Abstract
Ebf1 is a transcription factor essential for B cell fate specification and function and important for the development of olfactory sensory neurons. We show here that Ebf1 also plays an important role in regulating osteoblast and adipocyte development in vivo. Ebf1 mRNA and protein is expressed in MSCs, in OBs at most stages of differentiation, and in adipocytes. Tibiae and femora from Ebf1−/− mice had a striking increase in all bone formation parameters examined including the number of OBs, osteoid volume, and bone formation rate. Serum osteocalcin, a marker of bone formation, was significantly elevated in mutant mice. The numbers of osteoclasts in bone were normal in younger (4 week-old) Ebf1−/− mice but increased in older (12 week-old) Ebf1−/− mice. This correlated well with in vitro osteoclast development from bone marrow cells. In addition to the increased osteoblastogenesis, there was a dramatic increase in adipocyte numbers in the bone marrow of Ebf1−/− mice. Increased adiposity was also seen histologically in the liver but not in the spleen of these mice, and accompanied by decreased deposition of adipose to subcutaneous sites. Thus Ebf1-deficient mice appear to be a new model of lipodystrophy. Ebf1 is a rare example of a transcription factor that regulates both the osteoblast and adipocyte lineages similarly.
Keywords: B Cells, Cell Differentiation, Transcription Factors, Transgenic/Knockout Mice
Introduction
Osteoblasts (OBs) are mesenchymal in origin and one of their major functions is to make bone. These cells secrete osteoid (non-mineralized bone matrix), which over time mineralizes to form new bone. OBs regulate the differentiation of osteoclasts (OCs), bone resorbing cells, which are hematopoietic in origin [1, 2]. Cells in the OB lineage also support hematopoietic cell differentiation in endosteal niches [3]. Adipocytes or fat cells originate from mesenchymal stem cells (MSC) and are more closely related to OBs than other mesenchymal cells such as myoblasts or chondrocytes.
Several transcriptional regulators have been shown to control the OB and adipocyte lineages. Runx2 is expressed in OBs at all stages of development and its targeted disruption abolishes the OB lineage [4–6]. Also, inactivation of Osterix by gene targeting results in a block in OB development downstream of Runx2 [7]. Conversely, PPARγ is crucial for adipocyte differentiation and function [8], and members of the C/EBP family of transcription factors have been implicated in controlling aspects of adipocyte biology [8].
Growth and differentiation of either the adipocyte or OB lineages at the expense of the other has been demonstrated in studies that over-express lineage specific transcription factors and/or use specific growth conditions. Cell lines exist that can be differentiated into either adipocytes or OBs by changing the media components [9, 10]. This inverse relationship is exemplified by increased bone marrow adipogenesis and decreased osteoblastogenesis in age-related osteoporosis [11, 12]. Increased expression of PPARγ2 causes increased adipocyte and decreased OB formation [10, 13, 14]. Conversely, over-expression of ΔFosB, a naturally occurring truncated form of FosB, in transgenic mice leads to increased bone formation resulting in osteosclerosis [15], and ΔFosB inhibits adipogenesis both in vitro and in vivo [15]. Collectively, these findings have resulted in an “either/or” model of differentiation whereby one of these two lineages expands or differentiates at the expense of the other. However, the factors that regulate commitment at the branching point between OBs and adipocytes, the degree of plasticity between the two cell lineages, and unrecognized regulatory interactions remain unclear.
Ebf1 is the founding member of a small multigene family of novel helix-loop-helix proteins with defined roles in cellular differentiation and function. This factor was cloned simultaneously in two different studies and named olfactory factor 1 (Olf-1), and early B cell factor-1 (Ebf1), and sometimes referred to as O/E1 [16, 17]. Ebf1 is expressed in B cells, and mice deficient in Ebf1 contain only early progenitor B cells [17, 18]. Ebf1 and subsequently discovered family members, Ebf2, 3, and 4 control nervous system development and olfactory receptor gene expression [16,19]. Importantly, Ebf1, 2, and 3 are expressed in mouse adipocytes [17, 20] and appears to induce adipocyte differentiation with similar timing and efficiency as PPARγ2 [21].
Ebf1 is a member of a cascade of transcription factors that are required for B cell lineage development that includes PU.1, Ikaros, E2A, Ebf1 and Pax5 sequentially. We have reported that Pax5−/− mice are severely osteopenic resulting from a 3–5-fold increase in the number of OCs in bone while the number of OBs is indistinguishable from wild-type controls [22]. Because Ebf1 regulates Pax5 expression in B cells, and in light of the prominent bone phenotype in Pax5−/− mice, we examined whether Ebf1 regulated the skeleton. We report here that Ebf1−/− mice, in stark contrast to Pax5−/− mice, had increases in all bone formation parameters measured, with marked increases in OB numbers, osteoid volume, and the bone formation rate. In contrast, the number of OCs was similar to controls in young mice but more than doubled in older mice. Ebf1−/− mice also exhibited a dramatic increase in adipocytes that filled the medullary canal, and increased fat was also detected in the livers of mutant mice, while subcutaneous fat stores were decreased. These data demonstrate for the first time that Ebf1 is required for maintaining the regulation of both osteogenesis and adipogenesis, and suggest that Ebf1−/− mice are a new model of lipodystrophy. Our data indicate that not only does the loss of Ebf1 result in the arrest of B cell development but also the appearance of a unique bone phenotype.
Materials and methods
Mice
129X1/SvJ and C57BL/6 mice were obtained from Jackson Labs, Bar Harbor ME, and the National Cancer Institute respectively, and housed at Yale University School of Medicine. The Ebf1−/− were originally produced and provided by Dr. Rudolf Grosschedl, Max-Planck-Institute of Immunobiology, Freiburg, Germany [18]. We had difficultly generating Ebf1−/− mice on the C57BL/6 genetic background but were able to produce Ebf1−/− mice by breeding Ebf1+/− mice maintained on the C57BL/6 genetic background to 129X1/SvJ mice, resulting in an F1 generation. The Ebf1+/−F1 mice were then interbred (F2 generation). Ebf1−/− F2 mice were used for all experiments and no further breeding of any F2 generation mice was attempted. Ebf1−/− mice were always compared to age-matched wild-type littermate controls and were genotyped as previously described [23]. The Yale Medical School IACUC approved the purchase and use of all animals.
Histology, Histomorphometry, μCT, Immunohistochemistry
Tibiae and femora were stripped of soft tissue, fixed in 10% buffered formalin, dehydrated, and embedded in methyl methacrylate before being sectioned and stained with toluidine blue [24]. To label bone mineralization fronts, mice were injected with 30 mg/kg of calcein 8 days and 24 hours prior to sacrifice. Histomorphometric measurements were performed on a fixed region just below the growth plate corresponding to the primary spongiosa [25] and analyzed by Osteomeasure software (Osteometrics, Atlanta, GA).
To stain for lipid, bones were prepared and fixed as described above, and treated with 2% osmium tetroxide in water with 5% potassium dichromate [26]. Bones were processed in the absence of lipophilic agents, embedded in methyl methacrylate, and sectioned. Brain, heart, kidney, liver and spleen were collected, fixed in 4% paraformaldehyde, frozen sections cut, and stained with Oil-Red-O (Sigma). Tibia and femurs were dissected, the epiphyses removed, and the bone marrow flushed using a 25 gauge needle. Bone marrow smears were prepared on microscope slides in a manner similar to preparing blood smears, air dried, and stained with Oil-Red-O.
