Abstract
Evidence from autopsy, mouse-model and in vitro binding studies suggests that adhesion of erythrocytes infected with Plasmodium falciparum to the human host intercellular adhesion molecule (ICAM)-1 receptor is important in the pathogenesis of severe malaria. Previous association studies between polymorphisms around the ICAM1 gene and susceptibility to severe malarial phenotypes have been inconclusive and often contradictory. We performed genetic association studies with 15 single-nucleotide-polymorphisms (SNPs) around the ICAM1 locus. All SNPs were screened in a family study of 1071 trios from Gambia, Malawi and Kenya. Two key non-synonymous SNPs with previously reported associations, rs5491 (K56M or ‘ICAM-1Kilifi’) and rs5498 (K469E), were tested in an additional 708 Gambian trios and a case-control study of 4058 individuals. None of the polymorphisms were associated with severe malaria phenotypes. Pooled results across our studies for ICAM-1Kilifi were, in severe malaria, odds ratio (OR) 1.02, 95% confidence interval (CI) 0.96 – 1.09, P=0.54, and cerebral malaria OR 1.07, CI 0.97 – 1.17, P=0.17. We assess the available epidemiological, population genetic and functional evidence which links ICAM-1Kilifi to severe malaria susceptibility.
Keywords: ICAM-1, Plasmodium falciparum, Genetic association study
Introduction
Several lines of evidence implicate intercellular adhesion molecule (ICAM)-1 in the pathogenesis of severe Plasmodium falciparum malaria. The causes of severe disease are complex, but a key feature is the adherence of parasite infected red blood cells (iRBC) to components of the vascular space and particularly endothelium.1 Adhesion of iRBCs to brain microvasculature is suspected to contribute to the development of cerebral malaria (CM) a manifestation of severe disease associated with high rates of mortality.2,3 P. falciparum erythrocyte membrane protein 1 (PfEMP1) is central to this adhesive behaviour. PfEMP1 is expressed by the parasite onto the surface of iRBCs where it is subject to antigenic variation during the course of an infection.4 PfEMP1 adheres to a range of molecules expressed by the human host including ICAM-1,5 IRBCs bind the first N-terminal immunoglobulin-like domain of ICAM-1.6 Autopsy studies of patients with fatal CM and severe malarial anaemia (SA) found iRBC sequestration on brain vascular endothelia cells, with enhanced expression of adhesion molecules, including ICAM-1, in the areas of iRBC binding.7,8,9,10 Studies of parasite isolates have found high rates of in vitro ICAM-1 binding among wild strains, but reported correlations between ICAM-1 binding and disease severity have been inconsistent.11,12,13
ICAM-1 (CD54), a member of the immunoglobulin super-family, is typically expressed on endothelial cells, particularly in the brain where expression is strongly increased by pro-inflammatory cytokines.14 As a key component of the immune system ICAM-1's role in malaria susceptibility is not limited to direct interaction with the PfEMP1. ICAM-1 binds lymphocyte function-associated antigen (LFA)-1, a leukocyte cell surface glycoprotein,15 allowing leukocytes passage through the blood brain barrier. Interaction between LFA-1 and ICAM-1 activates natural killer cells during P. falciparum infection.16 ICAM-1 is also a receptor for plasma protein fibrinogen,17 Mac-118 and Rhinovirus.19
In 1997 Fernandez-Reyes et al. sequenced the N-terminal domain of ICAM-1 in asymptomatic Kenyan children and found a non-synonymous coding polymorphism.20 The new allele, designated ICAM-1Kilifi (rs5491), causes a lysine to methionine change at position 56 of the coding sequence (position 29 in the mature protein). This raised the possibility that the mutant allele had been selected for by endemic malaria. However (to their surprise) a case-control study of Kenyan children (260 severe malaria cases and 287 community controls) suggested that the ICAM-1Kilifi allele was associated with an increased susceptibility to CM. The authors noted ICAM-1Kilifi was common (~30%) in Kenyan and Gambian populations, but not found in a sample of Caucasians, leading the authors to speculate that ICAM-1Kilifi provided a compensatory selective advantage through an unknown mechanism. In contrast to the first study a paired case-control analysis from Gabon (100 severe malaria cases and 100 mild cases), published in 1999, reported a protective effect from ICAM-1Kilifi.21 No subsequent association study has replicated either of these initial findings. Case-control studies in the Gambia,22 Thailand,23 Senegal,24 Nigeria,25 and further case-control26 and longitudinal studies27 in Kenya reported no significant association between malaria phenotypes and ICAM-1Kilifi. Case-control analysis in Nigeria did suggest a marginal signal of association with susceptibility to another single-nucleotide-polymorphism (SNP) in exon 6 (rs5498).
While there is a strong rationale why genetic variation around the ICAM1 gene could affect susceptibility to severe malarial phenotypes, only a fraction of the common SNPs around the locus have been tested for disease associations, and the results for the closely studied ICAM-1Kilifi variant are inconclusive. We designed and conducted an experiment to screen common variation across ICAM1, while targeting the two non-synonymous ICAM1 SNPs (ICAM-1Kilifi and rs5498, those with previously reported associations) with well-powered tests.
Results
Family-based association analysis of ICAM1 SNPs with severe malaria phenotypes
Fifteen SNP makers around the ICAM1 gene were selected for genotyping in 1071 parent-offspring trios from Gambia, Kenya and Malawi. Each trio comprised a child affected by severe malaria and their parents. The non-synonymous SNPs ICAM-1Kilifi and rs5498 were also genotyped in 708 additional Gambian families. Further details of subjects and severe malaria phenotypes are given in the materials and methods section. Markers were tested for disease associations in additive, dominant and recessive models using the FBAT application. The results for the additive model in severe malaria, along with allele frequencies for parents and offspring in the family study are shown in Table 1 (the association results for sub-phenotypes and specific populations are documented in Supplementary Table S1).
