human), it is impossible to tell whether a difference between these two species is due to the human having the derived trait and the mouse the ancestral trait (as most biomedical researchers would assume), the human having the ancestral trait and the mouse the derived trait, or indeed both having different derived traits and the common ancestor another trait altogether. At the least one needs a third species (typically called the ‘out-group’), which is from a lineage that diverged from the lineage of interest before the evolution of the ancestor of interest. If this out-group shares the trait with one of the two extant species, then this trait is more likely to be the trait of the ancestor as well, as that would only assume one evolutionary change in this trait, and hence be the more parsimonious explanation. The more species we have information for, the clearer the patterns usually get. This is a crucial insight for the use of model animals in the endeavour to understand the human brain (Preuss 2000).
3. Understanding structure–function relationships
The quote with which I started this article makes the assumption that the study of brain evolution is only about what came before. But brain evolution is also about describing and understanding distribution patterns of traits across extant species. The mapping of traits onto cladograms allows us to distinguish between homologous traits and homoplasic traits. Homology refers to similar traits in related species that have been inherited from a common ancestor. These can be identified as traits that are present in all species that share one common ancestor. Examples from brain evolution are laminated cortex in mammals, and the existence of a corpus callosum in placental mammals (figure 1). Homoplasy refers to similar traits that may occur in distantly related species, which are not shared by other more closely related species. Homoplasy is the result of either parallel or convergent evolution. An example of parallel evolution is the evolution of gyri and sulci in the cortex of diverse mammalian lineages (figure 1). Cortex itself is homologous among these lineages, but the fact that it is folded evolved several times. An example of convergent evolution is the evolution of laminated structures in different, non-homologous parts of the brains of vertebrates; e.g. cortex in mammals, optic tectum in birds, and the torus semicircularis in certain fishes. For a detailed overview of all these issues, with many examples, I strongly recommend Striedter's (2004) book.
Biomedical neuroscientists tend to focus on homologies, because if a brain structure in a laboratory animal can be shown to be homologous to a structure in human brains, it might function as a good model for this human structure. However, this is not necessarily the case. Homology can work on many levels, and just because two structures are homologous (e.g. hippocampus in rats and humans), they need not function in exactly the same manner (Shimizu 2004). Whereas homology does not automatically mean identical functioning, non-homology does not necessarily mean that things work differently either. In fact, there is great power in the use of homoplasies for understanding brain function. When one compares just two species, they will differ in many aspects of brain and behaviour. Therefore, ascribing a particular difference in behaviour to a particular difference in the brain is difficult. However, by mapping a number of traits (both behavioural and neural) onto a cladogram, one can check whether trait A and trait B always map together (Smulders 2006). In such an analysis, it is important to look for the theoretical ancestral species in which trait A appears or disappears, and observe whether trait B appears or disappears at the same time. For example, if the evolution of flight always goes together with the evolution of wings, then it is a good bet that wings have something to do with flying.
The same reasoning can be made for neural traits. One example of this is the size of the song system in birds. This is a network of inter-connected brain nuclei that are involved in the learning and production of song (for a recent summary of song system anatomy, see Wild 2004). When mapping repertoire sizes of male songbirds onto a cladogram, it becomes clear that whenever repertoire size becomes larger in evolutionary history, the size of one of the song control nuclei in the brain (nucleus HVC) grows larger as well (DeVoogd et al. 1993; Szekely et al. 1996; DeVoogd 2004), suggesting that HVC size may be important for repertoire size. The same approach could be taken for any other combination of neural and behavioural traits.
4. Implications for biomedical neuroscience
It is clear from the arguments made above that every neuroscientist should be aware of how brains evolve. The current misconceptions are likely due to a lack of education about evolution in the training of most neuroscientists. It is important to know that brain evolution is not a linear event that culminated in the human brain, but instead a dynamic system that has led to the diversity in brains that exists today. Because every species' brain is unique, with its own specific adaptations, it is especially important to be aware of brain evolution when using non-human animals with the aim of understanding the human brain. Only by knowing the pattern across many species can we determine that if the trait is similar, whether it is homologous or homoplastic, or if the trait differs, which of the two lineages has the more derived version of it. Knowing distribution patterns also allows us to map differences in structure onto differences in function, both neural and behavioural, something that would be a pure speculation when based on only two species. Understanding the basics of brain evolution is therefore more than just an intellectually interesting exercise. It is fundamental to any attempt at understanding brain function and to generalizing findings from one animal system to other animal systems (including humans), which is done routinely in biomedical science.
Acknowledgments
I would like to thank two anonymous reviewers for constructive comments on an earlier version of this manuscript. This work was supported by BBSRC grant BB/C006186/1.
Footnotes
One contribution of 10 to a Special Feature on ‘Brain evolution’.
References
- DeVoogd T.J. Neural constraints on the complexity of avian song. Brain Behav. Evol. 2004;63:221–232. doi: 10.1159/000076783. doi:10.1159/000076783 [DOI] [PubMed] [Google Scholar]
- DeVoogd T.J., Krebs J.R., Healy S.D., Purvis A. Relations between song repertoire size and the volume of brain nuclei related to song—comparative evolutionary analyses amongst oscine birds. Proc. R. Soc. B. 1993;254:75–82. doi: 10.1098/rspb.1993.0129. doi:10.1098/rspb.1993.0129 [DOI] [PubMed] [Google Scholar]
- Hennig W. University of Illinois Press; Urbana, IL: 1966. Phylogenetic systematics. [Google Scholar]
- Northcutt R.G. Evolution of the vertebrate cental nervous system: patterns and processes. Am. Zool. 1984;24:701–716. doi:10.1093/icb/24.3.701 [Google Scholar]
- Preuss T.M. Taking the measure of diversity: comparative alternatives to the model-animal paradigm in cortical neuroscience. Brain Behav. Evol. 2000;55:287–299. doi: 10.1159/000006664. doi:10.1159/000006664 [DOI] [PubMed] [Google Scholar]
- Preuss T.M. What is it like to be a human? In: Gazzaniga M.S., editor. The cognitive neurosciences III. MIT Press; Cambridge, MA: 2004. pp. 5–22. [Google Scholar]
- Shimizu T. Comparative cognition and neuroscience: misconceptions about brain evolution. Jpn. Psychol. Res. 2004;46:246–254. doi:10.1111/j.1468-5584.2004.00256.x [Google Scholar]
- Smulders T.V. A multi-disciplinary approach to understanding hippocampal function in food-hoarding birds. Rev. Neurosci. 2006;17:53–69. doi: 10.1515/revneuro.2006.17.1-2.53. [DOI] [PubMed] [Google Scholar]
- Striedter G.F. Sinauer; Sunderland, MA: 2004. Principles of brain evolution. [Google Scholar]
- Szekely T., Catchpole C.K., Devoogd A., Marchl Z., Devoogd T.J. Evolutionary changes in a song control area of the brain (HVC) are associated with evolutionary changes in song repertoire among European warblers (Sylviidae) Proc. R. Soc. B. 1996;263:607–610. doi:10.1098/rspb.1996.0091 [Google Scholar]
- Wild J.M. Functional neuroanatomy of the sensorimotor control of singing. Ann. NY Acad. Sci. 2004;1016:438–462. doi: 10.1196/annals.1298.016. doi:10.1196/annals.1298.016 [DOI] [PubMed] [Google Scholar]
- Wu X., Liu W., Norton C.J. Endocasts—The direct evidence and recent advances in the study of human brain evolution. Prog. Nat. Sci. 2007;17:993–1002. [Google Scholar]