Abstract
The invasive nature of malignant gliomas is a clinical problem rendering tumors incurable by conventional treatment modalities such as surgery, ionizing radiation, and temozolomide. Na+/H+ exchanger regulatory factor 1 (NHERF-1) is a multifunctional adaptor protein, recruiting cytoplasmic signaling proteins and membrane receptors/transporters into functional complexes. This study revealed that NHERF-1 expression is increased in highly invasive cells that reside in the rim of glioblastoma multiforme (GBM) tumors and that NHERF-1 sustains glioma migration and invasion. Gene expression profiles were evaluated from laser capture-microdissected human GBM cells isolated from patient tumor cores and corresponding invaded white matter regions. The role of NHERF-1 in the migration and dispersion of GBM cell lines was examined by reducing its expression with small-interfering RNA followed by radial migration, three-dimensional collagen dispersion, immunofluorescence, and survival assays. The in situ expression of NHERF-1 protein was restricted to glioma cells and the vascular endothelium, with minimal to no detection in adjacent normal brain tissue. Depletion of NHERF-1 arrested migration and dispersion of glioma cell lines and caused an increase in cell-cell cohesiveness. Glioblastoma multiforme cells with depleted NHERF-1 evidenced a marked decrease in stress fibers, a larger cell size, and a more rounded shape with fewer cellular processes. When NHERF-1 expression was reduced, glioma cells became sensitized to temozolomide treatment resulting in increased apoptosis. Taken together, these results provide the first evidence for NHERF-1 as a participant in the highly invasive phenotype of malignant gliomas and implicate NHERF-1 as a possible therapeutic target for treatment of GBM.
Introduction
Glioblastoma multiforme (GBM) is the most frequent form of primary brain cancer and has an average life expectancy from time of diagnosis of only 9 months to 1 year. The highly lethal nature of this tumor partly originates from its invasive phenotype, which affords the tumor cells the ability to infiltrate adjacent brain tissue [1,2]. In eradicating this invasive disease, it is considered incurable using treatment modalities presently available. As a result, identifying and characterizing molecular mechanisms that drive the invasive behavior of GBM may serve as diagnostic and prognostic markers, as well as candidate therapeutic targets.
Na+/H+ exchanger regulatory factor 1 (NHERF-1; also SLCA9A3R1) was initially recognized as a scaffolding protein that recruits membrane transporters/receptors and cytoplasmic signaling proteins into functional complexes localized at or near the plasma membrane in epithelial cells [3,4]. Specifically, NHERF-1 has been shown to regulate several G protein-coupled receptors, including receptors for parathyroid hormone, κ-opioid, and β2-adrenergic receptors [5]. Na+/H+ exchanger regulatory factor 1 interacts with specific growth factor receptors such as the epidermal growth factor receptor and platelet-derived growth factor receptor and modulates mitogenic signaling by these receptor tyrosine kinases [5]. Structurally, NHERF-1 contains two tandem PDZ domains (protein-binding domains conserved in the mammalian synaptic protein, PSD-95, Drosophila Dlg or discs large, and the adherens junction protein, ZO-1) that can oligomerize with other PDZ domains to enhance scaffolding function [6,7] as well as mediate other specific protein-protein interactions [8]. Previous studies have shown NHERF-1 to be upregulated in tumor tissue relative to its corresponding normal tissue in breast cancer [3], schwannoma [9], and hepatocellular carcinomas [10].
Whereas these studies indicate that NHERF-1 plays a role in the progression of several cancer types, the involvement of NHERF-1 in the pathogenesis of glioblastomas is unknown. Invasive cancer cells, including glioblastoma, are resistant to apoptosis [11–16]. However, decreasing the migratory capabilities of tumor cells can restore a certain level of sensitivity to cytotoxic insult [15,17]. To gain more insight into the functional role of NHERF-1, changes in the levels of this gene product in migrating primary human tumor cells were measured; NHERF-1 was depleted in GBM cell lines, then migration and dispersion responses were assessed, cell morphology was analyzed, and the activity of the cytotoxic, alkylating agent, temozolomide (TMZ), was tested.
Materials and Methods
Cell Culture Conditions and Extracellular Matrix Preparation
Human glioma cell lines SF767 [18] and T98G (American Type Culture Collection, Manassas, VA) were maintained in minimum essential medium (MEM; Invitrogen Corp., Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Inc., Logan, UT) in a 37°C and 5% CO2 atmosphere at constant humidity. Laminin from human placenta was obtained from Invitrogen.
Clinical Samples and Histology
Human glioblastoma tumor samples were obtained from patients who underwent primary therapeutic subtotal or total tumor resection performed under image guidance. All specimens (19 total) were collected and submitted to the study under institutional review board-approved protocols. No chemotherapy or radiotherapy was performed on the patients before resection. Portions of the specimen from the main tumor mass and the invasive rim were immediately frozen on dry ice. Another portion was fixed in paraformaldehyde and paraffin-embedded for histologic evaluation. Histologic diagnosis was made by standard light microscopic evaluation of hematoxylin and eosin (H&E)-stained sections. All tumor samples were classified as grade 4 GBM according to the World Heath Organization [19].
Laser Capture Microdissection
Laser capture microdissection (LCM) was performed as described previously [20–22]. Briefly, 1000 to 2000 tumor core and invasive rim cells were dissected from 8-µm sections cut from four flash-frozen glioblastoma (WHO grade 4) tumors. Cells in the tumor core were identified and captured; tumor cells immediately adjacent to necrotic areas, cortical areas, cells with small, regular nuclei, or that evidenced features of endothelial and blood cells were avoided. White matter-invading GBM cells were identified by means of their nuclear atypia and heteropyknotic staining, which was consistent with that of the cells within the tumor core. Reactive astrocytes (discriminated by their distinct starlike morphology with eosinophilic cytoplasm and large, acentric, round nuclei) were avoided.
