Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a water- and food-borne pathogen that causes hemorrhagic colitis. EHEC employs a type III secretion system (T3SS) to translocate effector proteins that subvert host cell function. T3SS-substrates encoded outside of the locus of enterocyte effacement are important to E. coli pathogenesis. We discovered an EHEC secreted protein, NleF, encoded by z6020 in O-island 71 of E. coli EDL933 that we hypothesized to be a T3SS substrate. Experiments are presented that probe the function of NleF and its role in virulence. Immunoblotting of secreted and translocated proteins suggest that NleF is secreted by the T3SS and is translocated into host cells in vitro where it localizes to the host cytoplasm. Infection of HeLa cells with E. coli possessing or lacking nleF and transient expression of NleF- GFP via transfection did not reveal a significant role for NleF in several assays of bacterial adherence, host cytoskeletal remodeling, or host protein secretion. However, competitive co-infection of mice with Citrobacter rodentium strains possessing or lacking nleF suggested a contribution of NleF to bacterial colonization. Challenge of gnotobiotic piglets also revealed a role for NleF in colonization of the piglet colon and rectoanal junction.
Keywords: EHEC, NleF, bacterial pathogenesis, type III secretion, gnotobiotic piglet
1. Introduction
Diarrheagenic strains of E. coli contribute to the enormous economic and health burden of food borne disease. Enteropathogenic E. coli (EPEC) is a frequent cause of infantile diarrhea, while enterohemorrhagic E. coli (EHEC) has emerged as an important cause of hemorrhagic colitis in developed countries (Garmendia, et al., 2005). EPEC and EHEC virulence proteins are translocated directly into host intestinal epithelial cells through a type III secretion system [T3SS; (Hueck, 1998)]. The T3SS and several effectors are encoded on a pathogenicity island known as the locus of enterocyte effacement (LEE; (McDaniel, et al., 1995)). Until recently, it was believed by many that the entire repertoire of T3SS-dependent effectors [Tir, EspF, EspG, EspH, Map, and SepZ; reviewed in (Garmendia, et al., 2005)] is encoded within the LEE.
Failure to reconcile this relatively small number of effectors with the diversity of host processes modulated by infection motivated a proteomic assay to identify novel secreted proteins in EHEC and Citrobacter rodentium, a murine-specific attaching/effacing (A/E) pathogen that also possesses the LEE (Deng, et al., 2004). These experiments identified over twenty secreted proteins from EHEC with a strong likelihood of being T3SS substrates. Bioinformatic analyses have suggested there may be greater than 60 putative effector genes (Tobe, et al., 2006). Discovery of these non-LEE-encoded effectors (Nles) suggested a larger armament of virulence proteins than was previously appreciated. Numerous descriptions of Nles encoded within cryptic prophages and pathogenicity islands have now been reported. These effectors include Cif (Marches, et al., 2003), TccP/EspFU (Campellone, et al., 2004), NleA/EspI (Gruenheid, et al., 2004), NleB (Kelly, et al., 2006), NleC and NleD (Marches, et al., 2005), NleE (Wickham, et al., 2007), EspJ (Dahan, et al., 2005), and EspK (Vlisidou, et al., 2006).
We discovered the NleF protein (Z6020) in a proteomic screen of EHEC and C. rodentium secreted proteins (Deng, et al., 2004) and found it is encoded within the same pathogenicity island (O-island 71) as nleA, an important EHEC and C. rodentium virulence factor (Gruenheid, et al., 2004). NleF contains no detectable homology to proteins of known function but does share some sequence similarity to several uncharacterized proteins from Yersinia pestis and Shigella dysenteriae. nleF contains a low GC nucleotide bias, suggesting it was acquired relatively recently through horizontal gene transfer. Thus, based on these preliminary observations, we undertook the study of NleF and its role in pathogenesis.
2. Materials and Methods
2.1. Bacterial strains, plasmids, and oligonucleotides
The bacterial strains and plasmids used in this study are described in Table 1. The oligonucleotides used in this study are described in Table 2.
Table 1.
Strains and plasmids utilized in this study.
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| C. rodentium DBS100 | C. rodentium ATCC 51459 | (Schauer & Falkow, 1993) |
| E. coli BL21(DE3) | E. coli F-ompT hsdSB (rB-mB-) gal dcm (DE3) | Novagen |
| EHEC EDL933 | wt E. coli O157:H7 isolate | Centers for Disease Control |
| EPEC E2348/69 | wt E. coli O127:H6 isolate | (Levine, et al., 1978) |
| s32 | EPEC E2348/69 ΔescN | J. Kaper |
| s231 | C. rodentium ΔnleF | This study |
| s342 | EHEC EDL933 ΔnleF | This study |
| s346 | p455 in BL21(DE3) | This study |
| s352 | p273 in EPEC E2348/69 | This study |
| s353 | p273 in EPEC E2348/69 ΔescN | This study |
| s467 | p558 in s342 | This study |
| s661 | p775 in wt EPEC E2348/69 | This study |
| s662 | p775 in EPEC E2348/69 ΔescN | This study |
| s666 | p765 in wt EPEC E2348/69 | This study |
| Plasmids | ||
| pBR322 | Plasmid cloning vector | New England Biolabs |
| PCR TOPO 2.1 | Subcloning of PCR products | Invitrogen |
| pCX340 | TEM-1 reporter plasmid | (Charpentier & Oswald, 2004) |
| pEGFP-C1 | Mammalian GFP fusion expression | Clontech |
| pET28a | Bacterial hexahistidine fusion expression | Novagen |
| pFLAG-CTC | Bacterial FLAG fusion protein expression | Sigma |
| pKD4 | Lambda red mediated mutagenesis | Barry Wanner |
| p273 | nleF in pFLAG-CTC | This study |
| p455 | nleF in pET28a | This study |
| p488 | nleF in pEGFP-C1 | This study |
| p558 | nleF in pBR322 | This study |
| p765 | nleF (AA 1-60) in pCX340 | This study |
| p775 | nleF in pCX340 | This study |
Table 2.