For μCT femora were stripped of soft tissue and stored in 75% EtOH at 4°C. The bones were scanned using a Scanco μCT-35 (Scanco, Brutissellen, Switzerland) and analyzed for numerous structural parameters at both the distal femur just below the growth plate (trabecular bone) and at the femoral midshaft (cortical bone).
For immunohistochemistry long bones from C57BL/6 mice were dissected free of soft tissue, decalcified in 4% EDTA, embedded in paraffin, 5 μm sections cut, and stained with anti-O/E1 antibody or rabbit IgG [27]. A secondary IgG-biotin antibody was used and visualized with a streptavidin-HRP (Vector Laboratories) with Nova Red as the chromagen to give a reddish-brown stain, and counterstained with H&E.
Cell Isolation and Culture
Murine calvarial OBs from mice less than 48 hrs old were prepared by sequential collagenase digestion as previously described [28] and cells collected from the third through fifth collagenase digestions were used. At preconfluent density (3 days in culture) <1% of the cells are CD45+ by FACS indicating very low levels of contaminating hematopoietic cells.
MC3T3-E1 is a continuously growing pre-OB cell line derived from C57BL/6 mice [29]. Cells were grown in α-MEM with glutamine, 10% FCS, and 1% Pen-Strep.
Mesenchymal stem cells (MSCs) were generously supplied by the Center for Gene Therapy at Tulane University Health Sciences Center (New Orleans, LA), and were originally isolated from C57BL/6 mice according to Dr. Darwin Prokop’s protocol [30]. They were maintained at low density (50–5000 cells/cm2) in MEMαsupplemented with 10% fetal calf serum, 10% donor equine serum, L-Gln, and 1% antibiotic-antimycotic. Cells were not used past passage 10. Osteoclast-like cells were induced from bone marrow flushed from tibiae and femurs of mutant or age matched control mice. The cells were plated in 48 well dishes (106/well) and treated with 30 ng/ml of rmM-CSF and 50 ng/ml rhRANKL (R&D Systems) as previously described [22]. After 7 to10 days the cells were fixed, and stained for TRAP using Leukocyte Acid Phosphatase 387-A kit (Sigma). TRAP+, multinucleated (>3) Ocs were counted.
T and B cells were isolated from spleens following red blood cell lysis using fluorescent-labeled αMac-1 (CD11b), αCD19, αCD4, and αCD8 antibodies (BD Biosciences) and flow cytometry. Mac-1− CD19+ (B cells), Mac-1− CD19− CD8− CD4+ (CD4 T cells), and Mac-1− CD19− CD4−CD8+ (CD8 T cells) populations were isolated on a FACSVantage flow cytometer and using CellQuest software (BD Biosciences) were found to be > 95% pure.
Subcutaneous fat was isolated from the pads surrounding the flank region. The wet weight of the adipose tissue was compared to the total weight of the animal and expressed as a percentage of the total body weight.
Cell Proliferation
Freshly isolated calvarial cells (5×103) were cultured in 96-well plates for prescribed times and proliferation was measured by adding CellTiter 96 Aqueous One Solution Proliferation Assay (Promega) directly to the media for 4 hours then measuring absorbance at 490 nm [31,32].
Measurement of Alkaline Phosphatase (ALP) and Osteocalcin
ALP and osteocalcin production was measured in OB cell lysates and sera respectively. ALP activity was determined using a commercially available kit (86R-1KT, Sigma) [22]. Osteocalcin levels in the sera were measured by a standard equilibrium radioimmunoassay using specific goat anti-mouse osteocalcin antibody [33].
RT-PCR
RNA was harvested using RNAzol B (Tel-Test) or Rneasy (Qiagen) as directed by the manufacturer. RNA was incubated with DNase I and subjected to reverse transcription (RT) using SuperScriptII (Invitrogen). Targets for PCR were amplified within a linear range, and products were separated on agarose gels stained with ethidium bromide. The PCR primer sequences can be furnished upon request.
Western Blot
Whole cell lysates were collected using a high salt buffer containing mild detergents and protease inhibitors. 30 μg of each sample was separated by SDS-PAGE on 10% acrylamide-bisacrylamide gels, transferred to Nitrocelluose membrane (Biorad), and probed for Ebf1 (Millipore #AB5949). HRP-conjugated secondary antibody (GE Healthcare) was activated with SuperSignal West Pico Chemiluminescent Substrate (Pierce), and visualized on radiographic film (Kodak).
Results
Expression of Ebf1 in osteoblasts and adipocytes
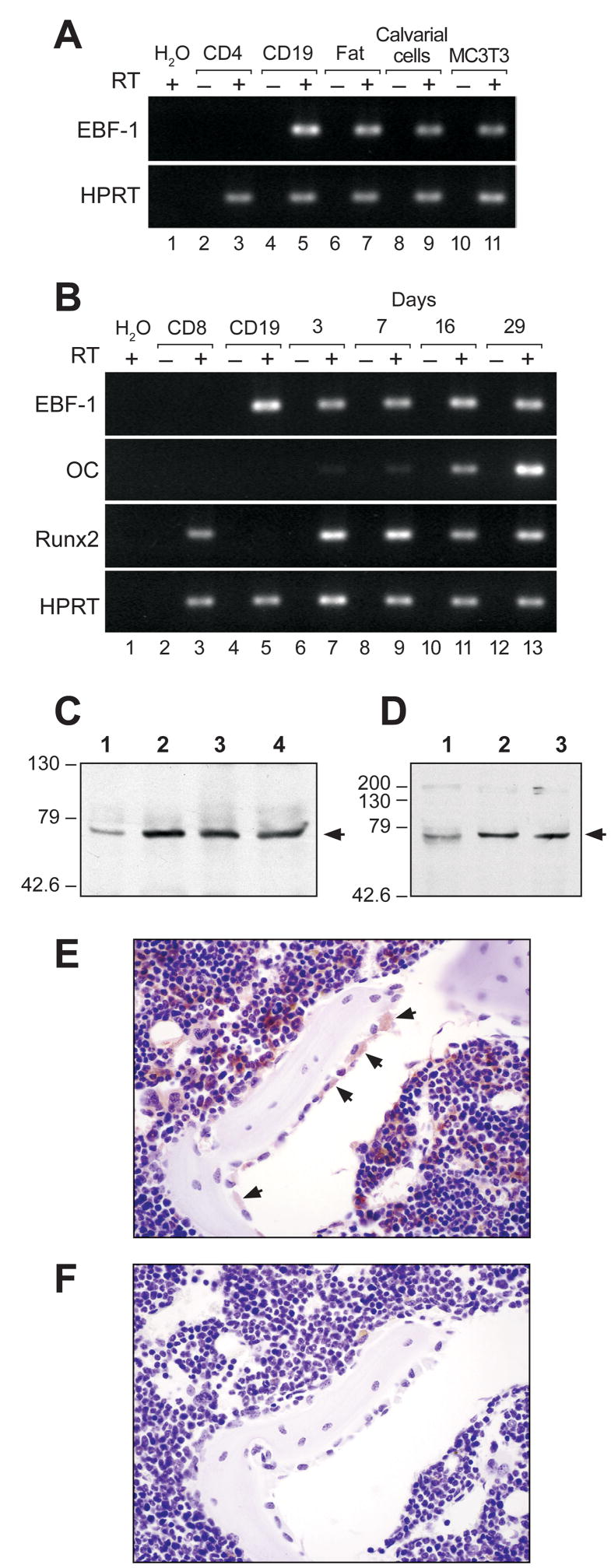
Ebf1 has been shown to play a role in adipocyte differentiation and function in vitro [20]. Due to the close relationship between adipocyte and OB lineage development, we hypothesized that Ebf1 may also be expressed in OB lineage cells. To test this idea, Ebf1 mRNA transcript levels were measured by RT-PCR using RNA isolated from B cells (CD19+), CD4+ T cells, inguinal fat pads, primary calvarial OBs and the pre-OB cell line MC3T3-E1. The adipose tissue was taken from Rag1−/− mice (which lack mature B and T cells) to assure that any detectable Ebf1 signal was not from contaminating B cells. Abundant Ebf1 transcripts were easily detected in both primary OBs and the MC3T3-E1 cell line at levels comparable to those found in B cells and fat. As expected, Ebf1 transcripts were readily detectable in B cells and adipose tissue, yet undetectable in CD4+ T cells (Fig 1A).