RNA Isolation and Amplification
Total RNA was isolated from LCM cells using the Paradise Reagent System (Arcturus, Mountain View, CA), and quantified by real-time reverse transcription-polymerase chain reaction (RT-PCR) performed with the LightCycler (Roche Diagnostics, Indianapolis, IN). This consisted of performing RT-PCR with Histone 3A primers using a serial dilution of cDNA of known concentrations as standards. The remaining RNA (approximately 10 ng) was amplified in two rounds with the RiboAmp RNA Amplification kit (Arcturus), yielding between 30 and 60 ng of copy RNA.
cDNA Microarray Analysis
The amount and quality of RNA preparations were evaluated on the Agilent 2100 Bioanalyzer with RNA 6000 Nano Reagents and Supplies (Agilent, Santa Clara, CA). The LCM analytes were hybridized to 44K Agilent Human Whole Genome oligo microarrays using Agilent SureHyb hybridization chambers; methods for microarray hybridization and washing were as described in the manufacturer's protocol. Hybridized DNA microarrays were scanned with a resolution of 5 µm on an Agilent Scanner G2505B workstation, and TIFF images were processed by Feature Extractor (Agilent) to measure intensity values, which were then exported into GeneSpring software version 7 (Silicon Genetics, Santa Clara, CA). Intensity values for each probe were divided by its control channel value in each sample; if the control channel was below 10 then 10 was used instead. The percentiles of all of the chips were calculated using only probes marked present. Intensity value for each probe was divided by the median of its measurements in all samples. Minimum Information About a Microarray Experiment-compliant raw data for this series of experiments have been deposited in the Gene Expression Omnibus (GEO) database maintained by the National Center for Biotechnology Information (accession no. GSE12689).1
Tissue Microarray
Five-micrometer sections from a glioma invasion tissue microarray (TMA) were subjected to the described staining methods using the anti-NHERF-1 antibody sc-51684 (Santa Cruz Biotechnology, Santa Cruz, CA). The TMA specifically contained WHO grade 4 glioma specimens (according to standardized criteria [22]) from 10 institutes. The glioma samples were obtained from patients who underwent primary therapeutic subtotal or total tumor resection performed under image guidance. The glioma invasion TMA is a consequence of an international consortium. All specimens (n = 31) were collected and submitted to the study under institutional review board-approved protocols. Patient information corresponding to the glioma samples on the TMA is available through an online database.2 The formalin-fixed paraffin-embedded tissue blocks were selected by both a neurosurgeon and a neuropathologist from each institution as listed on the online illumine database. In addition, each specimen block chosen met the criteria of nonnecrotic, nonirradiated, or chemotreated glioma tissue. Briefly, two separate face cuts were made for each of the specimens used to construct the TMA. H&E staining was performed on the face cuts to assist in the identification of the tumor cells. Two independent experienced neuropathologists (Dr. Ken Aldape, MD Anderson, and Dr. David Zagzag, New York University) reviewed the face cuts and designated the areas of core (center of the tumor) and rim (region distal to the edge but still containing notable tumor cells). The TMA was constructed from representative punches of tumor core and invasive rim using an indexed manual arrayer with attached stereomicroscope under the direction of Dr. Galen Hostetter, pathologist and Director of the TMA core facility at Translational Genomics Research Institute. Dr. Hostetter also reviewed and verified that the prescribed areas made by the neuropathologists were in agreement before punches were taken for the TMA paraffin block. Tissue microarray slides were stained by H&E every 50 sections to confirm that tissue morphology and diagnosis were consistent.
Immunohistochemistry-Fluorescence Staining
The TMA slide was baked at 65°C for 1 hour, deparaffinized in three xylene washes (2 minutes each), followed by a dehydration series of 100%, 95%, and 70% ethanol, then by rehydration in water (20 dips each). The slide was blocked, and antigens were retrieved using a sodium citrate-based buffer, pH 6.5, for 20 minutes (BondMax autostainer; Vision Biosystems, Norwell, MA). The primary monoclonal antibody, anti-NHERF-1 sc-51684 (Santa Cruz Biotechnology), was diluted at 1:100 and incubated for 30 minutes. For fluorescence staining, a 1:50 dilution of rabbit anti-glial fibrillary acidic protein (GFAP; Cell Signaling Technology, Danvers, MA) was used to stain the cytoplasmic portions of glial cells. A secondary antibody cocktail containing antirabbit Alexa 555 and anti-mouse-HRP was incubated for 30 minutes. Lastly, a tyramide signal amplification system using Cy5 (Perkin Elmer, Waltham, MA) was used to stain the primary target NHERF-1. The slide was coverslipped using ProLong Gold containing DAPI (Invitrogen). For immunohistochemistry (IHC) staining, the slide was incubated with a secondary antibody conjugated to HRP for 30 minutes followed by a diaminobenzidine (DAB) substrate. The slide was counterstained with hematoxylin and coverslipped for imaging.
Image Acquisition and Analysis
Fluorescence-based automated and quantitative analysis (AQUA; HistoRx technology [23,24]) measured alterations in the levels of expression of NHERF-1 within regions of interest defined by GFAP immunoreactivity within each spot of the TMA. The AQUA score is a numeric representation of fluorescence intensity in a user-defined area normalized by exposure time. Multiple, monochromatic, high-resolution (2048 x 2048 pixels) images were obtained from each TMA spot with an Olympus BX51RF microscope (Olympus, Melville, NY) [24] and analyzed with the AQUA software. For each TMA spot, areas of tumor are distinguished from stromal elements by creating an epithelial mask (GFAP) from the NHERF-1 protein signal, which was visualized through the Alexa 488 fluorophore. Positivity for NHERF-1 was determined by gating the pixels, in which an intensity threshold was set by visual inspection of TMA spots, and each pixel was recorded as “tumor” or “nontumor” by the software on the basis of the threshold. The DAPI image, used to designate the nuclei, was subjected to a rapid exponential subtraction algorithm that improves signal-to-noise ratio by subtracting the out-of-focus image from the in-focus image. After applying the exponential subtraction algorithm, the signal intensity of the target protein (NHERF-1), which was acquired under the Cy5 signal, was scored on a scale of 0 to 255. The AQUA score within the subcellular compartment of the cytoplasm was calculated by dividing the signal intensity by the area of the specified cytoplasmic compartment. Further scoring by a pathologist (J.M.E.) of NHERF-1 levels on the TMA was performed using a system for chromophore to capture the outcome: 1, negative to very weak; 2, moderate; 3, intense staining.
siRNA Preparation and Transfection
The siRNA oligonucleotide specific for GL2 luciferase was previously described [25]. The siRNA oligonucleotides specific for NHERF-1 were designed, validated, and purchased from Qiagen (Valencia, CA): NHERF-1-1 (494-514 bp; 5V-CTG CGG AAT GGA TCA CAC TGA) and NHERF-1-2 (2631-2651 bp; 5V-AAC TCA TTG GGT CAG CAA TTA). Transient transfection of siRNA was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were plated in a 60-mm plate at 8.0 x 105 cells per plate in 3 ml of Dulbecco's modified Eagle's medium, supplemented with 10% serum without antibiotics. Transfections were carried out according to the manufacturer's protocol after cells were fully adherent (6 hours after plating). Cells were transfected with NHERF-1 siRNA oligonucleotides or control GL2 luciferase siRNA oligonucleotides at a concentration of 20 nM for 16 hours. No cell toxicity was observed using this concentration of siRNA. Cells were assayed on either day 1 or day 2 after transfection.