Table 1.
ICAM-1 genotype frequencies and single marker family-based association analysis with severe malaria.
| SNP | Major Allele |
Minor Allele |
Parents | Cases | S | E(S) | Var(S) | P-value |
|---|---|---|---|---|---|---|---|---|
| MAF | MAF | |||||||
| rs5490 | A | C | 26.0 | 26.0 | 429 | 430.0 | 190.50 | 0.94 |
| rs5030340 | C | T | 5.5 | 5.5 | 100 | 105.0 | 52.00 | 0.49 |
| rs5030344 | G | A | 4.2 | 4.3 | 87 | 86.5 | 43.25 | 0.94 |
| rs5030351 | C | T | 24.6 | 24.8 | 414 | 417.5 | 183.75 | 0.80 |
| rs5491 | A | T | 23.9 | 23.7 | 687 | 688.5 | 303.75 | 0.93 |
| rs281432 | G | C | 24.8 | 24.7 | 383 | 398.5 | 180.25 | 0.25 |
| rs5496 | G | A | 9.5 | 9.7 | 176 | 179.5 | 88.25 | 0.71 |
| rs5498 | A | G | 11.4 | 11.0 | 346 | 368.0 | 182.00 | 0.10 |
| rs3093030 | C | T | 5.8 | 5.6 | 100 | 112.0 | 55.50 | 0.11 |
| rs281438 | T | G | 46.0 | 46.6 | 684 | 683.5 | 237.25 | 0.97 |
| rs2569693 | C | T | 5.5 | 5.2 | 87 | 89.5 | 43.75 | 0.71 |
| rs281439 | C | G | 38.8 | 38.6 | 583 | 589.5 | 236.25 | 0.67 |
| rs281440 | A | G | 31.9 | 31.0 | 461 | 478.5 | 202.75 | 0.22 |
| rs2075741 | G | C | 12.2 | 11.8 | 221 | 236.5 | 111.75 | 0.14 |
| rs11575074 | G | A | 9.7 | 9.6 | 172 | 182.0 | 89.00 | 0.29 |
Minor allele frequencies (MAF, %) for parents and affected offspring. FBAT statistic ‘S’, expected FBAT statistic ‘E(S)’, variance ‘Var(S)’ and P-value are reported under the additive model for transmissions of the minor allele.
No significant association was found between ICAM1 SNPs and severe malaria phenotypes, or between the SNPs and severe malaria in a specific study region (all P-values > 0.01). Given that 15 markers are tested for association with three phenotypes, in three genetic models, all P-values are substantially above relevant thresholds following correction for multiple testing. With specific reference to ICAM-1Kilifi this variant was not found to be associated with severe disease (N=1779 trios, p=0.93) or CM (N=842 trios, p=0.1) in our family-study.
Haplotype-specific association analysis
Using family trio data we performed haplotype-based association analysis with severe malaria phenotypes in FBAT. Linkage disequilibrium (LD) between ICAM1 SNPs was inspected using the HAPLOVIEW application. A haplotype block extending across 7 SNPs in the 5′ region of the gene was identified, common to all three study populations (Figure 1). This 5′ haplotype block spans across the exons coding for the first N-terminal domain which binds PfEMP1, LFA-1 and Rhinoviruses and is the location of ICAM-1Kilifi. FBAT analysis of common haplotypes (>5%) revealed no significant disease associations (Table 2).
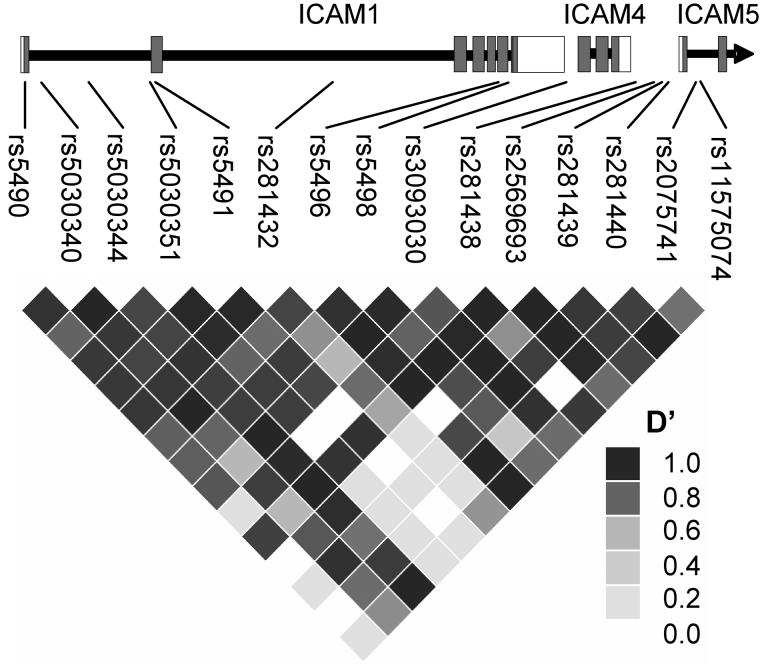
Figure 1.
Linkage disequilibrium at the ICAM1 locus in 612 Gambian family trios. An exonic model of the ICAM1 gene (along with the nearby ICAM4 and ICAM5 genes), demonstrating the relative positions of our genotyped SNPs. All SNPs were within 5kb of the ICAM1 gene. LD (D′) calculated using the HAPLOVIEW application. A region of relatively high D′ can be seen across the 5′ of the gene, a section coding for the PfEMP1 binding N-terminal of ICAM-1. This haplotype block (7 SNPs from rs5490 to rs5496) was used in our haplotype association analysis.