Figure 1.
Ebf1 expression in osteoblasts and adipocytes. (A) RT-PCR detection of Ebf1 and HPRT (loading control). RNA was from CD4+ T cells (lanes 2–3), CD19+ B cells (lanes 4–5), Rag1−/− fat tissue (lanes 6–7), calvarial OBs (lanes 8–9), and the pre-OB cell line MC3T3-E1 (lanes 10–11). (B) RT–PCR detection of Ebf1, osteocalcin (OC), Runx2, and HPRT. RNA was from CD8+ T cells (lanes 2–3), CD19+ B cells, and calvarial OBs at increasingly mature stages of differentiation (Days in culture, lanes 6–13). Lanes 6–7 contain the least mature samples and lanes 12–13 contain the most mature samples. (C) Western blot of Ebf1 expression in whole cell lysate from calvarial OBs. Lane 1 represents the least mature sample and lane 4 the most mature sample. (D) Western blot of Ebf1 expression in whole cell lysate from MSCs. Lanes 1–3 represent the least to most mature samples (3, 7, or 14 days in culture). (E) Immunohistochemical staining of proximal tibia with anti-Ebf1 antibody. Arrows indicate large stained OBs on bone surfaces. (F) Immunohistochemical staining of Ebf1 in bone with an isotype control.
Calvarial cells differentiate into mature OBs in culture [34]. To examine Ebf1 expression during OB differentiation, Ebf1 transcript levels were measured via RT-PCR using RNA isolated from primary wild-type calvarial cells after 3, 7, 16, and 29 days in culture. We detected equivalent levels of Ebf1 mRNA regardless of the differentiation state of the OBs (Fig 1B). Runx2, a regulator of OB development, was also expressed at all stages, while Osteocalcin, a marker of mature OBs, was induced as the cultures progressed confirming that each time point represented increasingly mature OB populations (Fig 1B).
Calvarial OBs were next used to measure Ebf1 protein levels at various stages of differentiation. When whole cell lysates were collected after 0, 3, 7, and 14 days in culture and evaluated by western blot, there appeared to be a small increase in Ebf1expression between the initial isolation and 3 days in culture (Fig 1C), but after this time, and consistent with the RNA observations shown above, Ebf1 expression was maintained constant independent of the differentiation stage. Similar results were seen using C57BL/6-derived MSCs. MSCs were seeded at low density, whole cell lysates collected at 3, 7, 14 days after seeding and assessed by western blot. These cells also differentiate along the osteoblast lineage in culture (default pathway), but unlike calvarial OBs, are not initially committed to the osteoblast lineage. While Ebf1 is present in the cells at all times tested, there appears to be an increase in the expression between 3 and 7 days in culture (Fig 1D).
To determine if Ebf1 was expressed by OBs in vivo, long bones were decalcified, and processed for immunohistochemistry using Nova Red to stain Ebf1+ cells a reddish-brown color. Distinct Ebf1 staining was seen in OBs attached to bone surfaces (Fig 1E, see arrows). Importantly, not all the OBs were stained, only the large cells. In contrast, none of the osteocytes were stained. Numerous bone marrow cells were Ebf1 positive, which is consistent with the large number of B lineage cells in bone marrow and serves as an internal positive control. Bone sections stained with an isotype control antibody (Fig 1F) were negative. These data confirm our gene expression data and demonstrate that Ebf1 is expressed in some but not all OBs in vivo. The fact that osteocytes are not stained may reflect either their state of activation or stage of differentiation.
Increased bone formation in Ebf1-deficient mice
We hypothesized that Ebf1 plays a role in skeletal biology due to its expression in OBs and examined Ebf1−/− mice for skeletal changes. All Ebf1−/− mice were runted when compared to age-matched controls (Fig 2A), and a portion of these mice died for unknown reasons at 3 to 5 weeks of age. We observed mice for as long as 8 months and the runting persisted, however it was more exaggerated at 1 month of age compared to mice over 6 months old. The Ebf1−/− mice had normal tooth development and were not blind, suggesting that functional OCs were present.
Figure 2.
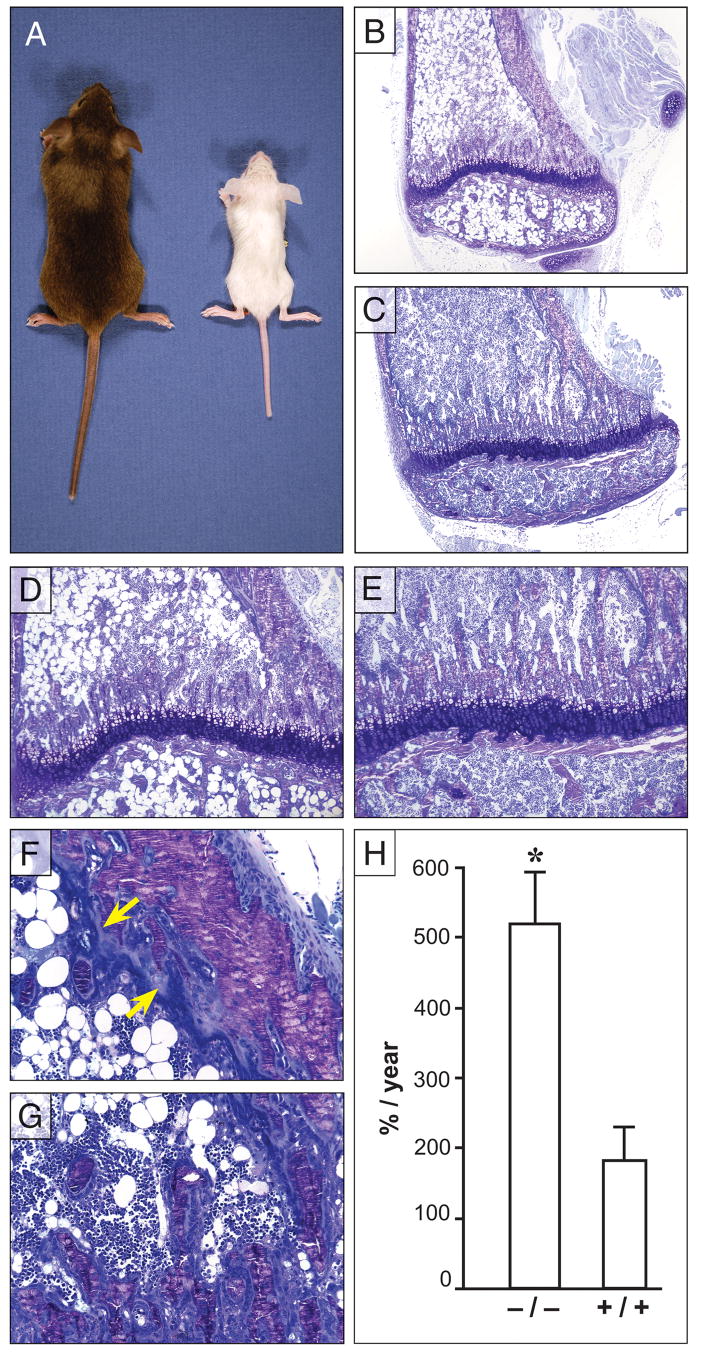
Physical and histological assessment of Ebf1−/− and wild-type littermate control long bones. (A) Comparison of 4-week-old Ebf1−/− (right) and age matched littermate control (left). Toluidine blue stained proximal tibiae from 4 week-old Ebf1−/− (B, D, F, G) and age-matched littermate wild-type control (C, E). (B) Ebf1−/− tibiae are striking smaller than control (C, 4x original magnification), characteristic of Ebf1 deficiency. (D) Although proportionally smaller, the growth plate in Ebf1−/− mice is similar to control (E) with orderly columns of developing chondrocytes (10x original magnification). (F) The interface between Ebf1−/− cortical bone and bone marrow shows increased osteoid thickness (Carolina blue line closest to bone, arrows) and the layers of OBs 2–3 cells thick adjacent to the osteoid (dark blue closest to the marrow). (G) Ebf1−/− OBs also surround individual trabeculae and fill the intratrabecular spaces at the growth plate (20x orginal magnification). Fat cell ghosts fill the medullary canal (B, D). (H) Comparison of the bone formation rate (BFR) between Ebf1−/− and littermate controls at 4 weeks of age. Values represent the mean ± SE. *Significant difference in BFR, p≥0.01.