Western Blot Analysis
Immunoblot analysis and protein determination experiments were preformed as previously described [26]. Briefly, monolayers of cells were washed in phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride and then lysed in 2x sodium dodecyl sulfate (SDI) sample buffer (0.25 M Tris-HCl, pH 6.8, 2% SDS, and 25% glycerol) containing 10 µg/ml aprotinin, 10 µg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Protein concentrations were determined using the BCA assay procedure (Pierce, Rockford, IL), with bovine serum albumin as reference. Fifteen micrograms of total cellular protein was loaded per lane, separated by 10% SDS-polyacrylamide gel electrophoresis, and then transferred to nitrocellulose (Invitrogen) by electroblotting. The nitrocellulose membrane was blocked with 5% nonfat dry milk in Tris-buffered saline (pH 8.0) with 0.1% Tween-20 before the addition of the primary antibody and then HRP-conjugated anti-mouse/rabbit IgG (Promega, San Luis Obispo, CA). Bound secondary antibodies were detected using a chemiluminescence system (NEN Life Science Products, Boston, MA). A mouse monoclonal antibody to NHERF-1 was obtained from Santa Cruz Biotechnology and used at a concentration of 1:1000. The rabbit polyclonal antibody to poly(ADP-ribose)polymerase (PARP) was obtained from Upstate Cell Signaling Solutions (Charlottesville, VA) and used at a concentration of 1:1000. A mouse monoclonal antibody for α-tubulin was obtained from Santa Cruz Biotechnology. All secondary antibodies were obtained from Promega.
Radial Cell Migration Assay
Quantification of cellular migration was performed using a microliter scale migration assay as described previously [26]. Briefly, 10-well slides were coated with 10 µg/ml human laminin at 37°C for 1 hour and washed with PBS to enhance cell attachment while promoting migration. Approximately 2000 cells were plated onto each well of slides using a cell sedimentation manifold (CSM, Inc., Phoenix, AZ) to establish a confluent 1-mm-diameter monolayer. Cells transfected with siRNA were treated in monolayer before transferring into manifolds. After sedimentation and adhesion, cells were allowed to disperse for 24 to 48 hours. Measurements of the area occupied by the cells were taken at regular intervals for 48 hours. The average radial migration rate of five replicates was calculated as the increasing radius of the entire cell population over time.
Three-dimensional Spheroid Dispersion Assay
Spontaneous multicellular spheroids, derived from T98G and SF767 cells, were sandwiched between 10 µl of collagen I gel below the spheroid and 25 µl of collagen I gel on top of the spheroid (Vitrogen; Angiotech, Palo Alto, CA) supplemented with MEM, 2% FBS in each well of a 384-well plate (Nunc, Rochester, NY), and overlain with 20 µl of MEM, 10% FBS. Eighteen hours after implantation, dispersion assays were monitored (Axiovert 40 CFL; Zeiss, Thornwood, NY) for up to 72 hours. Hanging drop culture [27] was used for all subsequent spheroid production where 2 x 105 cells/ml for T98G and SF767 lines were seeded in 20-µl droplets and cultured in a water-filled dish for 4 days. Cells transfected with siRNA were treated in monolayer before the formation of hanging drops.
Cell-Cell Adhesion Assay
Cells were grown to 75% confluency and treated with NHERF-1 siRNA or luciferase control overnight; the medium was changed and the cells were allowed to recover for 24 hours. The cells were washed with calcium/magnesium-free PBS and detached from the culture dishes with 4 mM EDTA in calcium/magnesium-free PBS to preserve cell surface expression of cadherin subtypes. Cells were separated to a single-cell suspension by trituration with a Pasteur pipette. After centrifugation, the cells were suspended in a final concentration of 0.5 million cells/ml in calcium-free suspension-modified Eagle's medium (Invitrogen) in the absence of serum, and 106 cells were maintained in suspension on 1 mg/ml poly-2-hydroxyethyl methacrylate-coated (Sigma, St. Louis, MO) 35-mm2 culture dishes to prevent cell attachment to the substrate. Images of cells, on the substrate of at least three fields per well, were taken after 60 minutes in culture using a Zeiss Axiovert 40 CFL. Cell aggregates were counted based on the number of cells per aggregate (see Results).
Immunofluoresence Microscopy
T98G and SF767 cells (n = 800) under control or siRNA NHERF-1 conditions were cultured for 24 hours then fixed with 4% paraformaldehyde. Cells were permeabilized with 0.5% Triton X-100 for 10 minutes, blocked with 2.5% FBS for 5 minutes, then stained with Alexa Fluor 546-labeled phalloidin antibody at a 1:140 dilution (Invitrogen) followed by PBS washes. Omission of primary antibody was used as a negative control. Slides were counterstained with DAPI and coverslipped using ProLong Gold antifade reagent with DAPI (Invitrogen). Cells were viewed using a 40x Zeiss oil immersion objective on a Zeiss LSM 510 inverted confocal microscope.
Cell Viability Assay
The Alamar Blue assay (Biosource, Camarillo, CA) was used to assess viability as described previously [28]. Briefly, 4000 cells of each population were seeded in quadruplicate wells of 96-well plastic plates in 200 µl of culture medium supplemented with 10% FBS. Cells were treated with various conditions, and the assay was developed 48 hours later. Alamar Blue was then added to the cells in a volume of 20 µl (10% of total volume) per well, and the plates were incubated for 6 hours at 37°C. The plates were read on a fluorescence plate reader (excitation, 560 nm; emission, 600 nm). Averages of the fluorescence values were calculated and normalized to controls without TMZ (Sigma; IC50 of 250 µM for T98G and 125 µM for SF767).
Results
NHERF-1 Expression Is Increased in Invasive Glioma Cells In Vivo
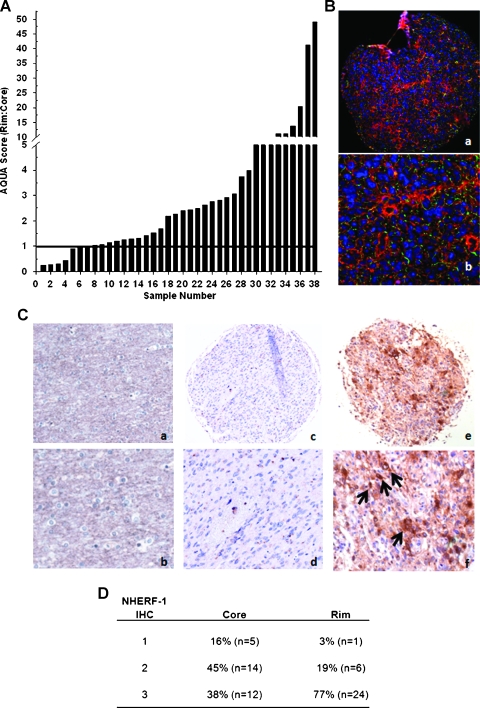
We examined whether increased expression of NHERF-1 coincided with the migratory behavior of invasive glioma cells in vivo. Two paired glioma subpopulations, from the tumor core or the invasive rim, were isolated from 19 specimens. Using gene expression profiling, followed by fold change analysis, we selected genes that were differentially regulated in the tumor core and the rim. One of the candidate genes identified to be overexpressed in the rim of more than 50% of the tumors was NHERF-1. In 10 of 19 glioblastoma core and rim biopsies, samples of cells invading at the rim showed 1.5- to nearly 6-fold induction of NHERF-1 compared with cells isolated from the core (Figure 1). Because of the heterogeneity of glioblastoma, it is not surprising that only a little more than 50% of the population exhibited an overexpression of NHERF-1 in the invasive rims of the tumors. Quantitative real-time polymerase chain reaction (qRT-PCR) validated the overexpression of NHERF-1 in 9 of the 10 GBM cases and validated the underexpression of NHERF-1 in 7 of the 9 glioma samples (data not shown).These findings substantiate that up-regulation of NHERF-1 accompanies migration of tumor cells at the rim of GBM tumor samples.