Table 2.
FBAT haplotype association analysis for the 5′ ICAM-1 haplotype block in severe malaria phenotypes
| Phenotype | Haplotype | Freq. | S | E(S) | Var(S) | P |
|---|---|---|---|---|---|---|
| All severe | ACGCAGG | 0.288 | 444.43 | 446.08 | 162.48 | 0.90 |
| - - - - - C - | 0.241 | 372.16 | 379.23 | 135.87 | 0.54 | |
| C - - T T - - | 0.210 | 328.81 | 326.72 | 122.41 | 0.85 | |
| - - - - - - A | 0.087 | 149.73 | 150.40 | 67.36 | 0.94 | |
| - T - - - - - | 0.053 | 95.87 | 90.13 | 39.40 | 0.36 | |
| CM | ACGCAGG | 0.279 | 248.30 | 247.77 | 92.88 | 0.96 |
| - - - - - C - | 0.234 | 212.09 | 221.48 | 74.32 | 0.28 | |
| C - - T T - - | 0.222 | 209.42 | 206.29 | 73.68 | 0.72 | |
| - - - - - - A | 0.085 | 91.86 | 87.62 | 37.08 | 0.49 | |
| - T - - - - - | 0.062 | 62.86 | 60.23 | 25.49 | 0.60 | |
| SA | ACGCAGG | 0.298 | 132.31 | 130.89 | 47.88 | 0.84 |
| - - - - - C - | 0.227 | 100.32 | 102.52 | 36.75 | 0.72 | |
| C - - T T - - | 0.206 | 83.27 | 93.48 | 33.74 | 0.08 | |
| - - - - - - A | 0.089 | 43.80 | 43.79 | 17.86 | 1.00 | |
| - T - - - - - | 0.052 | 27.00 | 23.91 | 10.41 | 0.34 |
Parental haplotype frequency derived from seven 5′ ICAM1 SNPs from rs5490 to rs5496, FBAT statistic ‘S’, expected FBAT statistic ‘E(S)’, variance ‘Var(S)’ and P-value are reported under the additive model for transmissions of the minor allele. Only haplotypes >5% frequency analyzed.
Population-based association analysis of ICAM-1Kilifi and rs5498 with severe malarial phenotypes
We genotyped the two non-synonymous SNPs, with previously reported malaria associations, in three additional population-based studies. The case-control studies were independent of the family study and in total comprised 2127 cases of severe malaria and 1931 population controls. Further details of subjects and severe malaria phenotypes are given in the materials and methods section. Both markers were tested for disease associations in additive, dominant and recessive models using logistic regression. Analysis of all case-control studies together suggests neither marker is significantly association with our severe malarial phenotypes (Table 3).
Table 3.
Case-control and pooled case-control/family data association analysis between ICAM-1Kilifi (rs5491), rs5498 and severe malaria phenotypes.
| SNP | Phenotype | OR | 95% CI. | P |
|---|---|---|---|---|
| Case-control studies a | ||||
| rs5491 | CM | 1.07 | 0.94 - 1.21 | 0.33 |
| - | SA | 1.10 | 0.95 - 1.28 | 0.21 |
| - | All Severe | 1.10 | 1 - 1.22 | 0.06 |
| - | Gambia | 1.23 | 1.01 - 1.5 | 0.04 |
| - | Kenya | 1.07 | 0.92 - 1.25 | 0.40 |
| - | Malawi | 1.05 | 0.87 - 1.27 | 0.62 |
| rs5498 | CM | 1.12 | 0.95 - 1.31 | 0.19 |
| - | SA | 0.97 | 0.79 - 1.18 | 0.75 |
| - | All Severe | 1.06 | 0.93 - 1.21 | 0.41 |
| - | Gambia | 1.10 | 0.86 - 1.42 | 0.45 |
| - | Kenya | 1.07 | 0.87 - 1.32 | 0.52 |
| - | Malawi | 0.99 | 0.78 - 1.26 | 0.94 |
| Pooled across case-control and family studies b | ||||
| rs5491 | CM | 1.065 | 0.97 - 1.17 | 0.17 |
| - | SA | 0.988 | 0.88 - 1.11 | 0.85 |
| - | All Severe | 1.021 | 0.96 - 1.09 | 0.54 |
| rs5498 | CM | 1.087 | 0.97 - 1.22 | 0.16 |
| - | SA | 0.924 | 0.79 - 1.09 | 0.34 |
| - | All Severe | 1.029 | 0.94 - 1.12 | 0.53 |
Results for severe malaria phenotypes across case-control studies and for severe malaria in regional case-control studies (P<0.05 underlined).
Results for severe malaria phenotypes pooled across case-control and family studies (using UNPHASED).
We specifically tested whether ICAM-1Kilifi homozygotes were at altered risk of cerebral malaria in individual regional populations. In the Gambia case-control study ICAM-1Kilifi homozygotes demonstrated a possible association with CM susceptibility (taking wild-type homozygotes as reference; ICAM-1Kilifi homozygotes odds ratio (OR) 2.5, 95% confidence interval (CI) 1.37 - 4.75, P=0.003), heterozygotes were not at significantly increased risk (OR 1.03, CI 0.75 - 1.42, P=0.85). This is reflected in the result for all severe cases in Gambia under the additive model (Table 3). A similar, but non-significant, trend was found in the family-study (ICAM-1Kilifi homozygotes OR 1.3, CI 0.75 - 2.24, P=0.35 and heterozygotes OR 1.0, CI 0.784 - 1.3, P=0.94 (performed by case-pseudo-control logistic regression conditioning on parental genotypes28)). This finding merits cautious interpretation, as (a) ICAM-1Kilifi homozygotes represent a relatively a small sub-group of the Gambians (6% of all Gambian cases, 50 individuals); (b) the trend was not followed in the other two populations, including Kenyans where a link with CM was originally reported 20; (c) no association between CM and ICAM-1Kilifi was reported in a previous Gambian study 22. No other significant region-and-sub-phenotype-specific associations were observed. Pooled family- and population-based association analysis was performed using the UNPHASED application (Table 3). Using this large combined dataset we have narrowed the confidence interval of the risk estimate for ICAM-1Kilifi, which does not appear to be associated with severe disease (OR 1.02, CI 0.96 to 1.09) or CM (OR 1.07, CI 0.97 – 1.17).
