Abstract
Cancer is frequently characterized histologically by the appearance of large cells that are either aneuploid or polyploid. Aneuploidy and polyploidy are hallmarks of radiation-induced mitotic catastrophe (MC), a common phenomenon occurring in tumor cells with impaired p53 function exposed to various cytotoxic and genotoxic agents. MC is characterized by altered expression of mitotic regulators, untimely and abnormal cell division, delayed DNA damage, and changes in morphology. We report here that cells undergoing radiation-induced MC are more plastic with regards to ploidy and that this plasticity allows them to reorganize their genetic material through reduction divisions to produce smaller cells morphologically indistinguishable from control cells. Experiments conducted with the Large Scale Digital Cell Analysis System (LSDCAS) are discussed that show that a small fraction of polyploid cancer cells formed via radiation-induced MC can survive and start a process of depolyploidization that yields various outcomes. While most multipolar divisions failed and cell fusion occurred; some of these divisions were successful and originated a variety of cell progeny characterized by different ploidy. Among these ploidy phenotypes, a progeny of small mononucleated cells, indistinguishable from the untreated control cells, is often seen. We report here evidence that meiosis-specific genes are expressed in the polyploid cells during depolyploidization. Tumor cells might take advantage of the temporary change from a pro-mitotic to a pro-meiotic division regimen to facilitate depolyploidization and restore the proliferative state of the tumor cell population. These events might be mechanisms by which tumor progression and resistance to treatment occur in vivo.
Keywords: radiation, mitotic catastrophe, polyploidy, depolyploidization, Large Scale Digital Cell Analysis System (LSDCAS), meiosis-specific genes, tumor cells, tumor progression, tumor resistance to treatment
Introduction
The fact that drug and/or ionizing radiation exposures trigger cell cycle checkpoints that inhibit entry into or progression through mitosis has been exploited in cancer treatment because these effects are considered major factors in these agents' toxicity (1; 2). However, cells lacking p53 function also lack G2 checkpoint function and will escape these constraints. This is an important fact for cancer treatment outcome because more than 50% of human tumors (especially solid tumors) are p53 non-functional (3; 4), and thus lack G2 checkpoint surveillance, and are often resistant to cytotoxic and genotoxic treatments. Thus, the ultimate goal of eradicating cancer by subjecting patients to surgical intervention and rounds of radiation and/or chemotherapeutic agent treatments might be compromised by the tumor response to these later treatments. Cancer cells are genetically very heterogeneous, containing aneuploid, tetraploid and polyploid populations, and the presence of polyploid cells in malignant tumors has been established in the literature (5; 6), but their origin and biological significance are still elusive. The question of whether selection of polyploid cells during radiation treatment might contribute to the development of resistance to such treatment was posed by Revesz and Norman in a paper published in 1960 (7), and polyploid tumor cells have been found to be more resistant to radiation treatment than their diploid counterpart (8), while various clinical trials carried out on patients treated with radiation therapy and/or chemotherapeutic agents for prostate cancer have pointed out that a high degree of aneuploidy in these patients is associated with reduced overall survival (9; 10). Reversion of extremely hyperploid cancer cells (metaphases with more than 1,000 chromosomes) to smaller pseudo-diploid cells over a period of 12 days was reported by Sandberg and collaborators (6) on peritoneal effusion cells from a patient with aggressive cancer of the colon. These earlier studies have raised questions as to the fate of polyploid tumor cells produced during anti-cancer treatment, the ability of these cells to undergo depolyploidization, and the role of these processes in tumor progression. That normal mammalian cells are able to undergo depolyploidization was reported by Zybina and Zybina in 2005 (11) who showed that whole genome segregation occurs in the giant nuclei (more than 100N) of the trophoblasts (the cells that form the outer layer of the blastocyst and mediate the implantation of the embryo into the endometrium) originating cells containing 2N, 4N and 8N chromosome complements. These results suggest that depolyploidization might be a common phenomenon and that cells may possess an internal control for segregation of entire genomes. Reduction division in somatic cells of higher organisms is considered a rare event (12); nevertheless, this might not be the case in tumor cells where a highly unstable genome is present (13). In fact, high frequency genetic exchange events occurring during reduction division (meiosis) may contribute to a genetic diversity that could render tumor cells more apt to survive after anti cancer treatments.
Depolyploidization in solid and circulating tumors following radiation-induced mitotic catastrophe (MC) (reviewed in ref. 14; 15-18) has been described by us in cytological studies (19), and, although MC is generally lethal (20), we and others have reported that a small fraction of cells which undergo radiation-induced MC and become polyploid can survive long enough to establish a growing population of cells (21-23). In the present paper, by employing the Large Scale Digital Cell Analysis System (LSDCAS), that allows for non-perturbing live cell imaging measurements, we show that polyploid tumor cells formed via radiation-induced MC are able to survive for many days post-irradiation and undergo multipolar divisions. Most of these divisions fail and cell fusion occurs, however, a fraction of these divisions marks the beginning of the process of depolyploidization, giving rise to smaller daughter cells that possess only one nucleus and whose size is indistinguishable from that of the untreated control cells. Some of these mononucleated smaller cells originating through reduction division events give rise to viable descendants. These cells will de facto have a disarrayed genomic composition; nevertheless, abnormal chromosome arrangements attributable to MC may endow the tumor cells with properties that not only differentiate them from normal somatic cells, but may also impart growth advantages over other cells and may make them more resistant to subsequent treatment.
Molecular analysis of the polyploid cells formed via radiation-induced MC demonstrates that meiosis-specific genes are activated during the depolyploidization process. Polyploid cells undergoing depolyploidization also present morphological features similar to those characteristic of meiotic prophase I. Thus, a small percentage of irradiated cells appears to acquire pseudo-meiotic properties enabling them to escape radiation-induced cell death. This fact has implications in the clinical setting; we speculate that tumors in vivo might take advantage of this switch from a pro-mitotic to a pro-meiotic division regimen to balance their disrupted genetic make-up and thus might acquire a more stable growth state and stabilize genetic alterations, eventually becoming resistant to subsequent cytotoxic treatment.
Materials and Methods
Cell Culture
HeLa S3 human cervical cancer cells were grown in Joklik's Minimum Essential Medium (S-MEM) (GIBCO); HeLa clone3 human cervical cancer cells were grown in Ham's F-10 Nutrient Mixture Medium (GIBCO); HCT116 human colon cancer cells were grown in McCoy's 5A Modified Medium (GIBCO); MDA-MB435 human breast cancer/melanoma cells were grown in high glucose Dulbecco's Modified Eagle's Medium (DMEM) (GIBCO). The origin of the latter cell line (MDA-MB435) has been under debate for some time (24, 25) as it is not clear the actual tissue from which it derives. This cell line is still a very good model for solid tumor response to radiation exposure regardless of its origin. We will mention this cell line by only its nomenclature from this point on without specifying the tissue of origin. All media were supplemented with 10% fetal bovine serum (Hyclone) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin; GIBCO). All cell lines were grown in 5% CO2 in air incubators at 37°C.
γ-Irradiation
Gamma-irradiation was delivered at room temperature using an 8,148 Ci 137Cs source at the dose rate of 0.92 Gy min-1.