Figure 1.
Na+/H+ exchanger regulatory factor 1 is overexpressed in invasive GBM cells. From 19 independent GBM specimens, 1000 to 2000 stationary (core) and invasive (rim) cells were harvested by LCM for microarray analysis. Relative NHERF-1 mRNA signal intensity was expressed as a ratio of rim to core.
Immunohistochemical Validation of NHERF-1 Protein Expression in a Brain Tumor TMA
The protein expression of NHERF-1 from a series of primary human glioblastoma specimens (independent sample set from the 19 specimens described previously) assembled on a TMA consisting of 38 tumor cases containing core and rim was examined by immunofluorescence staining. AQUA scores [23] of NHERF-1 within GFAP regions of interest were derived for matched samples. Greater levels of NHERF-1 expression (AQUA score > 1) were observed in the invasive rim compared with the core in 31 of 38 cases (Figure 2, A and B). Because AQUA is a relatively recent technology for quantifying immunofluorescence in tissue, data from Figure 2, A and B, were confirmed by DAB-IHC staining and manual visual scoring of NHERF-1 using sections from the same TMA. Of the 31 evaluable GBM cores, 5 were negative to weakly positive (16%, score = 1), 14 were moderately positive (42.4%, score = 2), and 12 were intensely positive (38%, score = 3). Of the 31 evaluable GBM rims, 1 was negative to weakly positive (3%, score = 1), 6 were moderately positive (19%, score = 2), and 24 were intensely positive (77%, score = 3; Figure 2D). The immunoreactivity of NHERF-1 was observed in both glioblastoma tumor cores (Figure 2C: c and d) and invading cells (Figure 2C: e and f). The highest amount of NHERF-1 protein expression was found in GBM cells at the invasive rims of these tumors. There are 3 (9.3%of matched samples) of the 31 cases where NHERF-1 expression is elevated in the cores compared with that in the rims (score of 3 in cores and 2 in rims). These contradicting results could be attributed to the heterogeneity of the disease. Examination of NHERF-1 levels in control nonneoplastic autopsy brain specimens (Figure 2C: a and b) showed negative to very weak staining of NHERF-1, which was present in some reactive astrocytes, as confirmed by a pathologist (J.M.E.).
Figure 2.
Quantitative analysis of NHERF-1 expression in 38 human glioma tumors. (A) AQUA scores of NHERF-1 protein levels were measured in matched sets of rim:core for each tumor; ratio of AQUA scores for each specimen are plotted, ranked from low to high. Samples with an AQUA score fold difference above “1” indicates significant up-regulation (indicated by horizontal bar). (B) Paraffin section of tumor core from specimen no. A13 on glioma TMA immunofluorescently stained with a monoclonal antibody against NHERF-1 and analyzed by a HistoRx imaging system. Red, NHERF-1 staining; green, GFAP for glial fiber staining; blue, DAPI nuclear staining. Tumor core original magnifications, x10 (a) and x20 (b). (C) Paraffin sections of tumor-free nonneoplastic brain (a, b), glioma core (c, d), and invasive rim (e, f) from TMA immunostained against NHERF-1. Original magnifications, x10 (a, c, e) and x20 (b, d, f). Arrows indicate representative NHERF-1 positive staining. (D) The DAB-IHC scoring of NHERF-1 expression of the TMA.
Depletion of NHERF-1 in Glioma Cells Inhibits Migration and Dispersion and Increases Cell-Cell Adhesion In Vitro
To examine the functional roles of NHERF-1, two independent sequences of siRNA designed specifically to inhibit NHERF-1 were introduced into T98G and SF767 glioma cells. Western blots were performed to assess the level of NHERF-1 in both cell lines transfected with either a luciferase control or two NHERF-1 siRNA. Figure 3, A and B, confirms the decrease of NHERF-1 protein levels in NHERF-1 siRNA-transfected T98G and SF767 cells relative to untransfected and luciferase controls. Furthermore, NHERF-1 mRNA was reduced by approximately 60% to 90% by both NHERF-1-1 and NHERF-1-2 siRNA as a confirmation by qRT-PCR (data not shown). Monolayer radial migration assays revealed that reduction of NHERF-1 in T98G and SF767 glioma cells significantly decreased in migration rates relative to mock-transfected cells (Figure 3, C and D).
Figure 3.
Na+/H+ exchanger regulatory factor 1 inhibits migration and dispersion in vitro. Reduction of NHERF-1 using siRNA decreases glioma migration/dispersion. (A) Western blot analysis (α-tubulin used as a loading control) to confirm reduction of NHERF-1 in the glioma cell lines T98G and (B) SF767 transfected with two NHERF-1-specific siRNA. Untransfected cells and cells transfected with a control siRNA (luciferase) are shown for comparison. Treatment with two separate NHERF-1 siRNA decreased migration of (C) T98G (luciferase: 7.7 ± 0.53 µm/h, NHERF-1-1: 2.2 ± 0.94 µm/h, NHERF-1-2: 0.82 ± 0.53 µm/h) and (D) SF767 cells in a radial migration assay when compared with untreated or luciferase-transfected cells (luciferase: 11.6 ± 0.46 µm/h, NHERF-1-1: 3.3 ± 0.65 µm/h, NHERF-1-2: 5.2 ± 1.4 µm/h). Values shown are the average of 5 ± SEM and representative of three independent experiments. For T98G cells: *P < .01, comparing luciferase-treated cells to NHERF-1-1-treated cells; **P < .01, comparing luciferase-treated cells to NHERF-1-2-treated cells. For SF767 cells: *P < .001, comparing luciferase-treated cells to NHERF-1-1-treated cells; **P < .001, comparing luciferase-treated cells to NHERF-1-2-treated cells; 2-tailed Student's t-test. (E) Dispersion of T98G cells from multicellular spheroid in 1% collagen is inhibited when NHERF-1 is depleted. Upper left panel indicates luciferase-transfected spheroid at 0 hour, upper right panel after 72 hours. Lower left panel indicates NHERF-1 siRNA-transfected (combination of the two sequences) spheroid at 0 hour, lower right panel after 72 hours. (F) Depletion of NHERF-1 causes SF767 cells to form large spheroids in 1% collagen culture. Upper left panel indicates luciferase-transfected spheroid at 0 hour, upper right panel after 72 hours. Lower left panel indicates NHERF-1 siRNA-transfected (combination of the two sequences) spheroid at 0 hour, lower right panel after 72 hours. Images shown are representative of three independent experiments.