Discussion
There is a strong rationale why genetic variation around the ICAM1 gene, particularly the ICAM-1Kilifi variant could affect susceptibility to severe malarial phenotypes. However genetic epidemiological studies of the ICAM-1Kilifi variant have demonstrated a pattern consistent with other candidate gene studies of complex human disease, in which early interesting results have not been replicated in subsequent studies29 (Table 4). We conducted an association screen for common variation across the ICAM1 gene, while targeting two non-synonymous SNPs with well-powered tests, using family- and population-based methods. This has led to a substantial refinement of the risk estimates for ICAM-1Kilifi and rs5498. Neither of these non-synonymous SNPs, nor any other genotyped SNP in the ICAM1 locus were significantly associated with severe disease or cerebral malaria in our study. We cannot rule out the possibility that ICAM-1Kilifi is associated with a small effect (e.g. an odds ratio of 1.1 or 1.05), particularly if it is limited to smaller subgroups e.g. a sub-phenotype of severe disease, or only the rare ICAM-1Kilifi homozygotes. However our results do not support the existence of the large effect sizes initially reported. Differences in clinical phenotype definition between study sites could confound results. In our study we have attempted to standardize phenotype definition using key clinical parameters (such as Blantyre Coma Score or Haemoglobin concentration), but it is possible that disparity in phenotype definition between studies is responsible for the diversity of findings reported in the literature.
Table 4.
Published risk estimates of severe malaria phenotypes associated with ICAM-1Kilifi.
| Study | Region | Design | Na | Phenotypeb | Allele/ Genotype |
OR | 95% CI. | Test | P |
|---|---|---|---|---|---|---|---|---|---|
| Fernandez-Reyes et al.,1997 20 | Kenya | Case-control | 444 | SA | T vs. A | 1.15 | 0.82 - 1.6 | Pearson's χ2 | 0.47 |
| - | - | - | 390 | CM | T vs. A | 1.55 | 1.17 - 2.06 | Pearson's χ2 | 0.0028 |
| - | - | - | 547 | Severe | T vs. A | 1.38 | 1.08 - 1.77 | Pearson's χ2 | 0.01 |
| Kun et al.,1999 21 | Gabon | Case-control | 200 | Severe vs. Mild | T vs. A | 0.54 | 0.34 - 0.84 | Pearson's χ2 | 0.0092 |
| Bellamy et al.,1998 22 | Gambia | Case-control | 616 | SA | T vs. A | 0.71 | 0.51 - 0.99 | Pearson's χ2 | 0.054 |
| - | - | - | 802 | CM | T vs. A | 0.89 | 0.69 - 1.14 | Pearson's χ2 | 0.37 |
| - | - | - | 474 | SA vs. Mild | T vs. A | 0.67 | 0.47 - 0.94 | Pearson's χ2 | 0.027 |
| - | - | - | 660 | CM vs. Mild | T vs. A | 0.83 | 0.63 - 1.09 | Pearson's χ2 | 0.19 |
| Ohashi et al.,2001 23 | Thailand | Case-control | - | Mild, non-cerebral severe & cerebral malaria |
- | - | - | - | NSc |
| Amodu et al.,2005 25 | Nigeria | Case-control | 200 | Severe vs. Asymptomatic Parasitaemia | T vs. A | 1.71 | 1.09 - 2.67 | Pearson's χ2 | 0.025d |
| - | - | - | 199 | Severe vs. Mild | T vs. A | 1.5 | 0.97 - 2.34 | Pearson's χ2 | 0.086 |
| Jenkins et al.,2005 27 | Kenya | Cohort | 1417 | Malaria-specific clinic visits |
AT vs AA | 0.89e | 0.74 - 1.08 | Poisson regression | 0.24 |
| - | - | - | 886 | Malaria-specific clinic visits |
TT vs AA | 0.91 | 0.68 - 1.21 | Poisson regression | 0.52 |
| Ndiaye et al.,2005 24 | Senegal | Cohort | 878 | Traits related to infection and carriage | - | - | - | - | NS |
| Ayodo et al.,2007 26 | Kenya | Case-control | 915 | Severe | T vs. A | 0.71 | 0.42 - 1.21 | Pearson's χ2 | 0.1 |
| Present study | Gambia, Kenya & Malawi |
Case-control & Family |
3920 | SA | T vs. A | 0.99 | 0.88 - 1.11 | UNPHASED | 0.85 |
| - | 5637 | CM | T vs. A | 1.07 | 0.97 - 1.17 | UNPHASED | 0.17 | ||
| - | 9395 | Severe | T vs. A | 1.02 | 0.96 - 1.09 | UNPHASED | 0.54 |
Using original data, were possible, we performed the allelic test (2×2, Pearson's χ2) deriving odds ratio (OR), 95% confidence interval and P-value.
Sample size (N) relates to the number of case and controls used in calculating association statistic.
Cases with this disease phenotype are compared with population controls, unless indicated otherwise.
Low allele frequency of ICAM-1Kilifi in Thailand (~2%) means very low power to detect association. NS = unavailable /unreported non-significant p-value.
Multivariate logistic regression adjusted for age and parasite density reported p=0.277.