Oligonucleotide primers utilized in this study.
| Primer | Sequence (5’-3’) |
|---|---|
| PRH-367 | GATCG2ATC2ATGT2AC2A2CA2GTG2T |
| PRH-499 | G2C3ATATGATGT2AC2A2CA2GTG2 |
| PRH-501 | G2C3TCGAGATGT2AC2A2CA2GTG2 |
| PRH-503 | C2G4TAC2AT2AT2AT3CTG2CACATG |
| PRH-509 | G2C3TCGAGA3TGT2AC2A2CA2GTG2 |
| PRH-533 | G2C3TCGAGTC2ACAT2GTA3GATC |
| PRH-543 | G2C5G3TC2ACAT2GTA3GATC2T3G |
| PRH-661 | G2ATC2G2ATCT2AT2AGTGTAG |
| PRH-663 | G2ATC2TCATC2ACAT2GTA3G |
| PRH-1110 | C2G4TAC2AG2AGA3GA3GTGAT2ATG |
| HZ6020-H1P1 | TGT2A2G5T4GATATGT2AC2A2CA2GTG2T2CT2CATGTAG2CTG2AGCTGCT2CG |
| HZ6020-H2P2 | A2CTCACAGAC2TCTA2TCATC2ACAT2GTA3GATC2T3CATATGA2TATC2TC2T2AG |
| HZ6021F | CG3ATC2TCA2TGT2G2TGTGA2TG |
| HZ6020R | GACG2ACGAGTCAGTA7GT |
| CZ6020-H1P1 | TCAG2A4GAGA2TC2T2CGACTC2G2A2TCAGTA2GTGA2TGTAG2CTG2AGCTGCT2CG |
| CZ6020-H2P2 | ATCA4G2ACTA4TCATCACTGCATC2T2CA2CGAG2CACATATGA2TATC2TC2T2AG |
| CZ6020-F | A2TCT3ACTCATG2ATGTAT |
| CZ6020-R | AGTA2GATC2G2CAC2T2A2T |
2.2. Purification of His-NleF
BamHI-XhoI compatible termini were appended by PCR, using primers PRH-367 and PRH-533, onto the E. coli EDL933 nleF coding region for cloning into pET28a (Novagen), an expression plasmid that encodes a C-terminal His6 fusion downstream from an IPTG-inducible promoter. His-NleF was expressed in 500 ml E. coli BL21(DE3) cells grown at 37 °C to an OD600 of 0.4, when IPTG was added to 1.0 mM and cells were incubated for an additional 5 h. Cells were pelleted by centrifugation (5,000 g, 20 min, 4 °C), resuspended in 20 ml cold lysis buffer (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 6 M guanidine) and shaken gently for 5 min. Cell extracts were clarified by centrifugation (12,000 g, 15 min, 4 °C) and the supernatant was removed to a fresh tube. A spin column (Clontech) was packed with 1.5 ml Ni-NTA agarose resin (Qiagen) and charged with 10 ml cold lysis buffer. Clarified extract was poured over the charged column three times. The column was washed three times with 20 ml wash buffer (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 6 M guanidine, 10 mM imidazole). Protein was eluted with 1 ml aliquots of elution buffer (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 6 M guanidine, 200 mM imidazole) and dialyzed into 25 mM sodium phosphate pH 7.5, 1 mM DTT, and 5 % glycerol.
2.3. Antisera preparation
Antiserum was prepared in female New Zealand white rabbits. 200μg of antigen was resuspended in 50 mM Tris HCl pH 7.5, 50 mM NaCl, mixed with Complete Freund’s Adjuvant, and administered into multiple sites subcutaneously along the back and intramuscularly in the hind limbs. Boost injections were made 3 times, every 2 weeks, using 200 μg antigen mixed with Incomplete Freund’s Adjuvent. After 10 weeks, rabbits were euthanized and exsanguinated. Whole blood was clotted at 20 °C for 2 h, chilled at 4 °C overnight, and processed into serum by centrifugation (3,000 g, 15 min, 4 °C).
2.4. Construction of nleF (Z6020) mutants of E. coli O157:H7 and C. rodentium
Non-polar mutations of nleF in E. coli O157:H7 EDL933 and C. rodentium DBS100 were generated by lambda red mediated mutagenesis (Datsenko & Wanner, 2000). Deletions replaced 175/189 and 127/189 nleF codons with a Kan resistance marker in E. coli and C. rodentium, respectively. Primers HZ6020-H1P1 and HZ6020-H2P2 (E. coli) and CZ6020-H1P1 and CZ6020-H2P2 (C. rodentium) and plasmid pKD4 were used to generate the deletion cassettes. Gene replacements were confirmed by PCR using primers HZ6021F and HZ6020R (E. coli) and CZ6020-F and CZ6020-R (C. rodentium). The growth kinetics of the nleF mutants did not differ from the parental strains.