In addition, cell spheroids that propagated three-dimensionally in collagen I gel indicate that inhibition of NHERF-1 by two independent NHERF-1 siRNA causes an evident decrease or total lack of cell dispersion of T98G glioma cells relative to mock-transfected cells (Figure 3E). Moreover, depletion of NHERF-1 expression in SF767 cells resulted in tight and robust spheroid formations in contrast to untreated cells that are typically incapable of spheroid formation and dispersion in collagen I gel (Figure 3F). In addition, when NHERF-1 levels are decreased, T98G and SF767 glioma cells showed a significant increase in cell-cell adhesion, through aggregate formation, compared with control cells treated with luciferase siRNA (Figure 4). Moreover, when NHERF-1 was depleted, T98G and SF767 glioma cells showed a significant increase in cell-cell matrix adhesion compared with control cells treated with luciferase plated on either bovine serum albumin or laminin (data not shown). These findings suggest that when NHERF-1 is downregulated, dispersion from a multicellular spheroid is compromised, and cells undergo a phenotypic switch causing an increase in cell-cell cohesion, likely due to the expression of one or more cell adhesion molecules.
Figure 4.
Depletion of NHERF-1 results in increased glioma cell aggregation. (A) Photomicrographs (phase contrast) of T98G and SF767 cells treated with NHERF-1 siRNA (combination of the two sequences) - or luciferase siRNA (control). Scale bar, 20 µm. (B) T98G (cell aggregates: [5–10 cell size] luciferase, 28.67 ± 4.16; NHERF-1 siRNA, 11.00 ± 1.00; [50–100 cell size] luciferase, 14.33 ± 5.51; NHERF-1 siRNA, 6.33 ± 1.15; [500+ cell size] luciferase, 3.33 ± 0.58; NHERF-1 siRNA, 35.67 ± 9.61) and (C) SF767 cell suspensions were scored visually for aggregated cells after 60 minutes in culture (cell aggregates: [5–10 cell size] luciferase, 32.00 ± 2.00; NHERF-1 siRNA, 15.33 ± 4.04; [10–50 cell size] luciferase, 12.33 ± 5.51; NHERF-1 siRNA, 21.33 ± 3.21; [100+ cell size] luciferase, 6.67 ± 2.08; NHERF-1 siRNA, 38.33 ± 0.58). Bars, the average number of cell aggregates after NHERF-1 siRNA. Values represent the mean and SD from three randomly selected fields. *P < .03, **P < .01, when comparing NHERF-1 siRNA-treated cell aggregates with corresponding luciferase-transfected control cell aggregates (2-tailed Student's t-test).
NHERF-1 Reduction Causes a Morphologic Change in Glioblastoma Cells Accompanied by Reorganization of the Actin Cytoskeleton
The resulting effects on motility and dispersion described above led us to examine the influence of NHERF-1 on T98G and SF767 cell morphology and cytoskeleton organization. Previous studies in a breast cancer model have shown that tumor cells that overexpressed NHERF-1 induced leading-edge pseudopodia and localized NHERF-1 to the pseudopodial tips, suggesting a role for NHERF-1 in tumor migration [29]. We therefore investigated the effects of NHERF-1 on actin using Alexa Fluor 546-labeled phalloidin, which binds to F-actin, and counterstained the nuclei with DAPI. The cells were transfected with NHERF-1 siRNA, or with the control siRNA specific for luciferase, for 24 hours under the same conditions as those used in the radial migration assay, stained, and then imaged. Fluorescent images of the stained cells (red, actin; blue, nucleus) are shown in Figure 5. In the control cells (left panels), F-actin was organized in stress fibers and morphologically showed small cell bodies with long extensions indicative of cell migration. In the NHERF-1 siRNA-treated T98G and SF767 cells, most of the F-actin was diffuse, and the cells exhibited a rounder and much larger morphology (right panels). The NHERF-1 knockdown resulted in a decrease in stress fibers and focal adhesions, suggesting that their assembly had been weakened, which could contribute to the cells' decreased migration and increased cohesion.
Figure 5.
Na+/H+ exchanger regulatory factor 1 inhibition causes morphological changes in glioma cells. T98G and SF767 cells with or without NHERF-1 siRNA treatment were seeded at a density of 1 x 104 per well on laminin-coated 10-well slides. After 24 hours, cells were fixed and stained with Alexa Fluor 546-labeled phalloidin (red fluorescence) and counterstained with DAPI (blue fluorescence). Confocal fluorescence images were taken (objective, 40x; scale bar, 10 µm).
NHERF-1 siRNA Sensitizes Glioma Cells to TMZ
There has been prior evidence indicating that apoptosis is suppressed when cells convey a migratory phenotype [30–33]. Because we have shown NHERF-1 to be a regulator of cell migration and adhesion, we evaluated whether depletion of NHERF-1 affected glioma cell sensitivity to TMZ treatment. Both T98G and SF767 cells were significantly more sensitive to cell death when NHERF-1 was depleted followed by 48 hours of treatment with 250 or 125 µM TMZ, respectively, compared with the luciferase controls treated with TMZ (Figure 6, A and B).
Figure 6.
Effect of NHERF-1 siRNA on vulnerability of T98G and SF767 cells to TMZ. (A) T98G and (B) SF767 cells were treated for 48 hours with NHERF-1 siRNA along with either 250 or 125 µM TMZ treatment, respectively. Bars, percentage cell survival normalized to the controls not containing TMZ. Values represent mean and SD from three independent experiments (T98G [% survival ± % SD]: NHERF-1-1 + TMZ: 23.17 ± 0.967, NHERF-1-2 + TMZ: 23.38 ± 0.948) (SF767 [% survival ± % SD]: NHERF-1-1 + TMZ: 32.65 ± 1.08, NHERF-1-2 + TMZ: 36.77 ± 0.235). For T98G cells: *P < .01, comparing luciferase-treated cells to NHERF-1-1-treated cells; **P < .01, comparing luciferase-treated cells to NHERF-1-2-treated cells. For SF767 cells: *P < .001, comparing luciferase-transfected cells treated with TMZ to NHERF-1-1-transfected cells treated with TMZ; **P < .001 comparing luciferase-transfected cells treated with TMZ to NHERF-1-2-transfected cells treated with TMZ (2-tailed Student's t-test). Data are representative of two independent experiments, each run with triplicate samples. (C) Western blot analysis to determine PARP cleavage. T98G and SF767 glioma cells were treated with nothing, luciferase, NHERF-1-1, or NHERF-1-2 siRNA with or without TMZ. After 24 hours, whole-cell lysates were prepared and immunoblotted for whole and cleaved PARP. Ponceau staining was used as a loading control. Data are representative of two independent experiments.