Incident rate ratio adjusted for sickle trait, age, sex, season, and ethnic group.
Variation in the prevalence of ICAM-1 binding strains between geographic regions (and in one region over time), could explain some of the differences between studies. Substantial research has been undertaken investigating the functional differences between wild type ICAM-1 and ICAM-1Kilifi. Laboratory isolates of P. falciparum adherent to ICAM-1 show differences in adhesive phenotype between reference ICAM-1 and ICAM-1Kilifi.30,31 Binding of the A4 parasite line to ICAM-1Kilifi is reduced under both static and flow conditions, while interestingly binding of the ItG line (derived from the same Brazilian isolate as A4 but with repeated selection on ICAM-1) is far less affected by the K29M residue change. ICAM-1 mutagenesis experiments have suggest that even closely related parasite strains can use different contact residues for adherence on ICAM-1.31 This raises the possibility that some wild African P. falciparum strains express PfEMP1 molecules adapted to preferentially bind ICAM-1Kilifi (or which shows little difference between ICAM-1 alleles). This could explain the absence of a significant disease association, despite the pronounced functional differences seen with laboratory strains.
The geographic distribution of the non-synonymous ICAM-1Kilifi has raised the suspicion of a selective event involving this variant. The ICAM-1Kilifi allele SNP is at a derived allele frequency of around 20-30% in many African populations (Gambia 19.6%, Kenya 28.4%, Malawi 29.6% (present study), Nigeria 31.6%, Gabon 27%,21 Senegal 17%24). However ICAM-1Kilifi is also present at frequencies around 5% in East Asia (Papua New Guinea 4.9%,21 Thailand 1.7%,23 Han Chinese 7.8%, Japanese 5.7% 32) and is uncommon but not absent from European-derived populations (Sweden 1.1%,33 North Carolina Caucasians 0.4%34). Although Caucasian allele frequencies might represent recent African admixture, the presence of ICAM-1Kilifi in several East Asian and Pacific Rim populations suggests that the allele has been common in humans for some time (potentially tens of thousands of years), and may have become rare in European-derived populations due to drift, demography or even selection.
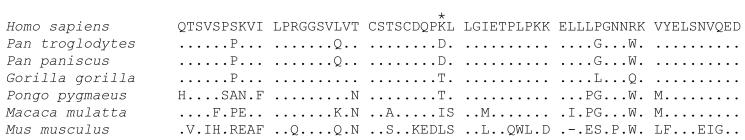
The ICAM1 locus has been screened in a number of genome-wide surveys for recent evolutionary selection (with metrics including heterozgosity, excess of rare alleles, high frequency derived alleles, population differentiation (FST) and long range haplotype tests) the locus has not been considered an outlier.32,35,36 There is evidence of selection at ICAM1 over longer timescales, as comparative sequence analysis has suggested the gene has been under selection in the human-chimp lineage.37 It should be noted that residue 29 is not conserved in primate evolution (Figure 2). If we include the ICAM-1Kilifi protein, the modern range of functional primate ICAM-1 molecules includes four diverse types of amino-acid side chain: positively charged (Lysine), negatively charged (Aspartic acid), polar uncharged (Threonine) and non-polar (Methionine). This may indicate that the residue, located in a coiled loop which projects from the surface of the N-terminal domain38 is subject to relatively few functional constraints, but would also be consistent with multiple changes to evade pathogen binding.
Figure 2.
ICAM-1 amino acid alignments. The first 60 residues of the mature ICAM-1 protein in Man, Common Chimpanzee, Pygmy Chimpanzee, Gorilla, Orangutan, Rhesus Monkey and Mouse (Genbank identifiers AF340038, AF340033, AF340042, AF340036, AF340041, AF340040 and NM_010493). The location of the K to M non-synonymous SNP rs5491 or ICAM-1Kilifi variant is indicated with an asterisk (position 29 of the mature protein).
The incidence of intermediate frequency non-synonymous mutations is probably greater than may often be considered. To date over 16 thousand polymorphic non-synonymous SNPs have been genotyped in a sample of Yoruba ethnicity individuals from Nigeria (YRI) as part of the International Haplotype Map (HapMap) project (release 22).32 For non-synonymous SNPs, polymorphic in YRI, 14.7% (2338/15883) have a Wright's FST between YRI and the CEPH samples (from Utah but of North-Western European origin) greater than 0.143 (the level of population differentiation seen at rs5491). Using published estimates of ancestral status (see online supplementary data associated with36 (http://hg-wen.uchicago.edu/selection/haplotter.htm)) we identified the derived allele in a subset of non-synonymous SNPs. 48.1% (4016/8344) of non-synonymous SNPs, polymorphic in YRI, have a derived allele frequency greater than 0.25 (the derived allele frequency of rs5491 in YRI). However, in the specific situation, where no derived alleles are found in the CEPH samples (as with rs5491), only 7.9% (86/1089) of non-synonymous SNPs, polymorphic in YRI, have a derived allele frequency greater that 0.25.