2.5. Secretion and translocation assays
XhoI compatible termini were appended by PCR, using primers PRH-501 and PRH-533, onto the E. coli EDL933 nleF coding region for cloning into pFLAG-CTC (Sigma), an IPTG-inducible expression plasmid. To assay NleF secretion, bacteria were grown overnight in LB broth at 37 °C, 200 rpm, subcultured 1:50 into Dulbecco’s modified Eagle’s medium (DMEM) containing 2.0 mM EGTA and grown 7 h to an OD600 of ~0.7 in a tissue culture incubator (with 5 % CO2) without shaking. Cultures were centrifuged twice (13,000 g, 10 min, 4 °C) to remove bacteria and the culture supernatants were precipitated with 10 % trichloroacetic acid (TCA), neutralized with saturated Tris, and dissolved in SDS-PAGE buffer. The bacterial pellet was dissolved in a volume of 2X SDS-PAGE buffer normalized to the OD600 of the cultures. Proteins were analyzed by 15 % SDS-PAGE and processed for Western blotting using α-NleF, α-DnaK (Stressgen), and α-Tir antibodies with standard ECL Western blotting protocols (Amersham Life Science).
To assay NleF translocation, HeLa monolayers were infected with EPEC strains (MOI ~100) for 6 h. Cells were washed three times in cold PBS and permeabilized with saponin lysis buffer (50 mM Tris pH 7.4, 0.2 % saponin, 1 mM EDTA). After 5 min incubation on ice, samples were centrifuged (12,000 g, 5 min, 4 °C) and the soluble cytoplasmic fraction was removed. The pellet was rinsed in PBS and the membrane proteins were separated from the insoluble components by addition of Triton lysis buffer (50 mM Tris pH 7.4, 1 % Triton X-100, 1 mM EDTA). Fractionation integrity was probed by Western blotting with α-NleF, α-DnaK, α-tubulin (Sigma), and α-calnexin (Stressgen) antibodies.
NleF-TEM-1 fusions were constructed by cloning nleF into the TEM-1 fusion cloning vector pCX340 (a kind gift from E. Oswald). NdeI and EcoRI or KpnI compatible termini were appended by PCR, using primers PRH-499 and PRH-1110 or PRH-503 onto full-length or the region of nleF encoding amino acids 1-60, respectively. Plasmids were introduced into wild type and ΔescN EPEC and used to infect HeLa cells as previously described (Charpentier & Oswald, 2004, Tobe, et al., 2006). After infections (3 h in DMEM supplemented with 1 mM IPTG), cells were washed and loaded with CCF2/AM (Invitrogen). Blue fluorescence was evaluated following illumination of cells at 409 nm.
2.6. Bacterial adherence assays
HeLa cells were infected with EHEC strains (MOI ~100) for 6 h, washed extensively with PBS to remove nonadherent bacteria, and treated with 0.25 % trypsin for 5 min at 37 °C. Samples were centrifuged (2,000 g, 5 min, 4 °C), resuspended in 1 ml PBS, serially diluted, and incubated on LB agar plates overnight at 37 °C. Colony forming units (CFUs) were enumerated, the geometric mean number of CFUs in each treatment group was calculated, and statistical comparisons between groups were performed with two-tailed Student’s t tests. Samples were stained with Hema 3 for detection of adherent bacteria at a magnification of 400x.
2.7. Transient expression of NleF in COS-7 cells
XhoI-XmaI compatible termini were appended by PCR, using primers PRH-509 and PRH-543, onto the E. coli EDL933 nleF coding region for cloning into pEGFP-C1 (Clontech), an expression plasmid that encodes a C-terminal GFP fusion for expression in mammalian cells. COS-7 cells were transfected with FuGene (Roche). After 48 h, the cells were fixed for 10 min in 4 % paraformaldehyde and processed for immunofluorescence microscopy using Alexa568-conjugated phalloidin (Molecular Probes) and 4’,6-diamidino-2-phenylindole [DAPI; Sigma]. Coverslips were mounted in Mowiol (Aldrich) and viewed at 350, 488, and 594 nm on a Zeiss Axiophot epifluorescence microscope.
2.7. SEAP secretion
Plasmids encoding secreted alkaline phosphatase (SEAP) (a gift from Craig Roy), NleF-pEGFP-C1, NleA-pEGFP-C1 (Gruenheid, et al., 2004) and pEGFP-C1 were isolated using GenElute HP Endotoxin Free Kit (Sigma) and cotransfected into CHO cells (4:1 SEAP:GFP) using FuGene. Cells were trypsinized after 48 h, sorted by FACS (BD Bioscience FACSAria) to select for cells displaying green fluorescence, and plated in 24-well dishes at 7.5 × 104 cells/well. After 3 h incubation, fresh media was added and SEAP activity was measured in triplicate wells 7 h later using the Phosphalight SEAP kit (Applied Biosystems). Brefeldin A (Bioshop) was used at 0.01 mg ml-1 where indicated.
2.8. Analysis of C. rodentium ΔnleF infection of mice
Five-week-old C3H/HeJ and C57BL/6 mice (Jackson Laboratory) were housed in the animal facility at the University of British Columbia (UBC) in accordance with guidelines drafted by UBC’s Animal Care Committee and the Canadian Council on the Use of Laboratory Animals. Wild-type (wt) and ΔnleF C. rodentium were grown in LB broth overnight in a shaker at 200 rpm, equal culture volumes were mixed, and 100 μL of the mixture was used to infect mice by oral gavage. Inocula were titered by serial dilution and plating. At indicated times after infection (4-14 d), mice were sacrificed, colonic tissues were homogenized in PBS using a Polytron tissue homogenizer, serially diluted, and plated on MacConkey agar. After overnight incubation, individual colonies were patched onto LB and LB+kanamycin (50 μg ml-1) plates. The competitive index was determined as the output ratio (ΔnleF:wt) divided by the input ratio (ΔnleF:wt).