Immunofluorescence
Harvested cells were suspended in 100% FCS, cytospun onto poly-L-lysine coated microscope slides and fixed either in 4% paraformaldehyde in PBS, pH7.4 (15 min). Slides were then washed twice in PBS at room temperature, and permeabilized for 10 min in PBS containing 0.25% Triton X-100 and then washed in PBS three times for 5 min each time. Slides were subsequently blocked for 30 min in 1% BSA in PBS containing 0.05% Tween-20 (PBST). Fifty μl of the appropriate dilution of antibody were applied to each sample and slides were incubated overnight at 4°C. Samples were then washed three times in PBST for 5 min each time, at which point 50 μl of the appropriate secondary antibody were applied at the appropriate dilutions and slides incubated at room temperature in dark for 1 h, and then washed three times in PBST for 5 minutes each time in the dark. Antibodies were diluted in 1% BSA in PBST. Samples were counterstained and mounted with DAPI Vectashield Mounting Media (Vector Laboratories) and evaluated under an Olympus B51 epifluorescence microscope.
Cytological Preparations
Harvested cells were suspended in 100% FCS, cytospun onto poly-L-lysine coated microscope slides and let dry overnight and fixed in Acetone:100% EtOH, 1:1 (v:v), at 4°C for 30 min. Slides were then allowed to dry for 15 min at r.t. Sample hydrolysis was performed by incubating the slides in 5N HCl at r. t. for 1 min, followed by 5 rinses in distilled H2O for 1 min each. Sample staining was performed on blot-dry slides by incubating the slides in 0.05% Tolouidine Blue in McIlvan's solution (pH 5) for 10 min at r.t. followed by a series of rinses in distilled H2O until removal of excess staining, at which point slides were blot-dry and dehydrated by incubating slides twice in butanol at 37°C for 3 min. Sample clearing was then performed by incubating slides twice in xylene for 3 min each at r. t.. Finally, cover slips were mounted with Cytoseal 60 Mounting Media and slides evaluated under an Olympus B51 epifluorescence microscope.
Large Scale Digital Cell Analysis System (LSDCAS)
LSDCAS is an automated live cell imaging microscope and image analysis system capable of analyzing thousands of living cells for a period of few days up to weeks in a single experiment (21; 26). LSDCAS is designed to allow quantitative study of cell populations grown under conditions identical to those used in routine biochemical/molecular investigations of a variety of phenomena and is useful for determining the kinetics of various cellular mechanisms on a cell-by-cell basis. For live cell imaging, 24 h prior to irradiation cells were plated at 1×105 cells in a T25 flask containing 3 ml of complete media. After irradiation, cells were fed 3 ml of warm CO2 equilibrated media and transported to the LSDCAS facility were 150 random fields were manually selected and acquisition initiated to run for 12 days. Image analysis was performed by using CasAnalyze, an in-house developed software package, which allows for the quantitation of division-related events (26).
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) Assay
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. cDNA was synthesized using SuperScript II RT (Invitrogen) according to the manufacturer's protocol. Reverse transcription was performed using 1 μg of total RNA and a mixture of oligo(dT) and random hexamer primers. One-twentieth of this reaction, or cDNA corresponding to 50 ng RNA was then used for PCR. PCR was performed using Immolase DNA Polymerase (Bioline) in 25 μl reactions containing, 1X Immolase buffer, 1.5 mM MgCl2, 200 μM each dNTP, 200 nM each primer, 200 nM probe, and 0.75 U Immolase. Primers and FAM-labeled probes were obtained from Integrated DNA Technologies. Reactions were performed in triplicate with the DNA Engine Opticon 2 Thermalcycler (MJ Research-Bio-Rad) using the following cycling parameters: 94°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The 60S acidic ribosomal protein P0 was used as a control to normalize for pipetting error. Data were analyzed using Opticon Monitor Analysis Software Version 2.02 (MJ Research). Samples were quantitated using a standard curve constructed of known copy number of p-GEM-T PCR cloning vector (Promega) containing a PCR amplicon in the multiple cloning region. This standard curve generated from total RNA is used to calculate the relative mRNA in the starting material. The average amount of mRNA for the gene of interest, calculated using the standard curve, is then normalized to the values for β-actin. Fold increases of mRNA levels in the treated cells are then calculated relative to control samples.
Superarray Assay
Total RNA extraction was performed as described above using an RNeasy Mini Kit (QIAGEN). The RT2 First Strand Kit (SuperArray cat #C-03) was used to eliminate genomic DNA and reverse transcribe 1μg of total RNA to cDNA. This template DNA was then added to the RT2 SYBR Green qPCR Master Mix (SuperArray cat. #PA-010) and 25μl were loaded into each well of the RT2 Profiler PCR Array (SuperArray cat. #CAPH-0492). The Superarray was custom designed to contain primers for the following genes: SYCP2, SYCP3, SPO11, REC8, DMC1, MOS, STAG3, and CCNB1 (cyclin B1). The PCR reactions was run in a DNA Engine Opticon 2 Thermalcycler (MJ Research-Bio-Rad) using the following cycling parameters: 40 cycles of 95°C for 15 seconds, 55°C for 30 seconds, and 70°C for 30 seconds, cycles were followed by a melting curve analysis to verify product purity. The data were analyzed using the 2-ΔΔCt method with two separate housekeeping genes, ACTB (β-actin) and RPLP0, and reported as fold increase over control.
Immunohistochemistry
Paraffin-embedded tissue sections from patients with cervical cancers were deparaffinized in xylene and re-hydrated through a graded alcohol series, applied to slides and washed in distilled water. Slides were incubated for 8 min with 3% hydrogen peroxide in distilled water to block endogenous peroxidase. At which point slides were rinsed in Dako Buffer, pretreated with proteinase K (Dako) for 10 min at r. t., and rinsed twice in Dako buffer for 5 min each time. Mouse anti-DMC1 monoclonal antibody (Abcam) was diluted 1:200 in Dako diluent, applied to slides, and incubated for 6 h at room temperature. Slides were then rinsed twice in Dako Buffer for 5 min each time and incubated in Dako Envision+ System labeled Polymer-HRP for 1 h at r. t. Slides were rinsed twice in Dako Buffer for 5 min each time and developed with Dako Dab Plus for 5 min, and again rinsed twice in Dako Buffer for 5 min each time. At which time Dako DAB Enhancer was applied and slides incubated for 3 min, rinsed in distilled water and counterstained with hematoxylin (Surgipate) for 3 min. Slides were finally washed in tap water, and tipped twice in ammonia water to enhance hematoxylin stain. Slides were dehydrated in distilled water, 70%, 95%, 100% ethanol, and xylene and mounted with Mount-Quick sealant.
Results
The polyploid cells formed via radiation-induced MC begin depolyploidization by forming metaphase plates and segregating nuclei
Following 10 Gy γ-irradiation HeLa S3 cells undergo MC and form polyploid cells (Figure 1, panel A). By the end of the first week post-irradiation, the majority of the cells that underwent MC die. At 7 to 9 days post-irradiation and later, the polyploid cells that have escaped death form metaphases plates (Figure 1, panel B), and initiate a depolyploidization process through which nuclei are segregated in what appear to be viable descendants (Figure 1, panel C). Previous work (19; 27) using DNA image cytometry revealed that when division is resumed the cells' DNA content (that had increased 6 to 8 times over the control values) is reduced to that of the untreated populations. Note that the segregated cells contain only one nucleus and are morphologically indistinguishable from control cells (Figure 1, panel D). Noteworthy to mention is the fact that, although informative, cytology data can be difficult to interpret when describing phenomena that are per se dynamic. The advantages of using a live cell imaging methodology, such as the Large Scale Digital Cell Analysis System (LSDCAS) are evident in the studies discussed here, as by employing this technology we are able to directly observe cellular behavior and phenomena that can only be inferred when analyzing a fixed cell population on a microscope slide. The LSDCAS image results (Figure 2 and video clip 1 in supplement) are discussed below, and demonstrate that a fraction of the polyploid cells formed via radiation-induced MC escape death and gives rise to a progeny of smaller cells (indistinguishable from the control cells) through a depolyploidization process.