Furthermore, validation of increased apoptosis by TMZ after the inhibition of NHERF-1 was confirmed by Western blot analysis of PARP cleavage after 24 hours of TMZ treatment. T98G and SF767 glioma cell lines manifest minimal levels, if any, of PARP cleavage when untreated, when treated with luciferase, or when in the presence of NHERF-1 siRNA alone. However, when both cell lines were first introduced with NHERF-1 siRNA, then treated with TMZ for 24 hours, there was a pronounced increase in the levels of PARP cleavage compared with cells treated with TMZ alone or in combination with luciferase siRNA (Figure 6C). These results indicate that when NHERF-1 is inhibited in glioma cells, they are sensitized to TMZ treatment evidenced by an increase in apoptotic cell death.
Discussion
We report that increased levels of NHERF-1 protein and mRNA expression associate with the invasive behavior of malignant glioma cells in vivo. In addition, immunohistochemical validation using a brain tumor TMA portrays the highest level of NHERF-1 protein expression in GBM cells at the invading rims of the tumors rather than at the tumor cores. Examination of NHERF-1 levels in non-neoplastic autopsy brain sections showed negative to very weak staining of NHERF-1, which occurred mainly in reactive astrocytes. Knockdown of NHERF-1 elicits antimigratory and antidispersive phenotypes in glioma cells as well as an increase in cell-cell adhesion. Moreover, a decrease in stress fibers and a morphological change were observed on NHERF-1 depletion in GBM cell lines. When NHERF-1 is inhibited, glioma cells become sensitized to cytotoxic effects of TMZ and undergo apoptotic cell death. These results suggest that NHERF-1 is a candidate therapeutic target of invasive glioma.
Na+/H+ exchanger regulatory factor 1 has an established function as a scaffolding protein that recruits membrane receptors/transporters and cytoplasmic signaling proteins into functional complexes localized at or near the plasma membrane [4]. In addition, NHERF-1 is overexpressed in several tumor types, including schwannoma [9] and hepatocellular carcinomas [10], and in breast cancer [3]. In the current study, NHERF-1 is reported to be differentially upregulated in migrating and invading human glioma cells, above elevated levels in the tumor core.
In a recent study, it was reported that NHERF-1 expression is found in varying amounts within regions of the normal rodent brain. Notably, neuronal elements (i.e., neuronal cell bodies visible in the granular layer of the dentate gyrus and the pyramidal cell bodies in the stratum pyramidale in the CA1 region of the hippocampus) did not contain NHERF-1; however, astrocytes surrounding such neurons were densely labeled [34]. In our study, NHERF-1 is strongly expressed in GBM cells located in the invading rim of human glioblastoma samples relative to tumor core and suggests that NHERF-1 may be a driver or contributor to glioblastoma invasion. Our laser-captured microarray data set is unique in its ability to decipher the expression of core and invasive rims of GBM tumors. We also accessed the REMBRANDT public expression data set, representative of brain tumor cores (http://caintegrator-info.nci.nih.gov/rembrandt). The REMBRANDT database expression of NHERF-1 was unchanged across tumor grade for the core biopsies available. Furthermore, in a recent breast cancer study, overexpression of NHERF-1 in MDA-MB-435 cells promoted a hypoxic or serum-deprived tumor microenvironment and induced leading-edge pseudopodia and corresponding redistribution of NHERF-1 to the pseudopodia tip [29]. Moreover, Cardone et al. [29] determined that escaping MDA-MB-435 cells from tumor lobules contained in three-dimensional Matrigel culture revealed NHERF-1 disproportionately localized to the pseudopodial tip of the escaping and invading tumor cells, suggesting a role for NHERF-1 in tumor cell invasion. These studies demonstrate that NHERF-1 is involved in cytoskeletal reorganization and motility. In support of these results, we find that glioblastoma cells lose their ability to migrate on laminin or to invade in a collagen-based environment when NHERF-1 is depleted.
Our experiments also show that SF767 cells, which typically are unable to form spontaneous spheroids, generate self-aggregating multicellular assemblies after depleting NHERF-1 by siRNA. It also seems that when NHERF-1 levels are decreased in T98G cells, the spheroids adopt very tight and well-defined borders. On the basis of these observations, we evaluated the propensity of single-cell suspensions of T98G and SF767 cells to develop cell-cell cohesion. Cell-cell cohesion assays clearly showed that NHERF-1 knockdown elicited stronger cell-cell aggregation when compared with luciferase-transfected control cells. A quantitative measurement of cell-cell adhesion after 60 minutes in culture, scored as aggregates as clumps of cells, revealed that glioma cells with decreased NHERF-1 expression had significantly higher numbers of aggregates of a larger size, compared with that in luciferase-transfected control cells. These increases in aggregation were consequent to depletion of NHERF-1, suggesting that an adhesion molecule(s) is likely involved in NHERF-1-related cell-cell adhesion. In addition, the presence of NHERF-1 induces a promotility phenotype resulting in actin cytoskeletal rearrangements that decrease the adhesive properties of the cells. Our data determined that when NHERF-1 is present in glioma cells, there are prevalent stress fibers observed within elongated motile cell bodies. However, when NHERF-1 is diminished, there is a visible decrease in stress fibers. Amore rounded cell is also observed along with what appeared to be focal adhesion complexes in many of the cells. These data suggest that NHERF-1 plays a major role in tumor cell migration and dispersion, and when inhibited, glioma cells cease to migrate and possibly increase the expression of cell adhesion molecules. On the basis of these phenotypic data, we speculate that NHERF-1 is involved in integrin signaling pathways leading to motility and stress fiber induction in glioma cells. Studies are underway to investigate the mechanism of regulation of adhesion- and migration-based molecules by changes in NHERF-1 expression, particularly the role of integrin-mediated signaling processes.