If we accept the geographical distribution of the ICAM-1Kilifi allele as evidence of an African selection event, particularly in light of the strong functional evidence – how could we tally selection with lack of a significant association with severe malaria phenotypes? An interesting explanation would be a frequency-dependant model; here an equilibrium exists between the human polymorphism frequency, and parasite strain frequency, with competing strains preferring either ICAM-1Kilifi or ICAM-1Ref binding. Change in host allele frequency is opposed by expansion in the relevant binding parasite strains, selecting against the expanded allele and returning the system to equilibrium. At close to equilibrium all individuals would have similar risk of life-threatening malaria irrespective of their ICAM-1 genotype. A simpler explanation is that ICAM-1Kilifi is under selection through processes unrelated to severe malaria. The ICAM-1 residue 29 is situated in the region binding LFA-139 and human rhinoviruses.40 ICAM-1Kilifi has reduced avidity for LFA-1, abolishes binding to soluble fibrinogen41 and prevents binding of some rhinovirus serotypes.42 A longitudinal study of Kenyan children has reported a significantly reduced incidence of non-malarial febrile illness among ICAM-1Kilifi homozygotes.27
Future research is likely to include increasingly large-scale epidemiological studies, based in developing countries, encompassing a broad range of infectious diseases. It will be interesting to see whether the report of an association between ICAM-1Kilifi and non-malarial infections is replicated in these experiments. The trend in genetic studies of complex disease has been a move from candidate-based to whole genome approaches. The possibility of host and parasite allele frequency-dependent models highlights the need to extend future work to capture information about the genome of the parasite as well. In conclusion, our analysis has demonstrated that variation in ICAM1, specifically ICAM-1Kilifi, does not have the substantial impact on malaria susceptibility reported in early studies. However, it is apparent that genetic variation in the ICAM1 gene can modulate immune function, and may be associated with susceptibility to non-malarial infections. Therefore ICAM-1 remains an important candidate molecule for future studies of human health in developing countries.
Materials and Methods
Subjects
Patient samples were collected during ongoing epidemiological studies of severe malaria at the Royal Victoria Hospital, Banjul, The Gambia; the Queen Elizabeth Central Hospital, Blantyre, Malawi; and Kilifi District Hospital, Kilifi, Kenya. P. falciparum malaria is endemic in these study sites, with most life-threatening disease occurring in children under the age of 5 years. All DNA samples were collected and genotyped following approval from the relevant research ethics committees and informed consent from participants. Controls were cord blood samples obtained from birth clinics in the same regions as the cases. Further demographics of the cases and controls including ethnicity, sex and age distribution have previously been published.43
Phenotype definition
All cases were children admitted to hospital with evidence of P. falciparum on blood film and clinical features of severe malaria.44,45 We used a Blantyre coma score of ≤2 as a criterion of cerebral malaria (CM), and hemoglobin <5g/dl or packed cell volume <15% as a criterion of severe malaria anaemia (SA). Some individuals had both CM and SA. Of the severe malaria cases that were not CM or SA by these criteria, most had lesser degrees of coma (Blantyre coma score 3) or anemia (Hb 5-6g/dl), or other complications such as respiratory distress. Our family-based study comprised:
i) 612 Gambian trios genotyped for all markers and an additional 708 Gambian trios used to genotype ICAM-1Kilifi and rs5498 (in total the Gambian trios included 512 CM cases and 343 cases of SA).
ii) 225 Malawi trios (216 CM, 39 SA).
iii) 234 Kenyan trios (114 CM, 85 SA).
The population-based study (used to genotype ICAM-1Kilifi and rs5498) comprised:
i) 701 Gambian cases and 624 controls (324 cerebral malaria, 217 severe malarial anaemia).
ii) 718 Malawian cases and 405 controls (640 cerebral malaria, 101 severe malarial anaemia).
iii) 708 Kenyan cases and 902 controls (216 cerebral malaria, 270 severe malarial anaemia).
Power calculations
Power calculations were performed using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/)46. Across all case-control studies we would expect 95% power (based on our sample size, allelic odd ratio of 1.2, high risk allele frequency of 0.25 and a type I error rate of 0.05), a single regional case-control study (for example 701 cases and 624 controls) would have 54% power. For a similar effect size across the total family trio study (1779 trios) we would have 92% power.
SNP selection
Data from HapMap release 19 (October 2005, www.hapmap.org) was used to identify 19 SNPs within 5 kb of the ICAM1 gene, polymorphic in the Yoruba ethnic group from Ibadan in South West Nigeria (YRI). Assays were developed for these and two further non-synonymous SNPs: rs5498 which was not typed in HapMap release 19, but has a reported association with severe malaria,25 and rs1799969 which was typed by HapMap but found to be monomorphic in the Yoruba. Following initial testing two assays (rs5030399, rs281437) were rejected on technical grounds, and four further assays were rejected due to low allele frequencies (less than 2 %) in Gambians (rs5030364, rs5030400, rs5030384, rs1799969).
Sample preparation and genotyping
Genomic DNA samples underwent whole genome amplification through either Primer Extension Pre-amplification (PEP)47 or Multiple Displacement Amplification (MDA)48, before genotyping on a Sequenom MassArray genotyping platform49. There were low rates of missing data and all population control genotypes/ untransmitted parental alleles were in Hardy-Weinberg equilibrium (P > 0.01) (Supplementary Table S2).
Statistical analysis
Family-based association analysis was performed using FBAT version 1.7.2 50,51. Case-control association analysis was performed by logistic regression using covariates of ethnic group, gender and Sickle status. DNA Sequenom genotyping for the Hemoglobin S (HbS) variant was performed for all samples as previously described.43 For logistic regression we utilized STATA (v9.2 for windows) and the genassoc package (http://www-gene.cimr.cam.ac.uk/clayton/software/) written for STATA by David Clayton. In both family- and population based studies we tested each marker in additive, dominant and recessive models, with three phenotypes (CM, SA and all severe cases) and tested for SNP-based associations with severe disease in each study region (Gambia, Malawi and Kenya). Pooling across all case-control and family-studies was performed using the UNPHASED application version 3.0 (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/) 52,53 which employs a retrospective likelihood framework for performing genetic association analysis, and can be used to combine data from nuclear families and unrelated subjects. Ethnic origin was found to be a significant confounder and was retained as a covariate in the UNPHASED analysis. Haplotype structure around the ICAM1 region, in each of our family datasets was examined using HAPLOVIEW version 3.32 54 (http://www.broad.mit.edu/mpg/haploview/), haplotype blocks were defined using a solid spine of LD (as defined by D′> 0.75). Haplotype disease association analysis was conducted within the FBAT framework using family data.