Figure 1. Nuclei segregation in polyploid cell formed via radiation-induced MC.
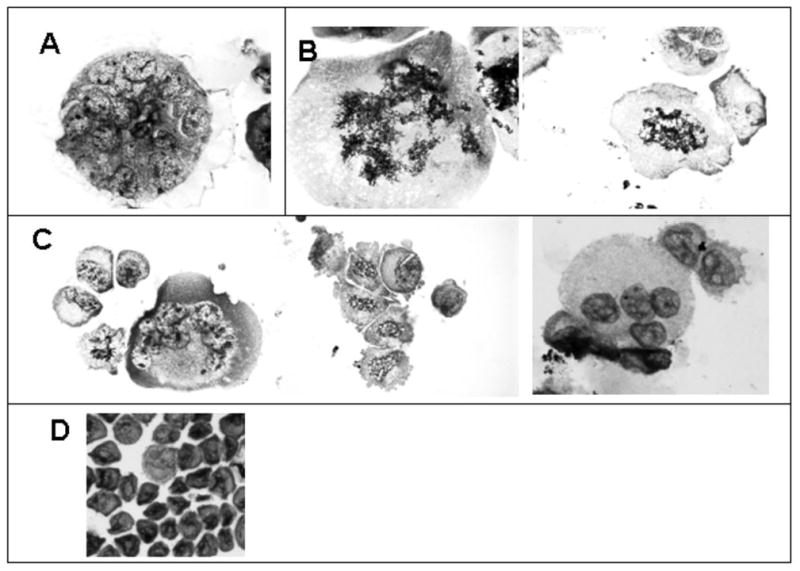
Photomicrographs of irradiated HeLa S3 cells (10 Gy, γ-rays), cytospin preparations. Panel A: a typical multinucleated MC cell (day 9 post-irradiation); Panel B: multiple metaphase plates at day 7 and 10 post-irradiation; Panel C: nuclei segregation (10 days post-irradiation); Panel D: sham-irradiated control.
Figure 2. Polyploid cells formed via radiation-induced MC undergo depolyploidization and originate small mononucleated cells that have the potential for long-term survival - LSDCAS live cell imaging (E5107Field99).
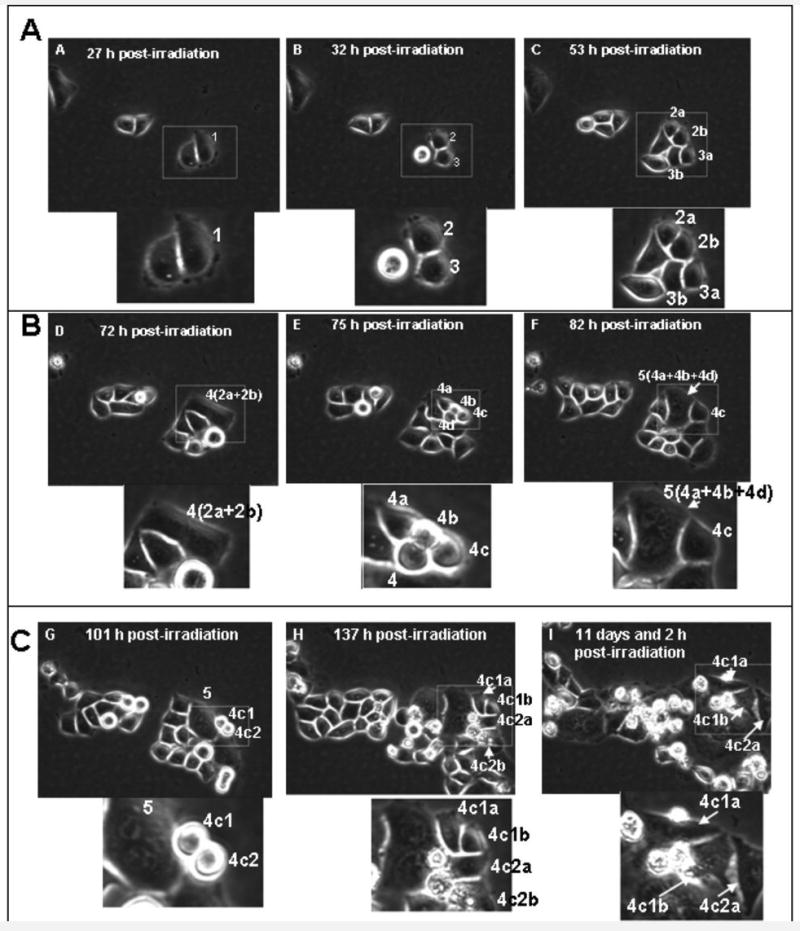
Panel A - 27 h post-irradiation: the cell 1 is the mother cell. It is mono-nucleated and of average size for HeLa Clone3 cell culture. - 32 h post-irradiation: cell 1 divided normally to produce two mono-nucleated daughter cells, cell 2 and cell 3. - 53 h post-irradiation: Cell 2 successfully completed a normal division to produce cell 2a and cell 2b. Cell 3 successfully completed a normal division to produce cell 3a and cell 3b. This image shows all second generation daughter cells in early-interphase, they are mono-nucleated and of a size similar to the untreated control cells. Panel B - 72 h post-irradiation: cells 2a and 2b fuse to form the binucleated cell 4. This panel depicts the very end of interphase as the cell rounds up for division two hours after this image was captured. - 75 h post-irradiation: Cell 4 unsuccessfully attempts to perform a multipolar division to form cells 4a, 4b, 4c and 4d. The image shows the four poles that were attempted during the division. - 82 h post-irradiation: Cells 4a, 4b and 4d fail to segregate and immediately fuse to form the large multi-nucleated cell 5. Cell 4c remains independent, is mono-nucleated, and of normal size. Panel C - 101 h post-irradiation: Cell 4c has just completed a normal division to produce the mono-nucleated daughter cells 4c1 and 4c2. The image shows that division proceeded normally and was not a failed multi-polar attempt. The multi-nucleated cell 5 from the previous division is still visible in this frame. - 137 h post-irradiation: Cell 4c1 has completed a successful division to produce cell 4c1a and cell 4c1b. Cell 4c2 has completed a successful division to produce cell 4c2a and cell 4c2b. The daughter cells are in early interphase in the above image. – 266 h post-irradiation: Cells 4c1a, 4c1b and 4c2a have stayed within the field of view and are still alive. Note: the area delimited by the rectangle is enlarged and reported below each still image.