2.9. Gnotobiotic piglet infections
Gnotobiotic piglets were delivered into germ-free incubators through sterile closed hysterotomy of pregnant sows in accordance with guidelines drafted by South Dakota State University’s Institutional Animal Care and Use Committee. Piglets were separated into individual compartments without regard to sex, fed individually with a sterile commercial piglet formula (SPF-Lac; PetAg, Inc.), and inoculated at 24 h of age with 3 ml tryptic soy broth containing 1*108 CFUs of wt EHEC, EHEC ΔnleF, or a complemented strain EHEC ΔnleF/p558 (prepared by cloning nleF into pBR322 using BamHI-compatible termini appended by PCR using primers PRH-661 and PRH-663). Piglets were observed every 4 h for signs of diarrhea, dehydration, and neurological signs of disease, for up to 48 h and then euthanized.
Distal ileum, spiral colon, cecum, and rectoanal junction specimens were collected at necropsy, split longitudinally, rinsed in PBS to remove feces, diluted 1:10 (w/v) in PBS, ground, serially diluted, and cultured to quantify the extent of intestinal colonization. Data were normalized by tissue weight. Histological specimens were fixed in 10 % neutral buffered formalin, stained with hematoxylin/eosin, and examined by light microscopy, in a manner blinded to individual animal treatment or clinical presentation.
3. Results
3.1. Characterization of the E. coli O157:H7 locus containing nleF
The EHEC O157:H7 EDL933 nleF gene (z6020) is encoded on O-island 71. This region contains the virulence factor nleA (Gruenheid, et al., 2004), as well as several phage structural proteins and putative transposases, and is absent from the genome of E. coli K-12 (Perna, et al., 2001). Bioinformatic analysis of the Sakai strain of E. coli O157:H7 has also permitted discovery of several other known or putative T3SS substrates within the phage-like element Sp9 of this strain, including nleF (ECs1815), nleA (ECs1812), and ECs1810/1, -1814, -1821, -1824, -1825, and -1828 (Tobe, et al., 2006). Thus, this region of the O157:H7 genome appears replete with potential virulence factors (Tobe, et al., 2006).
nleF encodes a predicted 21.4 kDa protein with a pI of 4.94. Analysis of the primary sequence of NleF revealed several predicted alpha-helical regions but no regions similar to protein domains of known function. PSI-BLAST revealed ~27 % identity over ~200 residues to two hypothetical proteins from Yersinia pseudotuberculosis (YPTB2540 and YpseI_02001372) and 58 % identity over 43 residues to the hypothetical protein SDY_P223 from Shigella dysenteriae. Homology to other proteins with an annotated function was not apparent. NleF is present with 100 % amino acid identity in other EHEC (Sakai) and EPEC (E2348/69), and an ortholog is also found in C. rodentium, with which it shares 85 % identity over 189 residues.
3.2. NleF is secreted and translocated by the T3SS
Based on previous proteomic data showing that C. rodentium NleF can be detected in the secretome of a ΔsepL mutant (Deng, et al., 2004), we hypothesized that NleF may be a substrate of the EHEC/EPEC T3SS. We first purified a C-terminally His-tagged version of NleF (His-NleF) to facilitate production of polyclonal antiserum against NleF (Fig. 1A). Western blotting with this antiserum demonstrated the presence of a ~ 20 kDa cross-reactive band in wt, but not ΔnleF EHEC (Fig. 1B). A similar cross-reactive band could be detected in ΔnleF/p558, suggesting successful complementation of the mutation. Similar results were obtained with the C. rodentium mutant (data not shown).
Figure 1. NleF secretion.

A. Denaturing SDS polyacrylamide gel analysis of recombinant His-NleF. Molecular weight standards (kDa), IPTG-induced bacterial lysate (lane 1) and eluate (lane 2) are indicated. B. Complementation of ΔnleF. Rabbit polyclonal antisera raised against His-NleF was used to detect NleF in whole cell lysates from wt EHEC (lane 1), ΔnleF (lane 2), and ΔnleF/p558 (lane 3). C. NleF is secreted by the T3SS. Western blot analysis of secreted proteins (left) and bacterial pellets (right) of wild-type and ΔescN EPEC expressing p273. Blots were probed with α-NleF, α-DnaK, and α-Tir antibodies.
To determine whether NleF is secreted by the T3SS, we cloned nleF into an IPTG-inducible plasmid and expressed this construct (p273) in both wild type (wt) and ΔescN EPEC, a strain deficient in T3S function. Although NleF was expressed at similar levels in wt and ΔescN EPEC, the protein was detectably secreted only by wt EPEC (Fig. 1C). NleF secretion was not detectably secreted in the absence of significant protein overexpression from p273. DnaK immunoblotting was used to validate the absence of non-secreted proteins in the secreted protein samples (Fig. 1C).
We then considered whether NleF might be detectably translocated into mammalian host cells during infection. wt and ΔescN EPEC expressing p273 were applied separately to HeLa cells for 6 h, when bacteria were removed and cell lysates were subjected to immunoblotting. NleF was detected in the host cytosol after a 6 h infection with wt, but not ΔescN EPEC (Fig. 2A). Western blotting with antibodies to proteins specific to each fraction (calnexin, membrane; tubulin, cytoplasm; DnaK, adherent bacteria) suggested little cross-contamination among fractions (Fig. 2A). In agreement with a previous report (Tobe, et al., 2006), we suggest that NleF may be translocated into HeLa cells in a T3SS-dependent manner.
Figure 2. NleF translocation.