A major characteristic of glioblastoma is the propensity to invade and become resistant to chemotherapeutic agents [15,26,35–39]. Temozolomide is an oral alkylating agent that readily crosses the blood-brain barrier, and it has been used for the treatment of brain cancer since 1999. It induces apoptosis in neoplastic cells by interfering with DNA replication [40]. However, a main cause of treatment failure, with TMZ treatment [41], is the ability of tumor cells to acquire resistance to apoptosis, which is necessary for tumor development and progression [37,42]. The absence of apoptosis resistance would otherwise induce tumor cell death when deprived of the support from neighboring cancer cells [15,37,42]. In this study, cell migration and dispersion are inhibited and an increased sensitization of glioblastoma cells to TMZ treatment is observed when NHERF-1 expression is depleted. Specifically, NHERF-1 down-regulation in conjunction with TMZ treatment showed an increase in apoptosis versus TMZ treatment alone. A possible molecular link between migration arrest due to NHERF-1 inhibition and enhanced TMZ-induced apoptosis is currently under investigation.
In summary, our current findings indicate an important role of NHERF-1 in the regulation of glioma cell migration and dispersion. Significantly, these data provide the first evidence that NHERF-1 is upregulated in migrating GBM cells in vitro and in vivo, and when silenced, tumor cells exhibit little to no migratory or dispersion activity leading to an increased susceptibility and vulnerability to chemotherapy. Specifically, when NHERF-1 is inhibited, glioma cells become sensitized to TMZ treatment. Further investigation of downstream signaling mechanisms of NHERF-1 related to adhesion molecules needs to be conducted to determine its role in glioma invasion. It is necessary to understand the biologic functions of NHERF-1 to allow innovative therapeutic interventions to be developed with the goal of targeting migratory and invasive glioma cells.
Supplemental Data
Supplementary Material
Supplemental Method: qRT-PCR
Real-time quantitative PCR was performed using a Light-Cycler (Roche Diagnostics) with SYBR green fluorescence signal detection. Briefly, total RNA was isolated from human brain tumor tissues using the Paradise Reagent System (Arcturus). cDNA was synthesized from 1 µg of DNase I-treated total RNA in a 20-µl reaction volume using the Retroscript kit (Ambion Inc., Austin, TX) for 60 minutes at 42°C. Quantitative real-time polymerase chain reaction was performed on 2.5 µl of the cDNA diluted 1:5 from final Retroscript reaction using the following primers: NHERF-1, sense 5′-CCT TCA CCA ATG GGG AGA TA-3′ and antisense 5′-GGT CGG AGG AGG AGG TAG AC-3′ (amplicon size, 207 bp); histone 3A, sense 5′-CCA CTG AAC TTC TGATTC GC-3′ and antisense 5′-GCG TGC TAG CTG GAT GTC TT-3′ (amplicon size, 215 bp; Operon, Huntsville, AL). The qRT-PCR data were analyzed with the LightCycler analysis software to calculate crossing point values. The difference in the cycle number was normalized to the housekeeping gene histone 3A (H) and then used to calculate the fold difference in copy number according to the formula: F = 2-(HsiRNA - HsiRNA)(HNT - HNT), where F is fold difference in NHERF-1 expression, H is histone 3A, N is NHERF-1 (gene of interest), siRNA is siRNA-treated cells, and NT is cells given no treatment (Figure W1).
Acknowledgments
The authors thank Stephen Coons, Roland Goldbrunner, David Zagzag, Ab Guha, Luigi Mariani, Joseph Megyesi, Tom Mikkelson, Andy Sloan, and John Trusheim for providing glioblastoma tissue samples and biopsy specimens; Aprill Watanabe for the assembly of the glioma invasion TMA; and Nhan Tran for review and helpful discussions of the manuscript. The technical and scientific contribution of Tim Demuth and participation by Dominique Hoelzinger and Linsey Reavie in collecting the analytes for the GBM core/rim expression profiling are also gratefully acknowledged.
Abbreviations
- NHERF-1
Na+/H+ exchanger regulatory factor 1
- GBM
glioblastoma multiforme
- TMZ
temozolomide
- TMA
tissue microarray
Footnotes
Supported by grants 5R01NS042262 and ABRC0806 and by Students Supporting Brain Tumor Research Fund.
This article refers to supplementary material, which is designated by Figure W1 and is available online at www.neoplasia.com.
Reviewer access to GBM Invasion Array GEO series: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lbafrgcqkqomcvw&acc=GSE12689.
Online database in Illumine may be accessed at https://illumine.5amsolutions.com/ with Username: reviewer, Password: glassbox2008; read-only access.
References
- 1.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 2.Keles GE, Berger MS. Advances in neurosurgical technique in the current management of brain tumors. Semin Oncol. 2004;31:659–665. doi: 10.1053/j.seminoncol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Stemmer-Rachamimov AO, Wiederhold T, Nielsen GP, James M, Pinney-Michalowski D, Roy JE, Cohen WA, Ramesh V, Louis DN. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas. Am J Pathol. 2001;158:57–62. doi: 10.1016/S0002-9440(10)63944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology (Bethesda) 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- 5.Weinman EJ, Hall RA, Friedman PA, Liu-Chen LY, Shenolikar S. The association of NHERF adaptor proteins with G protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- 6.Brone B, Eggermont J. PDZ proteins retain and regulate membrane transporters in polarized epithelial cell membranes. Am J Physiol Cell Physiol. 2005;288:C20–C29. doi: 10.1152/ajpcell.00368.2004. [DOI] [PubMed] [Google Scholar]
- 7.Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 8.Voltz JW, Brush M, Sikes S, Steplock D, Weinman EJ, Shenolikar S. Phosphorylation of PDZ1 domain attenuates NHERF-1 binding to cellular targets. J Biol Chem. 2007;282:33879–33887. doi: 10.1074/jbc.M703481200. [DOI] [PubMed] [Google Scholar]
- 9.Fraenzer JT, Pan H, Minimo L, Jr, Smith GM, Knauer D, Hung G. Overexpression of the NF2 gene inhibits schwannoma cell proliferation through promoting PDGFR degradation. Int J Oncol. 2003;23:1493–1500. [PubMed] [Google Scholar]
- 10.Shibata T, Chuma M, Kokubu A, Sakamoto M, Hirohashi S. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38:178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- 11.Bockhorn M, Roberge S, Sousa C, Jain RK, Munn LL. Differential gene expression in metastasizing cells shed from kidney tumors. Cancer Res. 2004;64:2469–2473. doi: 10.1158/0008-5472.can-03-0256. [DOI] [PubMed] [Google Scholar]