Polyploid cells formed via radiation-induced MC start a program of depolyploidization reverting to a morphology identical to that of control cells and have the potential for long-term survival
LSDCAS imaging data (Figure 2, and video clip 1, need QuikTime Player, in supplement) show that, as a consequence of a 5 Gy γ-irradiation exposure, HeLa clone3 cells undergo MC, become polyploid, and initiate rounds of multipolar divisions. Some of these multipolar divisions are only attempted divisions, as cytokinesis fails to complete and cell fusion occurs (Figure 2, panel B, 72 h and 82 h post-irradiation). Other multipolar divisions are successful (Figure 2, panel C, 101 h and 173 h post-irradiation). The end result is the production of a heterogeneous population of daughter cells containing different numbers of nuclear fragments. As shown in Supplemental Data, 305 cells were imaged in all the flask over a period of 266 h; in this time frame there were 210 multipolar divisions; of these divisions 118 produced at least one small mononucleated cell, while the total number of the small mononucleated cells produced by multipolar divisions was 198 (note that this number is different from the previous - 118 - because often in a single multipolar division more than one mononucleated cell is produced); 4 is the number of the mononucleated cells that are still alive at the end of the movie. In this query only the mononucleated cells (generated by a multipolar division) that have divided at least two times are counted as survivors; these cells are considered to have a potential for further normal divisions. In the still images reported in Figure 2 the generation of the smaller mononucleated cell is shown in panel B, 82 h post-irradiation (cell 4c); this cell is able to normally divide giving rise to two mononucleated daughter cells (Figure 2, panel C, 101 h post-irradiation, cells 4c1 and 4c2). Note that these cells contain only one nucleus and if they were to be observed in standard microscopic cell preparations they might be mistaken for unaffected cells that escaped MC cell killing, while, indeed, they originate from a polyploid MC cell. Both these daughter cells are able to divide successfully as well (Figure 2, panel C, 137 h post-irradiation, cells 4c1a, 4c1b and 4c2a, 4c2b). At 266 h (11 days and 2 h) post-irradiation (Figure 2, panel C), these 4 daughter cells are still alive. Multipolar divisions of irradiated cells that have undergone MC is not a sole characteristic of HeLa cells, but a quite common phenomenon in irradiated tumor cells, as demonstrated by image data obtained by us observing the human head and neck squamous cell carcinoma FaDu, the human ovary adenocarcinoma cell lines SKOV3, OVCAR and EG (data not shown), and the MDA-MB435 cells (video clip 2, need QuikTime Player, in supplement). Video clip 2 shows images of multipolar divisions occurring in the MDA-MB435 cells irradiated with a dose of 5 Gy of γ-irradiation. The MDA-MB435 cells are also able to produce smaller mononucleated cells through a reduction division process. These results indicate that a small fraction of cells with morphology identical to that of control cells originate from polyploid cells formed via radiation-induced MC.
Meiotic genes are expressed during depolyploidization of polyploid cells formed via radiation-induced MC
At day 7 and 9 post-irradiation morphological features reminiscent of the meiotic synaptonemal complexes are present in the HeLa S3 cells' nuclei (Figure 3). Synaptonemal complexes are meiosis-specific supramolecular tripartite proteinaceous structures that develop during pairing of homologous chromosomes in the late zygotene and pachytene stages of meiosis I. This structure forms between two homologous chromosomes and mediates chromosome pairing, synapsis, and crossover (28; 29), and consists of two parallel lateral regions and a central element and it comprises three specific components, the SC protein-1 (SYCP1), the SC protein-2 (SYCP2), and the SC protein-3 (SYCP3). A series of experiments were designed to determine if components of the synaptonemal complex are expressed in cells undergoing depolyploidization. To this end, we performed quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) experiments. Quantitative RT-PCR data of irradiated HeLa S3 cells reveals a 1 to 2.5-fold increase in SYCP3 mRNA expression at days 4-11 post-irradiation, with a recurring peak increase (∼1.5-fold) at days 18-20 post-irradiation (Figure 4, panel A). A second experiment performed for longer times confirmed the fold increases reported and revealed a 1.5-fold increase of SYCP3 mRNA at 24 days post-irradiation. The same experiment was performed using the human colon cancer cells HCT116-379.2, with mutated p53 gene – thus lacking p53 function (a kind gift of Dr. Bert Vogelstein at John Hopkins University). Expression of SYCP3 mRNA was also present in this cell line (Figure 4, panel B) with a 1.5 to 1.8-fold increase at 2-16 h post-irradiation and recurring peak increases at 32 and 48 h post-irradiation.
Figure 3. Morphological features reminiscent of synaptonemal complexes in irradiated HeLa S3 cells.
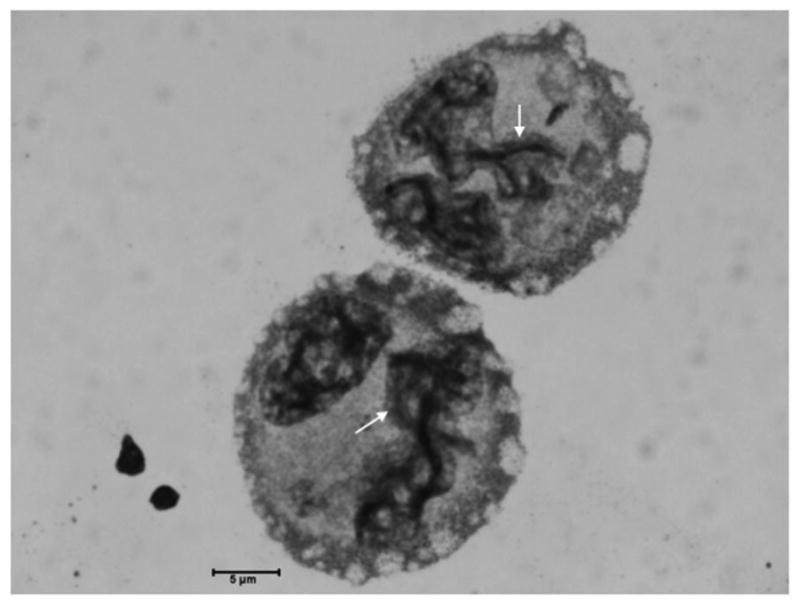
Photomicrographs of HeLa S3 cells at 7 days post 10 Gy of γ-irradiation. Morphological features reminiscent of the meiotic synaptonemal complexes are here marked by red arrows; bar = 5 μm; cytospin preparations.
Figure 4. mRNA and protein expression of meiosis-specific genes in various human tumor cell lines exposed to different doses on γ-irradiation.
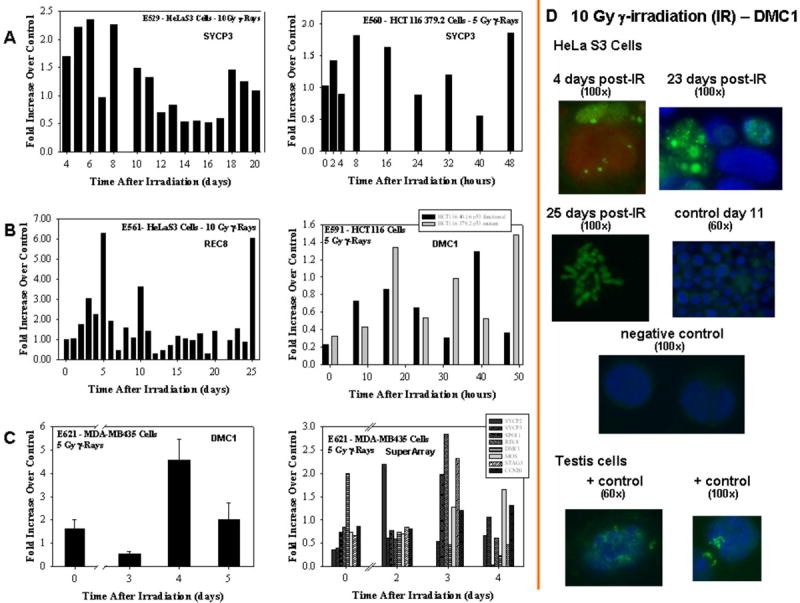
Panel A: SYCP3 mRNA expression in HeLa S3 and Hct-116 379.2 cells. Panel B: Rec8 and DMC1 mRNA expression in HeLa S3 and HCT-116 40.16 and HCT-116 379.2 cells. Panel C: DMC1 mRNA expression in MDA-MB435 cells and custom designed RT-PCR SuperArray showing mRNA expression of various meiosis-specific genes in MDA-MB435 cells. mRNA levels were assayed using qRT-PCR technique as described in Materials and Methods. Each sample (and the reference gene, β-Actin) was run in duplicate. Measurements were repeated at least two times, representative experiments are reported in Panels A and B. For the MDA-MB435 cells (Panel C left hand site) three repeats were performed. SuperArray data (Panel C right hand site) are semi-quantitative. Panel D: immunofluorescence for DMC1 protein in HeLa S3 cells. Focal staining of DMC1 (green) is visible at various times post-irradiation. Condensed chromosomes are also visible at 25 h post-irradiation. Positive controls: untreated mouse testis cells. Primary antibody: DMC1 (Abcam, 1:100 dilution); secondary antibody Goat anti-mouse FITC IgG (whole molecule) (Sigma, 1:100 dilution); counterstaining DAPI (red or blue).