Supplementary Material
Acknowledgments
This work was funded by a Wellcome Trust Clinical Research Training Fellowship (to A.E.F.) and the UK Medical Research Council (to D.P.K.). This manuscript is published with the permission of the director of the Kenya Medical Research Institute. T.W. was funded by the Wellcome Trust, MalariaGEN and the European Union Network 6 BioMalPar Consortium.
References
- 1.Newbold C, Craig A, Kyes S, Rowe A, Fernandez-Reyes D, Fagan T. Cytoadherence, pathogenesis and the infected red cell surface in Plasmodium falciparum. Int J Parasitol. 1999;29:927–937. doi: 10.1016/s0020-7519(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 2.Berendt AR, Tumer GD, Newbold CI. Cerebral malaria: the sequestration hypothesis. Parasitol Today. 1994;10:412–414. doi: 10.1016/0169-4758(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 3.van der Heyde HC, Nolan J, Combes V, Gramaglia I, Grau GE. A unified hypothesis for the genesis of cerebral malaria: sequestration, inflammation and hemostasis leading to microcirculatory dysfunction. Trends Parasitol. 2006;22:503–508. doi: 10.1016/j.pt.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Schlichtherle M, Wahlgren M. Molecular aspects of severe malaria. Clin Microbiol Rev. 2000;13:439–450. doi: 10.1128/cmr.13.3.439-450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagen T, et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci U S A. 2000;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ockenhouse CF, Betageri R, Springer TA, Staunton DE. Plasmodium falciparum-infected erythrocytes bind ICAM-1 at a site distinct from LFA-1, Mac-1, and human rhinovirus. Cell. 1992;68:63–69. doi: 10.1016/0092-8674(92)90206-r. [DOI] [PubMed] [Google Scholar]
- 7.Porta J, Carota A, Pizzolato GP, Wildi E, Widmer MC, Margairaz C, et al. Immunopathological changes in human cerebral malaria. Clin Neuropathol. 1993;12:142–146. [PubMed] [Google Scholar]
- 8.Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 9.Armah H, Dodoo AK, Wiredu EK, Stiles JK, Adjei AA, Gyasi RK, et al. High-level cerebellar expression of cytokines and adhesion molecules in fatal, paediatric, cerebral malaria. Ann Trop Med Parasitol. 2005;99:629–647. doi: 10.1179/136485905X51508. [DOI] [PubMed] [Google Scholar]
- 10.Silamut K, Phu NH, Whitty C, Turner GD, Louwrier K, Mai NT, et al. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am J Pathol. 1999;155:395–410. doi: 10.1016/S0002-9440(10)65136-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newbold C, Warn P, Black G, Berendt A, Craig A, Snow B, et al. Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am J Trop Med Hyg. 1997;57:389–398. doi: 10.4269/ajtmh.1997.57.389. [DOI] [PubMed] [Google Scholar]
- 12.Rogerson SJ, Tembenu R, Dobano C, Plitt S, Taylor TE, Molyneux ME. Cytoadherence characteristics of Plasmodium falciparum-infected erythrocytes from Malawian children with severe and uncomplicated malaria. Am J Trop Med Hyg. 1999;61:467–472. doi: 10.4269/ajtmh.1999.61.467. [DOI] [PubMed] [Google Scholar]
- 13.Heddini A, Pettersson F, Kai O, Shafi J, Obiero J, Chen Q, et al. Fresh isolates from children with severe Plasmodium falciparum malaria bind to multiple receptors. Infect Immun. 2001;69:5849–5856. doi: 10.1128/IAI.69.9.5849-5856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich JB. The adhesion molecule ICAM-1 and its regulation in relation with the blood-brain barrier. J Neuroimmunol. 2002;128:58–68. doi: 10.1016/s0165-5728(02)00114-5. [DOI] [PubMed] [Google Scholar]
- 15.Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 16.Baratin M, Roetynck S, Pouvelle B, Lemmers C, Viebig NK, Johansson S, et al. Dissection of the Role of PfEMP1 and ICAM-1 in the Sensing of Plasmodium falciparum-Infected Erythrocytes by Natural Killer Cells. PLoS ONE. 2007;2:e228. doi: 10.1371/journal.pone.0000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Languino LR, Plescia J, Duperray A, Brian AA, Plow EF, Geltosky JE, et al. Fibrinogen mediates leukocyte adhesion to vascular endothelium through an ICAM-1-dependent pathway. Cell. 1993;73:1423–1434. doi: 10.1016/0092-8674(93)90367-y. [DOI] [PubMed] [Google Scholar]
- 18.Diamond MS, Staunton DE, de Fougerolles AR, Stacker SA, Garcia-Aguilar J, Hibbs ML, et al. ICAM-1 (CD54): a counter-receptor for Mac-1 (CD11b/CD18) J Cell Biol. 1990;111:3129–3139. doi: 10.1083/jcb.111.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Reyes D, Craig AG, Kyes SA, Peshu N, Snow RW, Berendt AR, et al. A high frequency African coding polymorphism in the N-terminal domain of ICAM-1 predisposing to cerebral malaria in Kenya. Hum Mol Genet. 1997;6:1357–1360. doi: 10.1093/hmg/6.8.1357. [DOI] [PubMed] [Google Scholar]
- 21.Kun JF, Klabunde J, Lell B, Luckner D, Alpers M, May J, et al. Association of the ICAM-1Kilifi mutation with protection against severe malaria in Lambarene, Gabon. Am J Trop Med Hyg. 1999;61:776–779. doi: 10.4269/ajtmh.1999.61.776. [DOI] [PubMed] [Google Scholar]
- 22.Bellamy R, Kwiatkowski D, Hill AV. Absence of an association between intercellular adhesion molecule 1, complement receptor 1 and interleukin 1 receptor antagonist gene polymorphisms and severe malaria in a West African population. Trans R Soc Trop Med Hyg. 1998;92:312–316. doi: 10.1016/s0035-9203(98)91026-4. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Looareesuwan S, Tokunaga K. Absence of association between the allele coding methionine at position 29 in the N-terminal domain of ICAM-1 (ICAM-1(Kilifi)) and severe malaria in the northwest of Thailand. Jpn J Infect Dis. 2001;54:114–116. [PubMed] [Google Scholar]