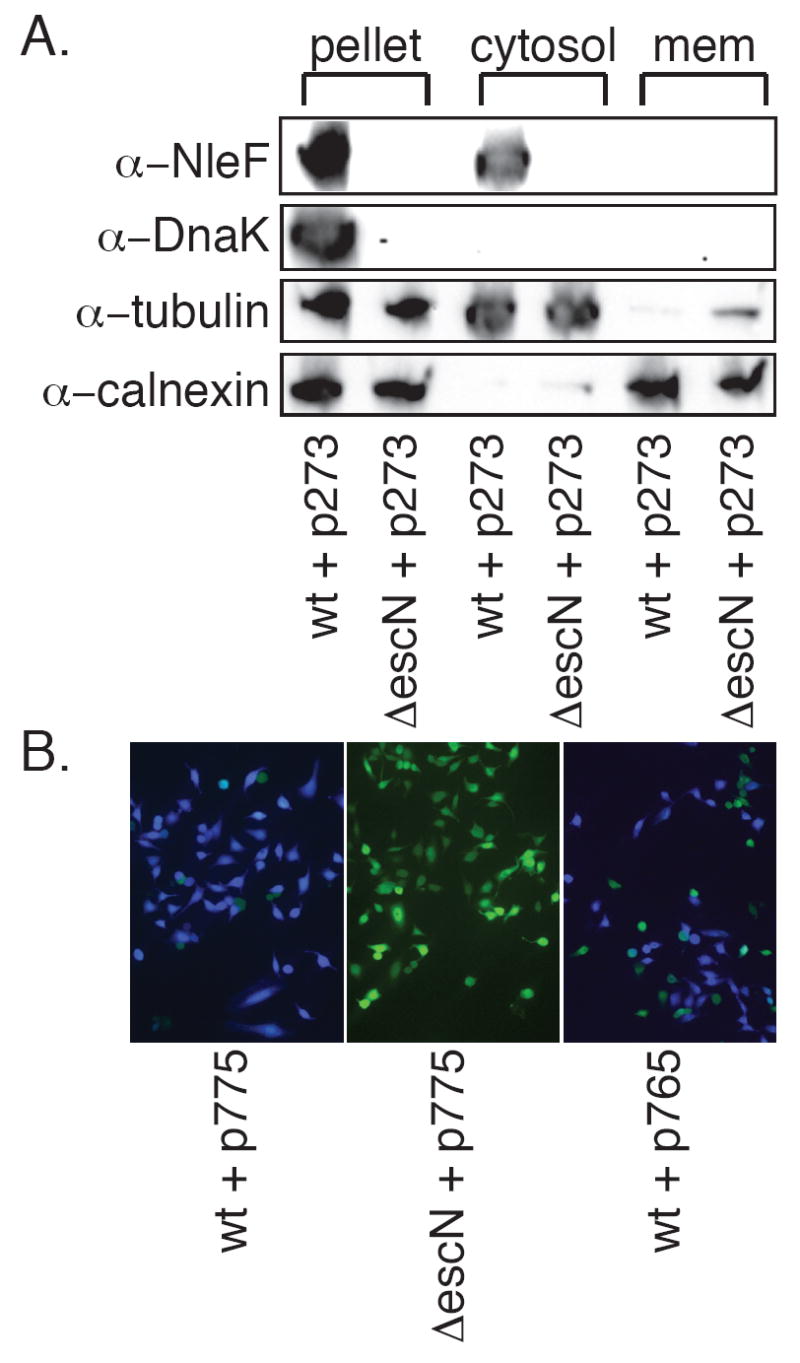
A. Subcellular fractionation. HeLa cells were infected with wild type and ΔescN EPEC expressing p273 and subjected to subcellular fractionation by differential centrifugation into low-speed pellet (bacteria, unbroken cells and cytoskeleton), host cytosol, and host membrane fractions. Blots were probed with α-NleF, α-DnaK, α-tubulin, and α-calnexin antibodies. B. NleF-TEM translocation. HeLa cells were infected with wild-type or ΔescN EPEC expressing full-length (p775) or NleF residues 1-60 (p765) fused to TEM-1. After infection, cells were loaded with CCF2/AM and fusion protein translocation was assayed by fluorescence microscopy.
Translocation was also assessed by TEM-1 reporter assays. NleF-TEM-1 constructs were introduced into wild type and ΔescN EPEC, used to infect HeLa cells, and analyzed by fluorescence microscopy following loading with CCF2/AM. In agreement with a previous report (Tobe, et al., 2006), we detected translocation of NleF-TEM-1 into HeLa cells infected with wt, but not ΔescN EPEC (Fig. 2B).
This reporter system also permitted preliminary assessment of the location of the translocation domain for NleF. In agreement with previous translocation experiments utilizing other T3SS effector fusions (Charpentier & Oswald, 2004), and with biochemical and bioinformatic identification of an N-terminal degenerate CesT-interacting domain (Thomas, et al., 2007), we found that fusion of amino acids 1-60 of NleF to TEM-1 appeared sufficient to promote translocation into HeLa cells (Fig. 2B).
3.3. NleF does not contribute to E. coli O157:H7 adherence or pedestal formation in HeLa cell culture
To investigate the function of NleF, we created a ΔnleF mutant in E. coli O157:H7 EDL933 through lambda red-mediated recombination (Datsenko & Wanner, 2000), and also complemented this mutant through expression of nleF from a pBR322-based plasmid (ΔnleF/p558). HeLa cells were separately infected with wt, ΔnleF, or ΔnleF/p558 for 6 h and monitored by quantitative bacterial adherence assays and Hema 3 staining. As shown in Fig. 3A, EHEC ΔnleF adhered to HeLa cells at similar concentrations compared to wt. Hema 3 staining confirmed the formation of microcolonies in similar abundance (Fig. 3B). Actin pedestals were evident in cells infected with either wt or ΔnleF (data not shown). These data suggest NleF is not required for E. coli O157:H7 adherence or actin pedestal formation.