Using these same cell lines we have also measured the expression of two other meiosis-specific genes, Rec8 and DMC1. Rec8 is a family member of meiotic cohesins that are a set of meiosis-specific proteins involved in sister chromatid cohesion and homology pairing during meiosis (30); DMC1 is essential for meiotic homologous recombination and for cell cycle progression (31; 32). Figure 4, panel B, reports the Rec8 mRNA expression in irradiated HeLa S3 cells and the DMC1 mRNA expression in the irradiated syngenic HCT116-379.2 (mutated p53) and HCT116-40.16 (wild type p53, also a kind gift of Dr. Bert Vogelstein at John Hopkins University) cells. For Rec8, mRNA expression increases between days 2 and 10 with an increase as high as 5-fold by day 5 post-irradiation, a second peak is also visible at day 25 post-irradiation. For DMC1, in both cell lines, mRNA expression increases in the range of 1.2- to 1.6-fold at 16, 40 and 48 h post-irradiation. It is noteworthy to mention that radiation exposure leads to increases in expression of DMC1 mRNA in both the wild type and the mutated p53 HCT116 cells. Thus, allowing for the speculation that certain types of solid tumors, albeit with functional p53, might attempt to activate the same mechanisms of survival as for the non-functional p53 tumors, thus escaping p53-mediated programmed cell death and gaining a survival advantage.
DMC1 mRNA expression was also measured in the MD-MBA435 cells (Figure 4, panel C) irradiated with 5 Gy of γ-rays. Increases in DMC1 mRNA are seen between zero and 5 days post-irradiation with a 5-fold peak increase at 4 days post-irradiation. Immunofluorescence studies, carried out with HeLa S3 cells confirm the translation of the DMC1 mRNA in the functional protein (Figure 4, panel D). DMC1 focal staining is noticeable in the polyploid γ-irradiated HeLa S3 cells at different points in time; representative time points of 4, 23 and 25 days post-irradiation are presented in Figure 4, panel D.
Note that the meiosis-specific genes were found activated, in all the cell lines reported, at times when polyploid MC cells have already formed in irradiated cells (LSDCAS data not shown, and LSDCAS data in ref. 27). These observations support our speculation that the activation of pseudo-meiotic pathways in irradiated tumors plays a role in the depolyploidization processes.
SuperArray custom designed assay reveals the activation of other meiosis-specific genes in irradiated MDA-MB435 cells
Intrigued by the results described above, we decided to analyse the possible expression of other meiosis-specific genes involved in homologous pairing, meiotic recombination and cohesion in the MDA-MB435 cells. An RT-PCR SuperArray was designed using primers for the following genes: SYCP2, SYCP3, SPO11, STAG3, DMC1, REC8, and MOS; CCMB1 (cyclin B1) was also enclosed in the array. SPO11 is a meiotic recombination protein that mediates DNA cleavage in double-strand breaks that initiate meiotic recombination (33) and is also involved in axial element and synaptonemal complex formation and in the maintenance of meiotic chromosome condensation and proper spindle formation (34). STAG3 is expressed specifically in testis and is involved in chromosome pairing and maintenance of the synaptonemal complex structure during the pachytene phase of meiosis in a cohesin-like manner. During anaphase I STAG3 dissociates from the centromeres allowing chromosome segregation (35). MOS regulates oocyte maturation by arresting the unfertilized cells in M phase and is destroyed before fertilization after exit from meiosis II (36). Cyclin B1 is the regulatory subunit of the maturation-promoting factor (MPF) and, when phosphorylated by the MPF catalytic subunit cdc2, it plays a role both during meiosis and mitosis. In meiosis, cyclin B1 is responsible for meiotic maturation of oocytes; while in mitosis, it is responsible for the cell cycle G2/M phase transition. The function of SYCP, REC8 and DMC1 genes have been described above. The results of the SuperArray assay are reported in Figure 4, panel C (right hand site). All the meiotic genes tested are expressed in MDA-MB435 cells after exposure to 5 Gy of γ-irradiation. Moreover, as previously reported by us (15) cyclin B1 mRNA is also increased at days 4 and 5 post-irradiation. The increases in mRNA expression seen in all the RT-PCR data presented in this paper are comparable to the increases shown by other investigators for mRNA expression of a variety of meiotic genes (37; 38). These data demonstrate that meiosis-specific genes involved in homologous pairing and recombination are activated in polyploid tumor cells formed via radiation-induced MC that have escaped death. This activation occurs in concert with the onset of a depolyploidization process as demonstrated by the LSDCAS data discussed in Figure 2.
Tissue specimens from human cervical cancer are positive for DMC1 staining
Immunohistochemistry performed with the meiosis-specific protein DMC1 on human cervical cancer tissues demonstrates that this protein is preferentially expressed in these specimens (Figure 5, panel C) as well as in the testis tissue (Figure 5, panel A) serving as a positive control, while the normal tissue presents little staining (Figure 5, panel B). To our knowledge, this is the first time that expression of a meiosis-specific gene is reported in human tumor tissue specimens.
Figure 5. Immunohistochemistry of paraffin embedded cervical cancer tissue with DMC1 antibody.
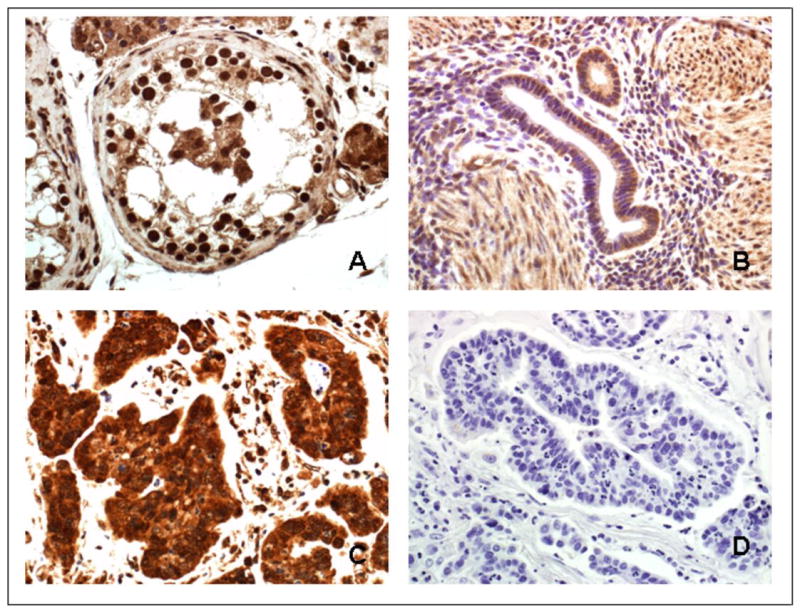
Tissue sections were pretreated with proteinase K and incubated with DMC1 antibody (Abcam, 1:200 dilution). Secondary antibody, Dako Envision + System labeled Polymer-HRP, anti-mouse; counterstaining, hematoxylin (Surgipate). Panel A: human normal testis specimen, positive control; Panel B: human lower uterine segment, normal specimen; Panel C: human adenocarcinoma of the cervix; Panel D: human adenocarcinoma of the cervix, negative control, same specimen than in Panel C. Magnification 40×.