- 12.Buttiglieri S, Deregibus MC, Bravo S, Cassoni P, Chiarle R, Bussolati B, Camussi G. Role of Pax2 in apoptosis resistance and proinvasive phenotype of Kaposi's sarcoma cells. J Biol Chem. 2004;279:4136–4143. doi: 10.1074/jbc.M306824200. [DOI] [PubMed] [Google Scholar]
- 13.Haga A, Funasaka T, Niinaka Y, Raz A, Nagase H. Autocrine motility factor signaling induces tumor apoptotic resistance by regulations Apaf-1 and caspase-9 apoptosome expression. Int J Cancer. 2003;107:707–714. doi: 10.1002/ijc.11449. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita K, Taupin DR, Itoh H, Podolsky DK. Distinct pathways of cell migration and antiapoptotic response to epithelial injury: structure-function analysis of human intestinal trefoil factor. Mol Cell Biol. 2000;20:4680–4690. doi: 10.1128/mcb.20.13.4680-4690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–2422. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 16.Sekharam M, Zhao H, Sun M, Fang Q, Zhang Q, Yuan Z, Dan HC, Boulware D, Cheng JQ, Coppola D. Insulin-like growth factor 1 receptor enhances invasion and induces resistance to apoptosis of colon cancer cells through the Akt/Bcl-x(L) pathway. Cancer Res. 2003;63:7708–7716. [PubMed] [Google Scholar]
- 17.Megalizzi V, Mathieu V, Mijatovic T, Gailly P, Debeir O, De Neve N, Van Damme M, Bontempi G, Haibe-Kains B, Decaestecker C, et al. 4-IBP, a sigma1 receptor agonist, decreases the migration of human cancer cells, including glioblastoma cells, in vitro and sensitizes them in vitro and in vivo to cytotoxic insults of proapoptotic and proautophagic drugs. Neoplasia. 2007;9:358–369. doi: 10.1593/neo.07130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berens ME, Rief MD, Loo MA, Giese A. The role of extracellular matrix in human astrocytoma migration and proliferation studied in a microliter scale assay. Clin Exp Metastasis. 1994;12:405–415. doi: 10.1007/BF01755884. [DOI] [PubMed] [Google Scholar]
- 19.Kleihues P, Sobin LH. World Health Organization classification of tumors. Cancer. 2000;88:2887. doi: 10.1002/1097-0142(20000615)88:12<2887::aid-cncr32>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 20.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani L, McDonough WS, Hoelzinger DB, Beaudry C, Kaczmarek E, Coons SW, Giese A, Moghaddam M, Seiler RW, Berens ME. Identification and validation of P311 as a glioblastoma invasion gene using laser capture microdissection. Cancer Res. 2001;61:4190–4196. [PubMed] [Google Scholar]
- 22.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 24.HistoRx, Inc., author [February 12, 2009];The Quantitative Tissue Biomarker Platform. 2005 Available at: http://www.historx.com.
- 25.Chuang YY, Tran NL, Rusk N, Nakada M, Berens ME, Symons M. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- 26.McDonough WS, Tran NL, Berens ME. Regulation of glioma cell migration by serine-phosphorylated P311. Neoplasia. 2005;7:862–872. doi: 10.1593/neo.05190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demuth T, Reavie LB, Rennert JL, Nakada M, Nakada S, Hoelzinger DB, Beaudry CE, Henrichs AN, Anderson EM, Berens ME. MAP-ing glioma invasion: mitogen-activated protein kinase kinase 3 and p38 drive glioma invasion and progression and predict patient survival. Mol Cancer Ther. 2007;6:1212–1222. doi: 10.1158/1535-7163.MCT-06-0711. [DOI] [PubMed] [Google Scholar]
- 28.Nakada M, Niska JA, Tran NL, McDonough WS, Berens ME. EphB2/R-Ras signaling regulates glioma cell adhesion, growth, and invasion. Am J Pathol. 2005;167:565–576. doi: 10.1016/S0002-9440(10)62998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardone RA, Bellizzi A, Busco G, Weinman EJ, Dell'Aquila ME, Casavola V, Azzariti A, Mangia A, Paradiso A, Reshkin SJ. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18:1768–1780. doi: 10.1091/mbc.E06-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SY, Klemke RL. Extracellular-regulated kinase activation and CAS/Crk coupling regulate cell migration and suppress apoptosis during invasion of the extracellular matrix. J Cell Biol. 2000;149:223–236. doi: 10.1083/jcb.149.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Demuth T, Ross KR, Berens T, Coons SW, Watts G, Trent JM, et al. Glioma cell motility is associated with reduced transcription of proapoptotic and proliferation genes: a cDNA microarray analysis. J Neurooncol. 2001;53:161–176. doi: 10.1023/a:1012253317934. [DOI] [PubMed] [Google Scholar]
- 32.Ridley AJ, Allen WE, Peppelenbosch M, Jones GE. Rho family proteins and cell migration. Biochem Soc Symp. 1999;65:111–123. [PubMed] [Google Scholar]
- 33.Senger DL, Tudan C, Guiot MC, Mazzoni IE, Molenkamp G, LeBlanc R, Antel J, Olivier A, Snipes GJ, Kaplan DR. Suppression of Rac activity induces apoptosis of human glioma cells but not normal human astrocytes. Cancer Res. 2002;62:2131–2140. [PubMed] [Google Scholar]
- 34.Lee A, Rayfield A, Hryciw DH, Ma TA, Wang D, Pow D, Broer S, Yun C, Poronnik P. Na+-H+ exchanger regulatory factor 1 is a PDZ scaffold for the astroglial glutamate transporter GLAST. Glia. 2007;55:119–129. doi: 10.1002/glia.20439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedlander DR, Zagzag D, Shiff B, Cohen H, Allen JC, Kelly PJ, Grumet M. Migration of brain tumor cells on extracellular matrix proteins in vitro correlates with tumor type and grade and involves alphaV and beta1 integrins. Cancer Res. 1996;56:1939–1947. [PubMed] [Google Scholar]
- 36.Lipinski CA, Tran NL, Menashi E, Rohl C, Kloss J, Bay RC, Berens ME, Loftus JC. The tyrosine kinase pyk2 promotes migration and invasion of glioma cells. Neoplasia. 2005;7:435–445. doi: 10.1593/neo.04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spruce BA, Campbell LA, McTavish N, Cooper MA, Appleyard MV, O'Neill M, Howie J, Samson J, Watt S, Murray K, et al. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004;64:4875–4886. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]
- 38.Eramo A, Pallini R, Lotti F, Sette G, Patti M, Bartucci M, Ricci-Vitiani L, Signore M, Stassi G, Larocca LM, et al. Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer Res. 2005;65:11469–11477. doi: 10.1158/0008-5472.CAN-05-1724. [DOI] [PubMed] [Google Scholar]
- 39.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert MR. New treatments for malignant gliomas: careful evaluation and cautious optimism required. Ann Intern Med. 2006;144:371–373. doi: 10.7326/0003-4819-144-5-200603070-00015. [DOI] [PubMed] [Google Scholar]
- 41.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 42.Decaestecker C, Debeir O, Van Ham P, Kiss R. Can anti-migratory drugs be screened in vitro? A review of 2D and 3D assays for the quantitative analysis of cell migration. Med Res Rev. 2007;27:149–176. doi: 10.1002/med.20078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.