Figure 3. Role of NleF in in vitro cellular adherence and host cell secretion.
A. Quantification of bacterial adherence to HeLa cells. HeLa cells were separately infected with wt, ΔnleF, or ΔnleF/p558 for 6 h, trypsinized, serially diluted, and plated on LB to enumerate adherent bacteria. Data are the average +/- standard deviation of three independent experiments. B. Hema 3 staining of HeLa cells infected with the indicated bacterial strains 6 h postinfection at a magnification of 400X. C. Transfection of nleF-eGFP in COS-7 cells 48 h post-transfection. D. Lack of influence of NleF on SEAP secretion. Shown is the % secretion of the SEAP reporter protein in CHO cells transfected with the indicated construct, relative to GFP-alone. NleA-eGFP (Kim, et al., 2007) and BFA treatments were used as positive controls.
3.4. Transiently transfected NleF localizes to the host cytoplasm
To determine the subcellular distribution of NleF in eukaryotic cells, NleF-GFP was transiently transfected into COS-7 cells. Immunofluorescence microscopy revealed diffuse cytoplasmic localization of NleF 48 h post-transfection (Fig. 3C). Obvious changes in the distribution of F-actin or tubulin were not detected (data not shown). Our α-NleF antiserum was unsuitable for localization of NleF in infected host cells via immunofluorescence microscopy. It is possible that the potential for targeting of NleF to a specific organelle may be masked by its high expression level or fusion to GFP in transient transfection assays.
Given the role of the E.coli effector NleA in the disruption of host protein secretion (Kim, et al., 2007), we sought to determine if NleF, encoded nearby nleA on O-island 71, also affects protein secretion in host cells. We transfected NleF-GFP into CHO cells and measured the secretion of an alkaline phosphatase reporter protein (SEAP). As shown in Fig. 3D, when compared to control cells transfected with GFP, cells expressing NleF-GFP showed no significant reduction in SEAP secretion (97 +/- 11 %). In contrast, cells treated with NleA-GFP (Kim, et al., 2007), or with brefeldin A, a drug that inhibits vesicle trafficking between the ER and the Golgi, showed a much greater reduction in SEAP secretion (47 % and 8 %, respectively). These data suggest that NleF does not modulate SEAP trafficking in transfected host cells.
3.5. NleF contributes to colonization of mice by C. rodentium
C. rodentium is a natural pathogen of mice that possesses highly homologous virulence determinants and pathogenic strategies with EHEC and EPEC, developing major hallmarks of E. coli-mediated disease, including diarrhea, A/E lesions, and colonic hyperplasia (Deng, et al., 2003). The C. rodentium nleF gene shares 85 % amino acid sequence identity with EDL933 nleF.
We utilized the C. rodentium model to assess how nleF contributes to virulence by constructing a non-polar nleF deletion mutant and comparing its ability to colonize mice, relative to wt C. rodentium in separate and competitive co-infections. Mice were infected with 50 μL overnight cultures of wt and ΔnleF via oral gavage. Mouse strains of differing inherent susceptibilities to C. rodentium [C3H/HeJ, susceptible; C57/BL6, resistant; (Vallance, et al., 2003)] were tested to permit detection of colonization defects and mouse survival at early vs. late time points, respectively.
Experiments in which mice were infected separately with wt or ΔnleF did not reveal a significant role for NleF in terms of mouse death, colonization, and pathology (colon weight and colonic hyperplasia; data not shown). However, competitive co-infection of mice permitted detection of a modest role for C. rodentium NleF in mouse colonization. In C3H/HeJ mice, wt consistently outgrew ΔnleF after 4 d post-inoculation (Fig. 4A; competitive index (CI) = 0.63; p = 0.02, Wilcoxon Rank Sum Test). In less susceptible C57BL/6 mice, this difference in colonization was also significant, even after 14 d (CI = 0.42, p = 0.05). Thus, nleF appears to contribute to C. rodentium colonization of mice.
Figure 4. Contribution of nleF to C. rodentium and E. coli virulence.