Discussion
During the last fifteen years, our interest has focused on the effects of radiation exposure on the cell cycle and on phenomena associated with cell cycle dysregulation. Under conditions that can compromise genome integrity, cells activate checkpoint control pathways that delay them from entering and exiting mitosis. The first set of checkpoints controls entry into mitosis and, in response to DNA damage or other detrimental effects, delays activation of the positive regulator of mitosis, cyclin B1/cdc2. Exit from mitosis is controlled by the spindle assembly checkpoint (SAC) that prevents the activation of the anaphase-promoting complex (APC) that is required to target securin and cyclin B1 for proteolysis. Destruction of these proteins allows for the separation of chromatids, thus marking anaphase, and exit of the cell from mitosis, marking telophase, respectively (39; and reviewed in ref. 40). A broad class of agents (41-46) induces a loss of regulation of the cell cycle with the end result that if the cell population does not immediately die, it often undergoes MC. We have demonstrated that radiation-induced MC occurs in a variety of mammalian cell lines with impaired p53 function and it is characterized by aberrant nuclear morphology observed following premature mitotic entry (42; 15; 47; 16-18; 14). Using live cell imaging we have also determined that irradiated HeLa cells undergo MC and are able to survive for many generation post-exposure (21); 50% of the surviving colonies exhibited MC that persisted throughout colony formation (21). Moreover, other live cell imaging experiments conducted with irradiated human-hamster hybrid GM10115 cells demonstrated the presence of a high frequency of surviving clones containing an elevated incidence of polyploid cells formed via MC. Specifically, 0.07% of the cell population underwent MC and continued to divide for up to 12 days following irradiation (6000 individual cells were studied for 10 days post-irradiation), making MC a possible contributor to radiation-induced genomic instability. Captivated by these results and by cytological studies recently published by us (19) in which we have reported the formation of large polyploid cells in irradiated HeLa cells, we initiated the series of experiments described in this paper to determine the mechanisms underlying the process of depolyploidization.
We report here LSDCAS imaging data showing that as result of γ-irradiation HeLa clone3 and MDA-MB435 cells undergo MC and become polyploid; within the first 5 days post-irradiation, the polyploid cells initiate a depolyploidization process. Mononucleated smaller cells are produced that are morphologically indistinguishable from the untreated cells; these cells are able to produce a cell progeny and are still alive 12 days post-irradiation. These results indicate that a small fraction of cells can survive MC. Both the morphological and the molecular results discussed in this paper delve into the possible mechanisms utilized by a tumor cell to escape death following radiation-induced MC. We find that irradiated tumor cells (colon, breast/melanoma, cervix) impaired in p53 function present morphological traits resembling features characteristic of meiotic prophase, and that at the time of the appearance of these morphological features, an induction of genes involved in meiotic pathways are observed. These data suggest that polyploid tumor cells conserve their original individual genomic integrity and can re-initiate cell division via reduction division. In a paper published in 1982 Dawkins (48) pointed out that meiosis can be envisioned as a mechanism that elaborate chemostats, such as diploid organisms, have evolved to maximize the fitness of the haploid organisms they carry, that is the gametes. Likewise, some tumor cells might attempt to switch to meiotic divisions to maximize their fitness and their chance at survival.
The results discussed here are in line with the data reported by Illidge and collaborators (49), Erenpreisa and collaborators (23), and Ivanov and collaborators (50) that found polyploid giant cells formation after irradiation of Burkitt's lymphoma cells. Our results are also in line with the data reported by Prieur-Carrillo and collaborators (22) that, using computerized video time-lapse techniques, found survivors of irradiated giant human bladder carcinoma cells up to a week post-irradiation. Taking into account the results reported above and our data discussed in this paper, it appears that polyploid cells, originated from MC, present a survival advantage over the remaining portion of the cell population. In the same paper referred to above (23), Erenpreisa and collaborators also showed the presence of chromosome double-loops in the giant cells by the second week post-irradiation. These formations are referred to as polyploid bouquets. The process of bouquet formation involves the clustering of telomeres (which are randomly distributed in pre-meiotic interphase and early leptotene) in a small region of the nuclear envelope during the zygotene stage. The bouquet coincides with homologous chromosome pairing and synapsis and it is suggestive of a role for telomeres in the homology search process (51). This telomere-mediated reorganization of the early meiotic prophase nucleus has the effect of reducing the search space required for chromosomes to find their partners. The fact that irradiated tumor cells present features resembling the bouquet formation suggests that this process might contribute to rendering the polyploid cells able to undergo a successful reduction division.
Using the same cell line, Ivanov and collaborators (50) have also reported that a portion of lymphoblastoid Namalwa cells exposed to severe genotoxic treatment undergo MC giving rise to polyploid cells some of which resemble cells in meiotic prophase. Homologous DNA recombinational repair occurs in these cells and they replicate their DNA for more than a few rounds of mitotic cycles producing descendants. This is an important aspect, as the signal for initiation of recombination is the production of DNA double strand breaks (dsb) mediated by the meiosis-specific endonuclease enzyme SPO11 (52), and the SPO11 gene is found activated in the MDA-MB435 cell line, as reported in this study. Thus, polyploid cells, originated from MC cells, might, through related mechanisms, gain a survival advantage over the remaining portion of the cell population.
The meiosis-specific genes SYCP2, SYCP3, DMC1, SPO11, REC8, STAG3, and MOS found activated in the tumor cell lines discussed in this paper can all be defined as cancer/testis antigens according to the classification reported by Simpson and collaborators (53); and these findings suggest that polyploid cells may have regulatory pathways in common with somatic reduction division and meiosis. In other studies (54-55), ectopic expression of meiotic genes in human cancers has been reported as being a feature of tumor progression. Expression of meiotic genes was also reported by Kalejs and collaborators (56) in lymphoma cell lines undergoing radiation-induced MC. These data suggest that a conserved mechanism of ordered genome assembly and disassembly exists in multi-genomic polyploid tumor cells, and that this process may take advantage of the activation of pro-meiotic pathways to initiate depolyploidization. These data also point towards the intriguing hypothesis that human tumors might utilize pseudo-meiotic pathways to reorganize their polyploid genome to attain a more stable state that might enable them to become more resistant to cytotoxic and/or genotoxic treatments, to escape death, and to promote progression. Thus, understanding the effects of anti-cancer treatment on polyploid tumor cell populations and the process of depolyploidization, might shed light into mechanisms of tumor resistance and progression, and might furnish new insight into treatment methodologies.