To evaluate bone remodeling, the femora and tibiae from Ebf1−/− mice and wild-type littermate controls were isolated, processed for histomorphometric analysis, and evaluated by light microscopy. Consistent with the runting of the animals, the bones from Ebf1-mutant mice (Fig 2B) were physically smaller than controls (Fig 2C). Although proportionally smaller, the growth plates of Ebf1−/− mice (Fig 2D) were orderly with normal appearing columns of developing chondrocytes that were similar to controls (Fig 2E). Additionally, both the proliferative and hypertrophic zones appeared similar. When Ebf1 expression was examined in primary growth plate chondrocytes or immortialized chondrocytic cell lines (ATDC5, RCJ3.1C5.18) Ebf1 mRNA was undetectable (data not shown). Taken together, these results suggest that the runting is not caused by a defect in chondrocytic differentiation or function.
Increases in osteoid volume (OV/TV) were apparent in histological sections (Fig 2F, see arrows) and by histomorphometry of Ebf1−/− bones (over 200% of wild-type) at both 4 and 12 weeks of age (Table 1). Osteoid thickness (OTh) in Ebf1−/− bones was also increased by 22% and 25% over controls at 4 and 12 weeks of age, respectively. The rate of bone formation (BFR/TV) in Ebf1−/− mice was 285% that of the control animals as measured by calcein fluorochrome double labeling (Fig 2H). Although the bone volume (mineral plus osteoid) was not different in histological samples between mutant and wild-type mice at 4 weeks of age, by 12 weeks the bone volume of Ebf1−/− mice was 75% greater than controls (Table 1). The increase in bone volume is consistent with the almost 50% reduction in trabecular spacing (TbSp) and a more than 66% increase in trabecular number (TbN) in 12 week old Ebf1−/− mice compared to controls (Table 1). Taken together, these data demonstrate an age-related increase in bone volume. The mineralization of the bone in these mice, however, appears to be delayed in the trabeculae. When the same parameters (BV/TV and TbN) were analyzed by μCT, which does not include nonmineralized osteoid, in 4 week-old animals, there was a significant lack of mineralized bone in the Ebf1−/− femora (Table 2).
Table 1.
Histomorphometry measurementsa
| Age | Mice | No of Mice | BV/TVC(%) | OV/TV(%) | OTh(μm) | NOb/TAR(/mm2) | NOc/TAR(/mm2) | TbSp(μ) | TbN(/mm) |
|---|---|---|---|---|---|---|---|---|---|
| 4 weeks | Ebf1 | 5 | 22±.3 | 4±1.0b | 2.2±0.1b | 2688±500b | 99±25 | 53±10 | |
| Control | 8 | 20±4 | 2±0.7 | 1.8±0.1 | 1454±217 | 138±21 | 50±10 | ||
| 12 weeks | Ebf1 | 6 | 28±2b | 2.5±0.1b | 2±0.1b | 1015±159b | 108±8b | 53±7b | 15±1b |
| Control | 6 | 16±1 | 1.1±0.1 | 1.6±0.1 | 536±63 | 44±6 | 99±2 | 9±0.2 |
Results are presented as mean ± SEM
At least p ≤0.05
BV/TV, bone volume/total volume; NOb/TAR, number of osteoblasts/tissue area; OV/TV, osteoid volume/total volume; OTH, osteoid thickness; NOc/TAR, number of osteoclasts/tissue area; TbSp, trabecular spacing; TbN, trabecular number.
Table 2.
μCT measurementsa
| Trabecular 3D Morphometry
|
Cortical Midshaft Morphometry
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | Mice | No of Mice | BV/TVC(%) | TbN(/mm2) | TbSp(mm) | TbT(mm) | BS/BV(%) | BV/TV(%) | CT(mm) | TD(HA/ccm) | pMOI(mm4) |
| 4 weeks | Ebf1 | 6 | 1.9±1.1b | 2.1±0.3b | 0.52±0.06b | 0.026±0.001 | 26.5±1.0 | 81.5±8.3 | 0.08±0.01 | 857.6±22.7b | 0.05±0.01b |
| Control | 6 | 4.7±0.4 | 3.3±0.3 | 0.32±0.04 | 0.030±0.002 | 16.9±4.9 | 88.7±1.1 | 0.12±0.01 | 927.5±19.7 | 0.19±0.03 | |
Results are presented as mean ± SEM
At least p ≤f5
BV/TV, bone volume/total volume; TbN, trabecular number; TbSp, trabecular spacing; TbT, trabecular thickness; BS/BV, bone surface/bone volume; CT, cortical thickness; TD, tissue density; pMOI, polar moment of inertia.
When the cortical bone of 4 week-old animals was analyzed by μCT, there was no difference in the BV/TV, bone surface/tissue volume (BS/TV) or cortical thickness (CT) (Table 2). A significant reduction in tissue density was recorded in the mutant mice as compared to controls. Although the bones were not subjected to mechanical loading or fracture analysis, the polar moment of inertia (pMOI) in 4 week-old animals was decreased by 72% in the Ebf1−/− animals compared to their age-matched controls (Table 2). These data taken as a whole, suggest that 4 week-old Ebf1 deficient animals possess a severe defect in bone strength.