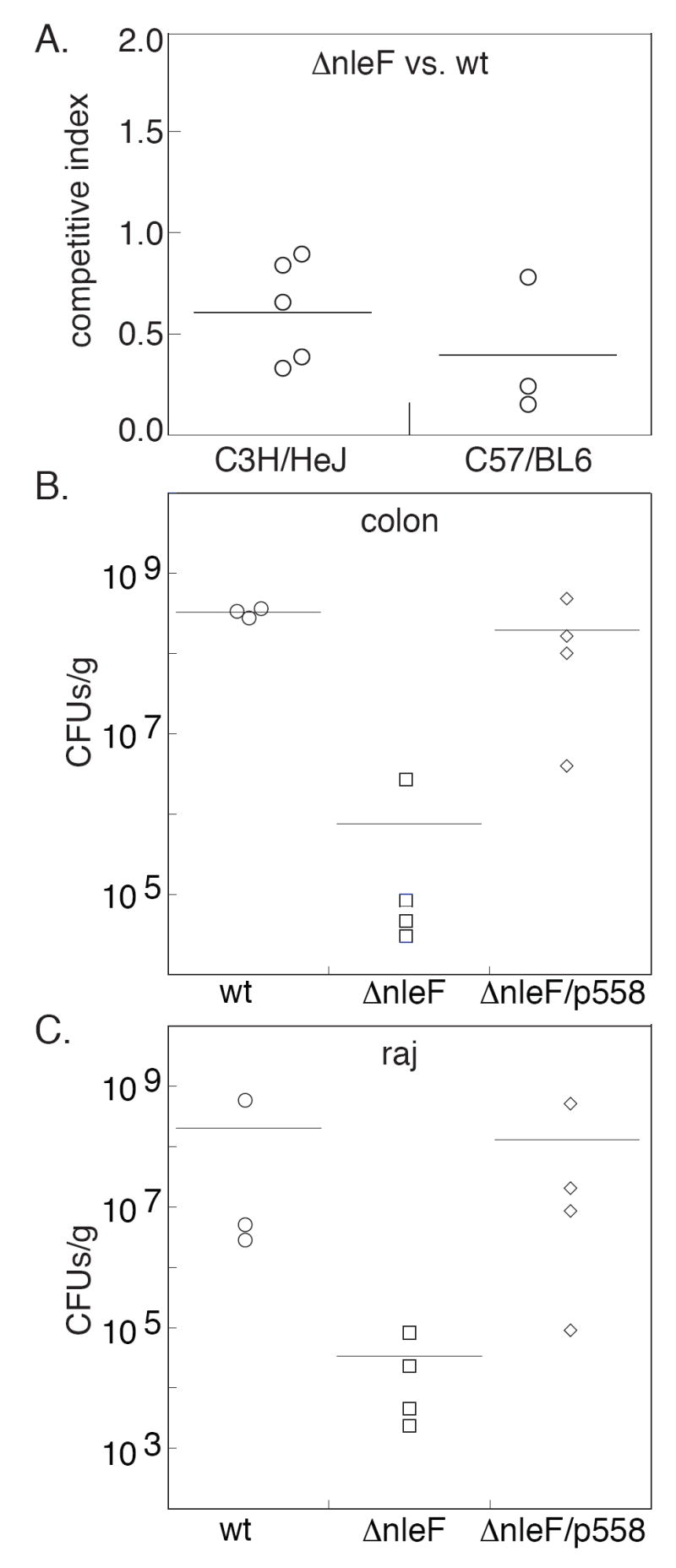
A. Competitive mouse infection experiments. The competitive index (CI) indicates the normalized bacterial colonization ratio of the C. rodentium ΔnleF mutant strain relative to the wt strain during a co-infection of the same mouse (equal colonization = 1.0). Data points represent individual mice and the horizontal line represents the mean. B. Colonization of the gnotobiotic piglet colon (CFUs/g) by E. coli EDL933 wt, ΔnleF, and ΔnleF/p558 48 h post-infection. Data points represent individual piglets and the horizontal line represents the mean. C. Colonization of the rectoanal junction (raj).
3.6. EHEC NleF contributes to gnotobiotic piglet colonization
EHEC is pathogenic for neonatal gnotobiotic pigs. Piglets challenged with EHEC develop diarrhea due to intimate attachment of bacteria to the mucosal surfaces of the terminal ileum and large bowel and develop fatal central nervous system (CNS) disease several days after inoculation (Francis, et al., 1989).
We infected gnotobiotic piglets with strains possessing or lacking NleF to evaluate the role of NleF in intestinal colonization, bacterial survival, and histopathological lesion formation. Piglets were infected orally at 24 h of age with 2*108 CFUs of wt (n = 3), ΔnleF (n = 4), or ΔnleF/p558 (n = 4). Animals were euthanized 48 h after inoculation, as this is the most sensitive time point at which to assess bacterial colonization and pathological changes in intestinal tissue in gnotobiotic piglets (McKee, et al., 1995).
Animals infected with wt EHEC had difficulty standing by the time of euthanasia. Two animals exhibited bladder hemorrhage, liver discoloration, and bloody diarrhea. The third animal had a stroke at 44 h post-infection and was euthanized. In contrast, piglets infected with ΔnleF had only mild, creamy diarrhea, with no evidence of blood in the stool. Piglets infected with ΔnleF/p558 displayed an intermediate phenotype, with two of four piglets having evidence of hemorrhagic colitis and liver discoloration, but without obvious neurological defects at euthanasia.
Tissue specimens were obtained from the distal ileum, spiral colon, cecum, and rectoanal junction for quantification of bacterial adherence. ΔnleF was attenuated in its ability to colonize both the piglet colon (Fig. 4B) and rectum (Fig. 4C), compared to wt (p = 0.06 and 0.03, respectively; Mann-Whitney test). Complementation with p558 partially restored colonization to wt levels. This intermediate phenotype was not unexpected, as antibiotics were not supplied to select for plasmid retention, nor was nleF expression properly regulated. No difference in colonization of the ileum or cecum was observed (data not shown).
4. Discussion
Much effort has recently been put towards discovery and characterization of Nles from A/E pathogens. Here we describe preliminary characterization of NleF, a protein encoded on O-island 71. We have demonstrated, as shown previously (Tobe, et al., 2006), that NleF is secreted by the T3SS and can be translocated into host cells, where it associates with the host cytoplasm. NleF appears to be secreted and translocated in very low concentration, as detection was achieved only after NleF overexpression. Tobe and coworkers analyzed the genome of the Sakai strain of E. coli O157:H7 for homologs of the C. rodentium NleA-F effectors (Tobe, et al., 2006). Their data also showed that NleF is secreted by the EHEC T3SS and indicated that NleF is translocated into host cells (Tobe, et al., 2006).
Despite the lack of an in vitro phenotype in cellular adherence and host cell secretion assays, a modest role in host colonization for nleF was detected in both C. rodentium infection of mice and in E. coli O157:H7 infection of gnotobiotic piglets. It remains to be determined what are the host binding partners and biochemical activities of NleF. It is possible that NleF may act, perhaps in concert with other effectors, to decrease the host response to infection. Although we specifically measured colonization, because in vivo disease associates with bacterial load, colonization defects resulting from an effector gene deletion might reasonably be interpreted as that gene playing a role in bacterial virulence.