Acknowledgments
This work was partially supported by the NIH Grants CA/GM94801 and CA86862, by the NASA Grant NRA NNJ06HH68G, by the Latvian-USA Governmental Exchange Grant enabling visits between Riga and Iowa City and vice versa, and by the Whitaker Foundation Special Opportunity Award. The authors are also indebted to Mrs. Jean L. Ross from the Central Microscopy Research Facility at the University of Iowa for her skillful assistance in collecting the cytology photographs, and to Mrs. Janis R. Rodgers from the Department of Pathology's Histology Research Laboratory at the University of Iowa for her expert assistance in tissue preparation and staining. We would also like to thank Mr. Paul J. Davis who contributed some of the analysis algorithms employed in the LSDCAS data imaging analysis.
References
- 1.Sudo T, Nitta M, Saya H, Ueno NT. Dependence of Paclitaxel Sensitivity on a Functional Spindle Assembly Checkpoint. Cancer Res. 2004;64:2502–2508. doi: 10.1158/0008-5472.can-03-2013. [DOI] [PubMed] [Google Scholar]
- 2.Jordan MA, Wilson L. Microtubules as a Target for Anticancer Drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang ZQ, Hainaut P. New Approaches to Understanding p53 Gene Tumor Mutation Spectra. Mut Res. 1999;431:199–209. doi: 10.1016/s0027-5107(99)00162-1. [DOI] [PubMed] [Google Scholar]
- 4.Soussi T, Beroud C. Assessing TP53 Status in Human Tumours to Evaluate Clinical Outcome. Nature Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham L, Griffin AC, Luck JM. Polyploidy and Cancer. J Gen Physiology. 1950;34:59–63. doi: 10.1085/jgp.34.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandberg AA, Ishihara T, Moore GE, Pickren JW. Unusually High Polyploidy in a Human Cancer. Cancer. 1963;16:1246–1254. doi: 10.1002/1097-0142(196310)16:10<1246::aid-cncr2820161004>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Revesz L, Norman U. Chromosome Ploidy and Radiosensitivity of Tumors. Nature. 1960;187:861–862. doi: 10.1038/187861a0. [DOI] [PubMed] [Google Scholar]
- 8.Castedo M, Coquelle A, Vitale I, Vivet S, Mouhamad S, Viaud S, Zitvogel L, Kroemer G. Selective Resistance of Tetraploid Cancer Cells against DNA Damage-Induced Apoptosis. Ann NY Acad Sci. 2006;1090:35–49. doi: 10.1196/annals.1378.004. [DOI] [PubMed] [Google Scholar]
- 9.Khoo VS, Pollack A, Cowen D, Joon DL, Patel N, Terry NH, Zagars GK, von Eschenbach AC, Meistrich ML, Troncoso P. Relationship of Ki-67 Labeling Index to DNA-Ploidy, S-Phase Fraction, and Outcome in Prostate Cancer Treated with Radiotherapy. Prostate. 1999;41:166–172. doi: 10.1002/(sici)1097-0045(19991101)41:3<166::aid-pros3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 10.Pollack A, Grignon DJ, Heydon KH, Hammond EH, Lawton CA, Mesic JB, Fu KK, Porter AT, Abrams RA, Shipley WU. Prostate Cancer DNA Ploidy and Response to Salvage Hormone Therapy after Radiotherapy with or without Short-Term Total Androogen Blockade: an Analysis of RTOG 8610. J Clin Oncol. 2003;21:1238–1248. doi: 10.1200/JCO.2003.02.025. [DOI] [PubMed] [Google Scholar]
- 11.Zybina TG, Zybina EV. Whole-Genome Chromosome Distribution in the Course of Nuclear Fragmentation of Giant Trophoblast Cells of Microtus Rossiaemeridionalis Studied with the use of Gonosomal Chromatin Arrangement. Cell Biol Int. 2005;29:1066–1070. doi: 10.1016/j.cellbi.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Nagl W. Endopolyploidy and Polyteny in Differentiation and Evolution. Amsterdam-New York-Oxford: North-Holland Publications; 1978. [Google Scholar]
- 13.Storchova Z, Pellman D. From Polyploidy to Aneuploidy, Genome Instability and Cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 14.Ianzini F, Mackey MA. Mitotic Catastrophe. In: Gewirtz DA, Holt SE, Grant S, editors. Apoptosis, Senescence and Cancer. Second. Humana Press; 2007. pp. 73–91. [Google Scholar]
- 15.Ianzini F, Mackey MA. Spontaneous premature chromosome condensation and mitotic catastrophe following irradiation of HeLa S3 cells. Int J Radiat Biol. 1997;72:409–421. doi: 10.1080/095530097143185. [DOI] [PubMed] [Google Scholar]
- 16.Mackey MA, Ianzini F. Enhancement of radiation-induced mitotic catastrophe by moderate hyperthermia. Int J Radiat Biol. 2000;76:273–280. doi: 10.1080/095530000138925. [DOI] [PubMed] [Google Scholar]
- 17.Ianzini F, Bertoldo A, Kosmacek EA, Phillips SL, Mackey MA. Lack of p53 Function Promotes Radiation-Induced Mitotic Catastrophe in Mouse Embryonic Fibroblast Cells. Cancer Cell International. 2006;6:11. doi: 10.1186/1475-2867-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ianzini F, Domann FE, Kosmacek EA, Phillips SL, Mackey MA. Human Glioblastoma U87MG Cells Transduced with a Dominant Negative p53 Adenovirus Construct Undergo Radiation-Induced Mitotic Catastrophe. Rad Res. 2007;168:183–192. doi: 10.1667/0033-7587(2007)168[183:HGUCTW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 19.Erenpreisa Je, Kalejs M, Ivanov A, Illidge TM, Ianzini F, Kosmacek EA, Mackey MA, Dalmane A, Cragg MS. Genomes Segregation in Polyploid Tumor Cells Following Mitotic Catastrophe. Cell Biol Int. 2005;29:1005–1011. doi: 10.1016/j.cellbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Mackey MA, Anolik SL, Roti Roti JL. Cellular mechanisms associated with the lack of chronic thermotolerance expression in HeLa S3 cells. Cancer Res. 1992;52:1101–1106. [PubMed] [Google Scholar]
- 21.Ianzini F, Mackey MA. Development of the Large-Scale Digital Cell Analysis System. Radiat Prot Dos. 2002;99:81–94. doi: 10.1093/oxfordjournals.rpd.a006787. [DOI] [PubMed] [Google Scholar]
- 22.Prieur-Carrillo G, Chu K, Lindqvist J, Dewey WC. Computerized Video Time-Lapse (CVTL) Analysis of the Fate of Giant Cells Produced by X-Irradiating EJ30 Human Bladder Carcinoma Cells. Radiat Res. 2003;159:705–712. doi: 10.1667/rr3009. [DOI] [PubMed] [Google Scholar]
- 23.Erenpreisa Je, Cragg MS, Fringes B, Sharakhov I, Illidge TM. Release of Mitotic Descendants by Giant Cells from Irradiated Burkitt's Lymphoma Cell Line. Cell Biol Int. 2000;24:635–648. doi: 10.1006/cbir.2000.0558. [DOI] [PubMed] [Google Scholar]
- 24.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JCF, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic Variation in Gene Expression Patterns in Human Cancer Cell Lines. Nat Gen. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 25.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 Cells are Derived from M14 Melanoma Cells – a Loss for Breast Cancer, but a Boon for Melanoma Research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 26.Davis PJ, Kosmacek EA, Sun Y, Ianzini F, Mackey MA. The Large Scale Digital Cell Analysis System: Open Source, Freely Available Software for Live Cell Imaging. J Microscopy. 2007;228:296–308. doi: 10.1111/j.1365-2818.2007.01847.x. [DOI] [PubMed] [Google Scholar]