Increased numbers of osteoblasts in the bones of Ebf1-deficient mice
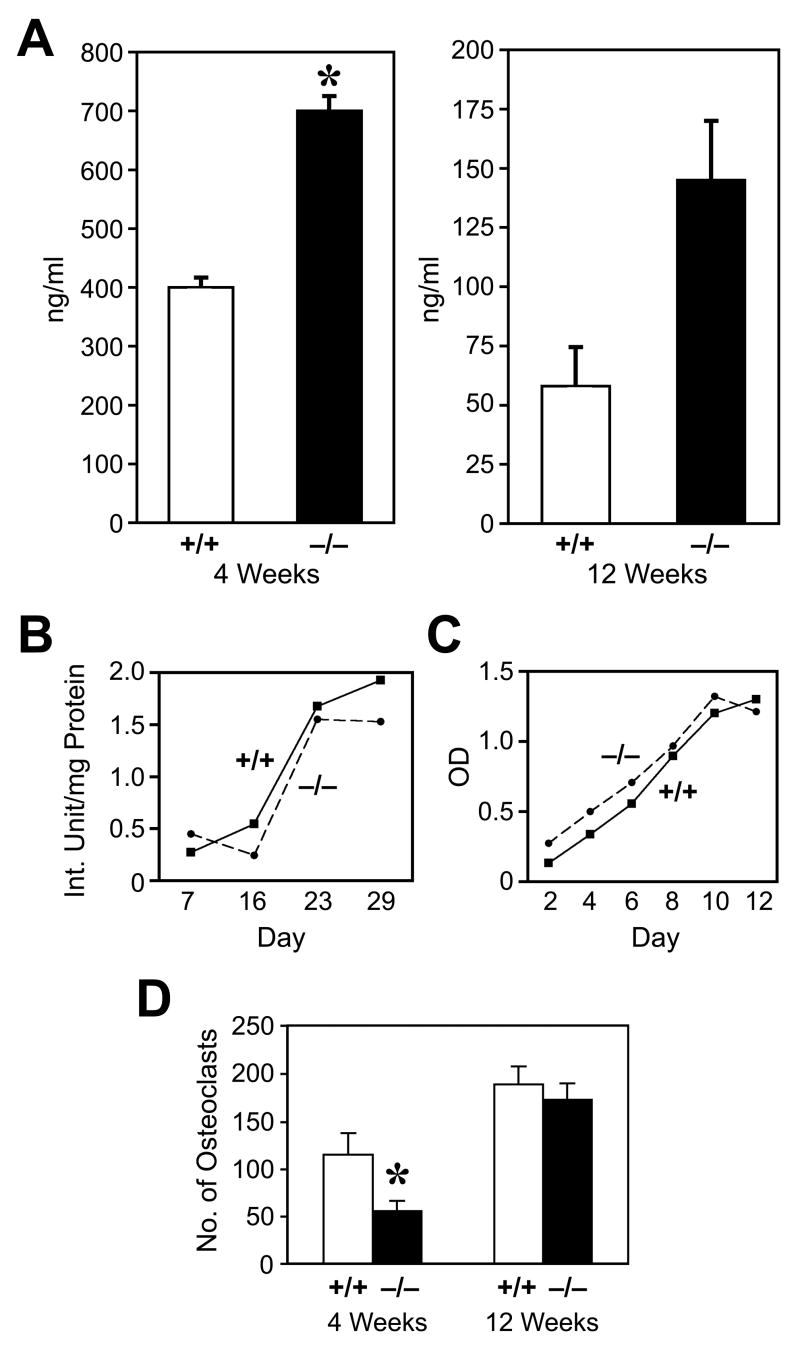
Osteoid is made by OBs and becomes bone following mineralization. Due to the increased osteoid volume and thickness in the bones of Ebf1−/− mice, we hypothesized that Ebf1 deficiency leads to an increase in OB numbers. Increases in the numbers of OBs in bone were apparent both qualitatively in histological sections, and quantitatively using histomorphometry. Multiple layers of OBs were seen attached to Ebf1−/− endosteal surfaces (Fig 2F, see arrows) as compared to controls (Fig 2E). The inter-trabecular spaces at the growth plate were filled with OBs, and OBs surrounded individual trabeculae (Fig 2G). Also, the number of OBs in 4 week-old Ebf1−/− mice was increased by 85% compared to controls (Table 1) as assessed by the number of osteoblasts/total area (NOb/TAR). Additionally, in 4 week old animals, most formation parameters were increased significantly: OBs on bone surfaces (Obs/BS) were increased 88%, and trabecular thickness was increased 21% (data not shown). This increase in OB numbers was also apparent in 12 week-old mice where the NOb/TAR was almost 90% above controls (Table 1). These data not only indicate a marked increase in the number of OBs in both 4 and 12 week-old Ebf1−/− mice but also suggest that they were functional. In fact, levels of serum osteocalcin, a highly specific secreted protein of mature OBs, in Ebf1−/− mice were increased by approximately 75% in 4 week-old mice, and by almost 130% in 12 week-old mice compared to age-matched controls (Fig 3A). These data correlate well with the observed increase in OB numbers in the mutant mice.
Figure 3.
Serum osteocalcin, osteoblast proliferation, alkaline phosphatase, and osteoclast development in Ebf1−/− mice. (A) Serum was collected from individual (>5) Ebf1−/− and +/+ 4 and 12-week-old mice and osteocalcin measured by specific RIA. (B) Both the production of ALP and the proliferative response (C) by cultured calvarial OBs were similar in Ebf1−/− and +/+ mice (SE were all ≥10%). (D) Bone marrow from Ebf1−/− and +/+ mice was cultured with M-CSF and RANKL and the number of large, TRAP+, multinucleated osteoclasts counted. Values represent the mean ± SE. *Significant decrease in Ebf1−/− mice, p ≤0.01.
To determine whether OBs from Ebf1−/− mice were functional, we next tested the ability of Ebf1−/− and wild-type control calvarial OBs to proliferate and differentiate into mature OBs in vitro. Expression of alkaline phosphatase (ALP), a marker of OB differentiation, was measured from lysates of cultured primary calvarial cells grown for 7, 16, 23, and 29 days. ALP expression by cultured Ebf1−/− OBs increased in a time-dependent manner and was similar to control cells (Fig 3B). Similarly, no differences were observed in the proliferative capacity of freshly-isolated Ebf1−/− and control OBs cultured for 2–12 days (Fig 3C).
Age-dependent increase in osteoclast numbers
Bone density is regulated through not only the anabolic actions of OBs, but also the catabolic actions of OCs. Unlike OBs, OCs are hematopoietic in origin arising from the myeloid lineage. While Ebf1−/− mice lack all but the earliest identifiable B cells, they exhibit normal numbers of T cells as well as Mac-1+ and Gr-1+ myeloid cells [18]. Having observed increases in bone density and OB number in the Ebf1−/− mice compared to their age-matched controls, we next examined whether OC numbers were also altered in the bones of Ebf1−/− animals. When OCs were assessed in decalcified bone sections, 12 week-old Ebf1−/− bones contained almost 2.5 times the number of OCs observed in control bones (Table 1 NOc/TAR). However, there was no statistically significant difference in the number of OCs in bones of 4 week-old Ebf1−/− mice compared to wild-type controls. When OC formation was evaluated in vitro, the number of OCs produced from 4 week-old Ebf1−/− bone marrow cells was reduced by 50% compared to control (Fig 3D). In contrast, bone marrow cells from 12 week-old mutant mice and controls produced similar numbers of OCs. These data suggest that loss of Ebf1 causes an early defect in OC progenitors that corrects with age.