- 24.Ndiaye R, Sakuntabhai A, Casademont I, Rogier C, Tall A, Trape JF, et al. Genetic study of ICAM1 in clinical malaria in Senegal. Tissue Antigens. 2005;65:474–480. doi: 10.1111/j.1399-0039.2005.00388.x. [DOI] [PubMed] [Google Scholar]
- 25.Amodu OK, Gbadegesin RA, Ralph SA, Adeyemo AA, Brenchley PE, Ayoola OO, et al. Plasmodium falciparum malaria in south-west Nigerian children: is the polymorphism of ICAM-1 and E-selectin genes contributing to the clinical severity of malaria? Acta Trop. 2005;95:248–255. doi: 10.1016/j.actatropica.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Ayodo G, Price AL, Keinan A, Ajwang A, Otieno MF, Orago AS, et al. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins NE, Mwangi TW, Kortok M, Marsh K, Craig AG, Williams TN. A polymorphism of intercellular adhesion molecule-1 is associated with a reduced incidence of nonmalarial febrile illness in kenyan children. Clin Infect Dis. 2005;41:1817–1819. doi: 10.1086/498156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- 29.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 30.Adams S, Turner GD, Nash GB, Micklem K, Newbold CI, Craig AG. Differential binding of clonal variants of Plasmodium falciparum to allelic forms of intracellular adhesion molecule 1 determined by flow adhesion assay. Infect Immun. 2000;68:264–269. doi: 10.1128/iai.68.1.264-269.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tse MT, Chakrabarti K, Gray C, Chitnis CE, Craig A. Divergent binding sites on intercellular adhesion molecule-1 (ICAM-1) for variant Plasmodium falciparum isolates. Mol Microbiol. 2004;51:1039–1049. doi: 10.1046/j.1365-2958.2003.03895.x. [DOI] [PubMed] [Google Scholar]
- 32.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J, Mollsten A, Falhammar H, Brismar K, Dahlquist G, Efendic S, et al. Genetic association analysis of the adiponectin polymorphisms in type 1 diabetes with and without diabetic nephropathy. J Diabetes Complications. 2007;21:28–33. doi: 10.1016/j.jdiacomp.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Register TC, Burdon KP, Lenchik L, Bowden DW, Hawkins GA, Nicklas BJ, et al. Variability of serum soluble intercellular adhesion molecule-1 measurements attributable to a common polymorphism. Clin Chem. 2004;50:2185–2187. doi: 10.1373/clinchem.2004.036806. [DOI] [PubMed] [Google Scholar]
- 35.Walsh EC, Sabeti P, Hutcheson HB, Fry B, Schaffner SF, de Bakker PI, et al. Searching for signals of evolutionary selection in 168 genes related to immune function. Hum Genet. 2006;119:92–102. doi: 10.1007/s00439-005-0090-0. [DOI] [PubMed] [Google Scholar]
- 36.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wildman DE, Uddin M, Liu G, Grossman LI, Goodman M. Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: enlarging genus Homo. Proc Natl Acad Sci U S A. 2003;100:7181–7188. doi: 10.1073/pnas.1232172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casasnovas JM, Stehle T, Liu JH, Wang JH, Springer TA. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci U S A. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staunton DE, Dustin ML, Erickson HP, Springer TA. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 40.Register RB, Uncapher CR, Naylor AM, Lineberger DW, Colonno RJ. Human-murine chimeras of ICAM-1 identify amino acid residues critical for rhinovirus and antibody binding. J Virol. 1991;65:6589–6596. doi: 10.1128/jvi.65.12.6589-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig A, Fernandez-Reyes D, Mesri M, McDowall A, Altieri DC, Hogg N, et al. A functional analysis of a natural variant of intercellular adhesion molecule-1 (ICAM-1Kilifi) Hum Mol Genet. 2000;9:525–530. doi: 10.1093/hmg/9.4.525. [DOI] [PubMed] [Google Scholar]
- 42.Xiao C, Tuthill TJ, Bator Kelly CM, Challinor LJ, Chipman PR, Killington RA, et al. Discrimination among rhinovirus serotypes for a variant ICAM-1 receptor molecule. J Virol. 2004;78:10034–10044. doi: 10.1128/JVI.78.18.10034-10044.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fry AE, Griffiths MJ, Auburn S, Diakite M, Forton JT, Green A, et al. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum Mol Genet. 2008;17:567–576. doi: 10.1093/hmg/ddm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization, Division of Control of Tropical Diseases Severe and complicated malaria. Trans R Soc Trop Med Hyg. 1990;84(Suppl 2):1–65. [PubMed] [Google Scholar]
- 45.Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 46.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez JM, Portillo MC, Saiz-Jimenez C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ Microbiol. 2005;7:1024–1028. doi: 10.1111/j.1462-2920.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 49.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 50.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 51.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 52.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 53.Dudbridge F. Technical Report 2006/5. MRC Biostatistics Unit; Cambridge, UK: 2006. UNPHASED user guide. [Google Scholar]
- 54.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.