- 27.Erenpreisa Je, Ivanov A, Wheatley SP, Kosmacek EA, Ianzini F, Anisimov AP, Mackey MA, Davis PJ, Plakhins G, Illidge TM. Endopolyploidy in Irradiated p53 Deficient Tumour Cell Lines: Persistence of Cell Division Activity in Giant Cells Expressing Aurora B-Kinase. Cell Biol Int. 2008;32:1044–1056. doi: 10.1016/j.cellbi.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moses MJ. Chromosomal Structures in Crayfish Spermatocytes. J Biophys Biochem Cytol. 1956;2:215–218. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zickler D. From Early Homologue Recognition to Synaptonemal Complex Formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 30.Parisi S, McKay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. Rec8p, A Meiotic Mecombination and Sister Chromatid Cohesion Phosphoprotein of the Rad21p Family Conserved from Fission Yeast to Humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a Meiosis-Specific Yeast Homolog of E. coli RecA Required for Recombination, Synaptonemal Complex Formation, and Cell Cycle Progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 32.Neale MJ, Keeney S. Clarifying the Mechanics of DNA Strand Exchange in Meiotic Recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keeney S, Giroux CN, Kleckner N. Meiosis-Specific DNA Double-Stranded Breaks are Catalyzed by Spo11, a Member of a Widely Conserved Protein Family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 34.Celerin M, Merino ST, Stone JE, Menzie AM, Zolan ME. Multiple Roles of Spo11 in Meiotic Chromosome Behavior. EMBO J. 2000;19:2739–2750. doi: 10.1093/emboj/19.11.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pezzi N, Prieto I, Kremer L, Perez Jurado LA, Valero C, Del Mazo J, Martinez AC, Barbero JL. STAG3, a Novel Gene Encoding a Protein Involved in Meiotic Chromosome Pairing and Location of STAG3-Related Genes Flamking the Williams-Beuren Syndrome. FASEB J. 2000;14:581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- 36.Sagata N, Oskarsson M, Copeland T, Brumbaugh J, Vande Woude GF. Function of c-MOS Proto-Oncogene Product in Meiotic Maturation in Xenopus Oocytes. Nature. 1988;335:519–526. doi: 10.1038/335519a0. [DOI] [PubMed] [Google Scholar]
- 37.Calenda A, Allenet B, Escalier D, Bach JF, Garchon HJ. The Meiosis-Specific Xmr Gene Product is Homologous to the Lymphocyte Xlr Protein and is a Component of the XY Body. EMBO J. 1994;13:100–9. doi: 10.1002/j.1460-2075.1994.tb06239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olesen C, Nyeng P, Kalisz M, Jensen TH, Møller M, Tommerup N, Byskov AG. Global Gene Expression Analysis in Fetal Mouse Ovaries with and without Meiosis and Comparison of Selected Genes with Meiosis in the Testis. Cell Tissue Res. 2007;328:207–221. doi: 10.1007/s00441-006-0205-5. [DOI] [PubMed] [Google Scholar]
- 39.Hagting A, Den Elzen N, Vodermaier HC, Waizenegger IC, Peters JM, Pines J. Human Securin Proteolysis is Controlled by the Spindle Checkpoint and Reveals when the APC/C Switches from Activation by Cdc20 to Cdh1. J Cell Biol. 2002;157:1125–37. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasmyth K, Peters JM, Uhlmann F. Splitting the Chromosome: Cutting the Ties that Bind Sister Chromatids. Science. 2000;288:1379–85. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 41.Mackey MA, Anolik SL, Roti Roti JL. Cellular Mechanisms Associated with the Lack of Chronic Thermotolerance Expression in HeLa S3 Cells. Cancer Res. 1992;52:1101–1106. [PubMed] [Google Scholar]
- 42.Swanson PE, Carroll SB, Zhang XF, Mackey MA. Spontaneous Premature Chromosome Condensation, Micronucleus Formation, and Non-Apoptotic Cell Death in Heated HeLa S3 Cells. Am J Pathol. 1995;146:963–971. [PMC free article] [PubMed] [Google Scholar]
- 43.Lock RB, Stribinskiene L. Dual Modes of Death Induced by Etoposide in Human Epithelial Tumor Cells Allow Bcl-2 to Inhibit Apoptosis Without Affecting Clonogenic Survival. Cancer Res. 1996;56:4006–4012. [PubMed] [Google Scholar]
- 44.Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An Indolocarbazole Inhibitor of Human Checkpoint Kinase (Chk1) Abrogates Cell Cycle Arrest Caused by DNA Damage. Cancer Res. 2000;60:566–572. [PubMed] [Google Scholar]
- 45.Hirose Y, Berger MS, Pieper RO. Abrogation of the Chk1-Mediated G(2) Checkpoint Pathway Potentiates Temozolomide-Induced Toxicity in a p53-Independent Manner in Human Glioblastoma Cells. Cancer Res. 2001;61:5843–5849. [PubMed] [Google Scholar]
- 46.Yoshikawa R, Kusunoki M, Yanagi H, Noda M, Furuyama JI, Yamamura T, Hashimoto-Tamaoki T. Dual Antitumor Effects of 5-Fluorouracil on the Cell Cycle in Colorectal Carcinoma Cells: A Novel Target Mechanism Concept for Pharmacokinetic Modulating Chemotherapy. Cancer Res. 2001;61:1029–1037. [PubMed] [Google Scholar]
- 47.Ianzini F, Mackey MA. Delayed DNA Damage Associated with Mitotic Catastrophe Following X-Irradiation of HeLa S3 cells. Mutagenesis. 1998;13:337–344. doi: 10.1093/mutage/13.4.337. [DOI] [PubMed] [Google Scholar]
- 48.Dawkins R. The Extended Phenotype. Oxford: Oxford University Press; 1982. [Google Scholar]
- 49.Illidge TM, Cragg MS, Fringes B, Olive P, Erenpreisa Je. Polyploid Giant Cells Provide a Survival Mechanism for p53 Mutant Cells After DNA Damage. Cell Biol Int. 2000;24:621–633. doi: 10.1006/cbir.2000.0557. [DOI] [PubMed] [Google Scholar]
- 50.Ivanov A, Cragg MS, Erenpreisa Je, Emzinsh D, Lukman H, Illidge TM. Endopolyploid Cells Produced After Severe Genotoxic Damage Have the Potential to Repair DNA Double Strand Breaks. J Cell Sci. 2003;116:4095–4106. doi: 10.1242/jcs.00740. [DOI] [PubMed] [Google Scholar]
- 51.Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, Cande WZ. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- 52.Keeney S. The Mechanism and Control of Meiotic Recombination Initiation. Curr Topics Develop Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 53.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/Testis Antigens, Gametogenesis and Cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 54.Tureci O, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Identification of a Meiosis-Specific Protein as a Member of the Class of Cancer/Testis Antigens. Proc Natl Acad Sci USA. 1998;95:5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scanlan MJ, Simpson AJ, Old LJ. The Cancer/Testis Genes: Review, Standardization, and Commentary. Cancer Immun. 2004;4:1–15. [PubMed] [Google Scholar]
- 56.Kalejs M, Ivanov A, Plakhins G, Cragg MS, Emzinsh D, Illige TM, Erenpreisa Je. Upregulation of Meiosis-Specific Genes in Lymphoma Cell Lines Following Genotoxic Insult and Induction of Mitotic Catastrophe. BMC Cancer. 2006;6:6. doi: 10.1186/1471-2407-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