Altered adipocyte populations in Ebf1−/− mice
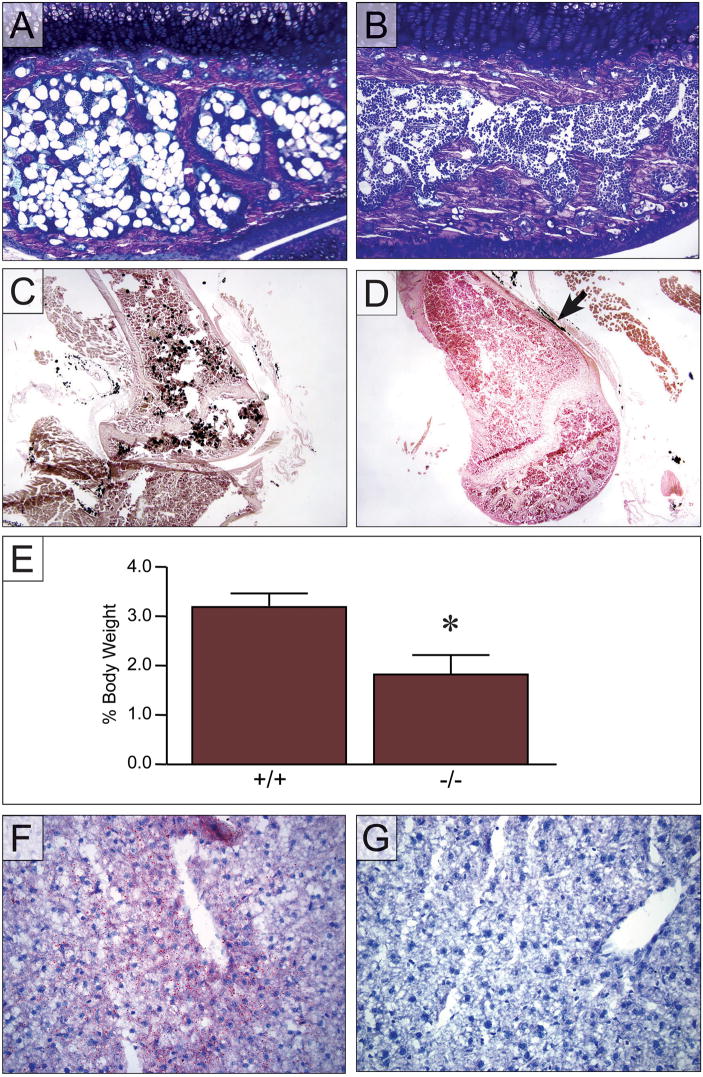
The most striking phenotype of Ebf1−/− bones was the massive increase in adipocytes that filled the medullary canal (Fig 2B-C) and marrow space in the secondary center of ossification (Fig 4A-B). Our interpretation that these cells were adipocytes was based on their location, morphology, and that they were devoid of toluidine blue stain. These structures were not fixed cells but fixed cell ghosts that appear white because lipophilic agents (acetone and ethanol) extract the lipid from the cells during sample preparation. To confirm the presence of increased adipocytes in the medullary canal, tibia from 4 week-old mice were isolated and then stained for lipid using osmium. Extensive osmium staining was seen above and below the growth plate and observed to extend into the diaphesis of the Ebf1−/− bones (Fig 4C). No staining was observed in the control bone (Fig 4D), however, staining was seen outside of the bone, in fatty tissue adjacent to the periosteum, serving as an internal positive control (Fig 4D see arrow). These data confirm our interpretation that the cell ghosts seen in the histologic sections were adipocytes.
Figure 4.
Fat distribution in Ebf1−/− mice. Toluidine blue stained proximal tibiae from 4 week-old Ebf1−/− and age-matched littermate control mice. (A) Adipocyte ghosts fill the secondary center of ossification (above the growth plate) in Ebf1−/− mice as compared to controls (B). (C) Increased adipogenesis in Ebf1−/− bone revealed by extensive osmium staining in the bones of 4-week-old mutant bone. (D) No osmium staining was seen in control bone. Extra-osseous fat serves as a control (arrow). (E) White adipose deposition in the subcutaneous fat pads surrounding the flank region is decreased in 4 week old Ebf1−/− (n=10) compared to controls (n=9). Each point represents the mean ± SE. Statistical significance was determined by ANOVA.
*Significant decrease in Ebf1−/− mice, p ≤0.01. (F) Deposition of fat, indicated by Oil-red-O staining, in liver from Ebf1−/− mice as compared to age-matched littermate control (G).
Similar to the increased number of OBs, the increase in bone marrow adipocytes was observed at both 4 and 12 weeks of age in Ebf1−/− mice. Oil-red-O staining of freshly prepared bone marrow smears also confirmed this increase in bone marrow fat (data not shown). As mentioned earlier, the Ebf1−/− mice were runted compared to their WT littermates and the animals had a mildly cachectic look. When the subcutanueous fat pads were removed from the flanks of these mice and weighed there was a significant decrease in the percent body weight of these deposits for the Ebf1−/− mice compared to their WT counterparts at 4 (Fig 4E) and 12 weeks of age (data not shown). Because other organs express low levels of Ebf1 mRNA [17], we examined brain, heart, kidney, liver, and spleen from Ebf1−/− mice for changes in fat distribution. Increased fat deposition, visualized by Oil-red-O staining, was seen histologically in the livers of some Ebf1−/− mice (Fig 4F) but never in controls (Fig 4G). No staining was observed in the other tissues of mutant or wild-type mice (data not shown). Thus, while fat deposition to the bone marrow and liver is increased in the Ebf1−/− mice, subcutaneous deposition of white adipose tissue was decreased.
Discussion
We have shown that Ebf1 is expressed at multiple stages of in vitro OB differentiation ranging from immature proliferating cells to mature osteoid secreting OBs. Ebf1 is also expressed in the MC3T3-E1 osteoblastic cell line, confirming the primary cell expression. Immunohistochemical localization showed Ebf1 expression in some, but not all, OBs on bone surfaces (the cells that were larger than those that were adherent but not positive), which is consistent with cellular activation. In contrast, osteocytes were negative for Ebf1 staining. Whether this is because they are embedded in bone or because they are the most mature cells in the OB lineage is unknown. The OB-specific expression of Ebf1 suggests that Ebf1-based regulation is, in part, cell intrinsic. It has been reported that Ebf1 is expressed in the stromal cell line OP9, which is highly efficient in supporting hematopoiesis, but not in cells that do not support hematopoiesis [35]. It may be that Ebf1 regulates hematopoiesis in part through an indirect effect on the supporting stromal cells and OBs. Ebf1 is expressed in MSCs, osteoblasts, adipocytes, and some stromal cells, but not in chondrocytes, suggesting that it is regulated during mesenchymal lineage allocation.
Our histologic and histomorphometric data show significantly increased OB numbers in the bones of Ebf1−/− mice explaining the increase in bone formation rate, trabecular number, osteoid volume and thickness and serum osteocalcin. Ebf1−/− OBs were indistinguishable from wild-type cells in their ability to proliferate and express ALP in culture, yet we found increased numbers of OBs in the bones of Ebf1−/− mice at both 4 and 12 weeks of age while an increase in bone volume was only detected by 12 weeks. This delay is most likely due to the time that is needed for osteoid to mineralize into new bone. Using μCT BV/TV, and TbSp were reduced in the primary spongiosa of Ebf1−/− mice. Histomorphometric measurements count both mineral and osteoid, while μCT determinations are made on mineralized bone only. This could account for the discrepancy between the two analyses, especially considering the large amount of unmineralized osteoid present in the Ebf1−/− mice. In addition, mutant and control mice are not different in trabecular thickness supporting this idea. These differences support our rationale for a delay in mineralization coupled with accelerated bone turnover in the trabecular and cortical compartments of the Ebf1−/− mice, and is supported by the dramatic increases in osteoid and bone formation rate in the trabecular compartment and the decrease in tissue density in the cortical compartment.