Zhang and colleagues used microarray-based comparative genomic hybridization to provide evidence for genetic divergence of E. coli O157:H7 into two distinct lineages. Notably, nleF and the adjacent effector gene nleH1-2 are found only in lineage I, which is more commonly associated with human disease than lineage II (Zhang, et al., 2007). Additionally, seropathotype classification of non-O157 STEC suggests that nleH1-2 and nleF are associated with epidemic potential and severe human disease (M. Wickham, personal communication). Other analyses of non-O157 EHEC strains by microarray and whole genome PCR scanning have shown that NleF is present in 90 % of the tested strains (Ogura, et al., 2007).
The ability of ΔnleF to colonize the piglet colon and rectoanal junction was significantly attenuated relative to wild type. While the terminal rectum is a primary site of E. coli colonization in cattle (Naylor, et al., 2003), perhaps because of the limited histopathology caused by EHEC in the rectum (Tzipori, et al., 1995), relatively few studies have examined the role of EHEC virulence factors in colonization of the porcine rectum in detail (McKee, et al., 1995). Measurement of rectal colonization in the piglet model might potentially be used to understand better the role of individual effector proteins in host colonization.
Acknowledgments
We thank Eric Brown, Amber Johnson, and Kristina Mateo for assistance with gnotobiotic piglet infections. We thank Jean-Philippe Nougayrède and Eric Oswald (Ecole Nationale Vétérinaire de Toulouse) for the generous donation of their TEM-1 reporter plasmid. This work was supported by NIH grant AI070339-01 (PRH) and by grants from the Canadian Institutes of Health Research and the Howard Hughes Medical Institute (BBF). MEW was a CJ Martin Fellow of the National Health and Medical Research Council of Australia, and was supported by postdoctoral fellowships from the Canadian Institutes of Health Research, The Michael Smith Foundation for Health Research, and The Killam Trusts. BBF is an HHMI International Research Scholar and the UBC Peter Wall Distinguished Professor.
Abbreviations
- A/E
attaching/effacing
- EHEC
enterohemorrhagic Escherichia coli
- EPEC
enteropathogenic Escherichia coli
- HUS
hemolytic uremic syndrome
- T3SS
type III secretion system
References
- 1.Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahan S, Wiles S, La Ragione RM et al. EspJ is a prophage-carried type III effector protein of attaching and effacing pathogens that modulates infection dynamics. Infect Immun. 2005;73:679–686. doi: 10.1128/IAI.73.2.679-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng W, Vallance BA, Li Y, Puente JL, Finlay BB. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol Microbiol. 2003;48:95–115. doi: 10.1046/j.1365-2958.2003.03429.x. [DOI] [PubMed] [Google Scholar]
- 6.Deng W, Puente JL, Gruenheid S et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis DH, Moxley RA, Andraos CY. Edema disease-like brain lesions in gnotobiotic piglets infected with Escherichia coli serotype O157:H7. Infect Immun. 1989;57:1339–1342. doi: 10.1128/iai.57.4.1339-1342.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garmendia J, Frankel G, Crepin VF. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun. 2005;73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruenheid S, Sekirov I, Thomas NA et al. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2004;51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 10.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly M, Hart E, Mundy R et al. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect Immun. 2006;74:2328–2337. doi: 10.1128/IAI.74.4.2328-2337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Thanabalasuriar A, Chaworth-Musters T et al. The bacterial virulence factor NleA binds and disrupts function of the mammalian COP II complex. Cell Host and Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 14.Marches O, Ledger TN, Boury M et al. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol Microbiol. 2003;50:1553–1567. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 15.Marches O, Wiles S, Dziva F et al. Characterization of two non-locus of enterocyte effacement-encoded type III-translocated effectors, NleC and NleD, in attaching and effacing pathogens. Infect Immun. 2005;73:8411–8417. doi: 10.1128/IAI.73.12.8411-8417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O’Brien AD. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor SW, Low JC, Besser TE et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogura Y, Ooka T, Asadulghani et al. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 2007;8:R138. doi: 10.1186/gb-2007-8-7-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perna NT, Plunkett G, 3rd, Burland V et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 21.Schauer DB, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;61:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas NA, Deng W, Baker N, Puente J, Finlay BB. Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J Biol Chem. 2007;282:29634–29645. doi: 10.1074/jbc.M706019200. [DOI] [PubMed] [Google Scholar]
- 23.Tobe T, Beatson SA, Taniguchi H et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzipori S, Gunzer F, Donnenberg MS, de Montigny L, Kaper JB, Donohue-Rolfe A. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect Immun. 1995;63:3621–3627. doi: 10.1128/iai.63.9.3621-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallance BA, Deng W, Jacobson K, Finlay BB. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun. 2003;71:3443–3453. doi: 10.1128/IAI.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlisidou I, Marches O, Dziva F, Mundy R, Frankel G, Stevens MP. Identification and characterization of EspK, a type III secreted effector protein of enterohaemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett. 2006;263:32–40. doi: 10.1111/j.1574-6968.2006.00410.x. [DOI] [PubMed] [Google Scholar]
- 27.Wickham ME, Lupp C, Vazquez A et al. Citrobacter rodentium virulence in mice associates with bacterial load and the type III effector NleE. Microbes Infect. 2007;9:400–407. doi: 10.1016/j.micinf.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Laing C, Steele M et al. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]