Increased bone volume at 12 weeks takes on additional significance because the bones of null mice are smaller than controls. The increased bone formation rate and the trabecular number (from histomorphometric analyis), as well as our data indicating that Ebf1−/− OBs are highly proliferative and express ALP in a differentiation-dependant manner, support this conclusion. Therefore, the increase in osteoid, and ultimately bone, is most likely due to the increased number of functional OBs in Ebf1−/− mice.
An alternative possibility to account for the increased bone is that the OCs in the Ebf1−/− mice were defective, but this seems unlikely. Although the trabecular spacing, measured by histomorphometry, was reduced by almost 50% in 12 week-old mutant bones the marrow space was not occluded. In fact, the medullary canal in 4 and 12 week-old Ebf1−/− mice was similar to wild-type controls. Also, Ebf1−/− mice had normal tooth eruption and the mice were not blind, both of which require functional OCs. Histomorphometry indicated that OCs were present in 4 week-old Ebf1−/− bones, and in vitro differentiation of OCs from Ebf1−/− bone marrow precursors was reduced compared to wild-type controls at 4 weeks of age, but corrected by 12 weeks. It is unclear why this reduction in OCs occurred. One possibility was that although there were increased numbers of OBs in bone at 4 weeks they were less able to support OC differentiation. This could be due to reduced expression of RANKL or M-CSF or increased OPG production by the OBs. However, by 12 weeks the number of OCs in vivo was almost 2.5 times that of controls, yet in vitro differentiation was equivalent. The most likely explanation for the partial recovery in OCs at 12 weeks is the induction by the increased numbers of mature OBs. Just as it takes time for the increase in bone volume to become apparent, it also takes time for OB-mediated induction of OCs. Alternatively, the reduction in OCs may be due to reduced numbers of OC progenitors. It must be remembered that Ebf1−/− bone marrow is missing all B cell lineage cells, with the exception of the very earliest B220+ population, which normally represent 20–30% of total bone marrow [36]. Therefore, the number of OC progenitors may be concentrated in Ebf1−/− bone marrow. Because the number of OCs differentiated in vitro was similar in mutant and control bone marrow from 12 week-old mice this suggests that the OC progenitors were reduced in the mutant mice. This would also explain the more than 50% reduction in OCs differentiated from 4 week-old bone marrow cells. The reduction cannot be explained by the lack of RANKL or M-CSF because these cytokines are added exogenously to the cultures. However, reduced receptor expression by the OC progenitors could account for the reduced numbers of OCs.
It is possible that the loss of B cells could account for the increase in OBs and adipocytes in the bone marrow. However, this seems unlikely because mice lacking most B cells due to the loss of Rag1 or μMT heavy chain, do not have a bone phenotype or increased marrow adipogenesis [22]. It has been reported that μMT heavy chain deficient are osteopenic due to increased bone resorption, which is caused by a decrease of B cell-secreted OPG [37]. In contrast, Ebf1−/− mice, which lack all but the very earliest population of pro-B cells, have increased bone mass with increased OCs.
Ebf2 was recently shown to be expressed in OBs, and while Ebf2−/− mice do display bone defects, the bone phenotype is very different from that of Ebf1−/− mice. The size and weight of Ebf2−/− mice was reduced compared to controls but not to the degree of Ebf1−/− mice [38]. Runting or delayed development appears common to these mutants, but whether this is a direct effect of the loss of the genes on skeletal cells or an indirect mechanism is not clear at this time. Ebf1 and 2 are expressed during brain development, which could affect skeletal development. The Ebf2-deficient mice were osteopenic with a 49% reduction in trabecular and a 65% reduction in cortical bone. Ebf2-deficient mice also had normal numbers of OBs and their bone formation rate was similar to that of controls, while the number of OCs in the bones of Ebf2−/− mice was, increased. This was accounted for by an observed increase in RANKL and decrease in OPG expression by Ebf2−/− OBs compared to controls. These data indicate that although Ebf2 has important effects on the skeleton, they are distinct from those exerted by Ebf1.
We have observed that many of the transcription factors shown to be important for B cell development and function, PU.1, Pax5 and Ebf1 [39, 40], also have roles in skeletal development [41]. PU.1 null mice fail to develop macrophages and osteoclasts [42] while Pax5−/− mice are severely osteopenic, missing more than 60% of their bone mass, due to a 3–5-fold increase in the number of osteoclasts while the number of osteoblasts indistinguishable from wild-type controls [22]. Now, we report that Ebf1 is another such regulator, and unlike PU.1 or Pax5, Ebf1 effects bone formation. We should note that the roles of these transcription factors in bone biology appear to be both direct and indirect, and in a manner similar to their regulation of B cell function, they may control bone-specific networks that can now be elucidated.
One of the most striking aspects of the Ebf1−/− phenotype is the altered distribution of adipocytes. Adipocytes were dramatically increased in the medullary canal and the secondary center of ossification and Ebf1−/− mice had fatty livers while other organs, such as spleen, were similar to control, but subcutaneous white adipose deposits were decreased in size. We detected Ebf1 mRNA in peripheral fat of wild-type or Rag-deficient mice suggesting that Ebf1-dependent regulation of adipocytes is at least partially cell intrinsic. These data confirm that Ebf1, and as reported previously Ebf2 and 3, are expressed in mouse and human adipocytes and the adipogenic cell line 3T3-L1 [17,20] where they have been implicated in adipogenesis. Forced expression of Ebf1 in the adipocyte precursor cell line 3T3L-1 and in fibroblasts induces adipogenesis [20], while reduction of Ebf1 or 2 by siRNA in 3T3-L1 cells completely abolishes the ability of those cells to differentiate into adipocytes [38]. Ebf1 activates the promoters of genes encoding C/EBPαand PPARγ1, transcription factors important for adipocyte differentiation, and PPARγ is required for Ebf1-mediated induction of adipocytes [38]. Positive regulation of PPARγ not only induces adipocyte differentiation, but also inhibits OB differentiation by increasing aP2, and adipsin synthesis and fat accumulation, while at the same time it suppresses Runx2, α1(l)procollagen, and osteocalcin synthesis [10].
Our in vivo results, showing a large increase in bone marrow adipocytes in the absence of Ebf1, appear to contradict many of the previously published in vitro findings. This dichotomy is more pronounced in light of the increased hepatic and reduced subcutaneous fat indicative of a lipodystrophic phenotype. We expected that the loss of Ebf1 would result in a bone and fat deficiency. However, our results indicate that the opposite occurred. While not the main focus of their study, the authors who investigated the role of Ebf2 in bone also noted increased adipocytes in bone marrow of Ebf2−/− mice [43]. It is reasonable to hypothesize that Ebf1 both positively and negatively regulates both the adipocyte and OB lineages through separate or similar mechanisms. This varied regulation may occur at different times during differentiation of these lineages. This would reconcile the in vivo adipocyte data on Ebf1 (our study) and Ebf2 [43] with the in vitro data discussed above. It is also possible that the use of transformed cell lines does not reveal all aspects of Ebf1 and 2 function in adipocytes, or that culture in vitro of single cell types does not correctly mimic the cell-cell interactions present within the bone marrow microenvironment. While most evidence points to Ebf1 being a positive regulator of transcription, it has been implicated in repression via p300/CBP and the repression of Glut4 expression in fat cell cultures [44, 45]. Ebf1 may activate a negative regulator of OB function or development. In fact, a novel transcription factor family of zinc finger proteins (Zfp) with 30 Krüppel-like zinc finger domains has been identified. One member, Zfp423/OAZ (O/E associated Zfp), binds to and negatively regulates Ebf1 [46], while another, Zfp521 (also known as EBF-associtaed Zfp, Ebfaz) is highly expressed in primary calvarial OBs, and transgenic mice over-expressing Zfp521 exhibit increased bone volume with increased OB number and osteoid volume [47] similar to the bone phenotype in the Ebf1−/− mice.
An additional possibility is that the loss of Ebf1 differentially regulates fat depending on its origin. White or brown fat have different anatomical locations, gene expression programs, temporal expression patterns, and lipid storage capacities [48, 49]. Within white fat, subsets of cells appear to be present, which function differently. Fat can be divided into central or visceral fat and subcutaneous and abdominal fat [50]. In humans, adipose tissue contains adipose-derived adult stem cells [51]. Although these cells can differentiate into multiple mesenchymal lineages, similarly to bone marrow derived mesenchymal stem cells it is unclear if they are regulated in the same way. It may be that the loss of Ebf1 regulates marrow adiposity differently than visceral or subcutaneous fat. Alternatively, fat accumulation in the BM may be the result of the lipodystrophic phenotype and not an effect of the loss of Ebf1 on adipocyte differentiation. Regardless of how the adipocytes are increased in the bones of Ebf1−/− animals, their presence adds another level of complexity to understanding the microenvironment of these animals. Adipocytes, in addition to storing fat, carry out other functions including the secretion of adipocytokines such as leptin, TNFαand IL-6, all of which are bone active [52].
The molecular requirements for cell fate specification to the OB and or adipocyte lineages have been only partially delineated. Therefore, the identification of new regulatory molecules is central to our understanding of the complex set of interactions that regulate the link between osteoblastogenesis and adipogenesis. The striking early increase in OBs and the unexpected concomitant accumulation of adipocytes in the bones of Ebf1−/− mice identifies Ebf1 as a heretofore-unrecognized transcription factor required for the regulation of OB and adipocyte development.
Acknowledgments
The authors are grateful to Dr. Rudolf Grosschedl, Max-Planck-Institute of Immunobiology, Freiburg, Germany for providing the Ebf1−/− mice and his review of the manuscript. The authors thank Nancy Troiano for her expert histologic and histomorphometric analysis. The authors thank Dr. Caren Gundberg for the serum osteocalcin analysis. The authors thank Dr. Randall R. Reed, Johns Hopkins University School of Medicine for providing the anti-O/E1 antibody. The authors also thank Dr. Marie Demay, Massachusetts General Hospital and Harvard Medical School, Boston, MA, for providing the chondrogenic cells, and Dr. Ernestina Schipani, Massachusetts General Hospital and Harvard Medical School, Boston, MA, for the growth plate chondrocytes. Authorship: D. H. designed and performed research, analyzed data and helped write; J. F. performed research and helped write; Y. X. performed research; T. N. performed research; S. Z. performed research; J. L. contributed analytic tools and helped write; D. S. analyzed data and helped write; M. H. designed experiments, performed research, analyzed data, helped write. This work was supported by grants from the NIH including; Training Grant AI07019 to D.G.T.H; grant AI32524 to D.G.S who is an investigator of the Howard Hughes Medical Institute; grants AR049190 and AR047342 to M.C.H.; grant AR048714 to J.A.L; the Yale Core Center for Musculoskeletal Disorders (AR046032); and the Department of Orthopaedics and Rehabilitation, Yale University School of Medicine.
Abbreviations
- Ebf
Early B cell factor
- OB
osteoblast
- OC
osteoclast
- Zfp
zinc finger protein
- PPARγ
peroxisome proliferator-activated receptor gamma
- Pax5
paired box gene 5
- ALP
alkaline phosphatase
- TbSp
trabecular spacing
- TbTh
trabecular thickness
- BV/TV
bone volume/total volume
- OV/TV
osteoid volume/total volume
- OTh
osteoid thickness
- NOb/TAR
number of osteoblasts
- NOc/TAR
number of osteoclasts
- TbN
trabecular number
Footnotes
CI Disclosures: The authors state that they have nothing to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz MC, Lorenzo JA. The origins of osteoclasts. Curr Opin Rheumatol. 2004;16:464–468. doi: 10.1097/01.bor.0000127825.05580.eb. [DOI] [PubMed] [Google Scholar]
- 3.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 4.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 5.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sata M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 6.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 8.Rosen ED, Spiegelman BM. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37741. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 9.Thompson DL, Lum KD, Nygaard SC, Kuestner RE, Kelly KA, Gimble JM, Moore E. The derivation and characterization of stromal cell lines from the bone marrow of p53−/− mice: new insights into osteoblast and adipocyte differentiation. J Bone Min Res. 1998;13:195–204. doi: 10.1359/jbmr.1998.13.2.195. [DOI] [PubMed] [Google Scholar]
- 10.Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARγ2. J Cell Biochem. 1999;74:357–371. [PubMed] [Google Scholar]
- 11.Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci. 1992;102:341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 12.Burkhardt R, Kettner G, Bohm W, Schmidmeier M, Schlag R, Frisch B, Mallmann B, Eisenmenger W, Gilg T. Changes in trabecular bone, hematopoiesis and bone marrow vessels in aplastic anemia, primary osteoporosis, and old age: a comparative histomorphometric study. Bone. 1987;8:157–164. doi: 10.1016/8756-3282(87)90015-9. [DOI] [PubMed] [Google Scholar]
- 13.Nuttall ME, Gimble JM. Controlling the balance between osteoblastogenesis and adipogenesis and the therapeutic implications. Curr Opin Pharmacol. 2004;4:290–294. doi: 10.1016/j.coph.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi J-Y, Pockwinse SM. Runx2 control of organization, assembly, and activity of the regulatory machinery for skeletal gene express. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- 15.Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. Overexpression of ΔFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med. 2000;6:985–990. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 16.Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 17.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 18.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 19.Wang SS, Leewcock JW, Feinstein P, Mombaerts P, Reed R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2003;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 20.Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akerbald P, Månsson R, Lagergren A, Westerlund S, Basta B, Lind U, Thelin A, Gisler R, Liberg D, Nelander S, Bamberg K, Sigvardsson M. Gene expression analysis suggests that EBF-1 and PPARgamma2 induce adipogenesis of NIH-3T3 cells with similar efficiency and kinetics. Physiol Genomics. 2005;23:206–216. doi: 10.1152/physiolgenomics.00015.2005. [DOI] [PubMed] [Google Scholar]
- 22.Horowitz MC, Xi Y, Pflugh DL, Hesslein DGT, Schatz DG, Lorenzo JA, Bothwell ALM. Pax5 deficient mice exhibit early onset osteoporosis with increased osteoclast progenitors. J Immunol. 2004;173:6583–6591. doi: 10.4049/jimmunol.173.11.6583. [DOI] [PubMed] [Google Scholar]
- 23.Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 24.Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar S, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, Cheng L, Donovan PJ, Peschon JJ, Bartlett PF, Willis CR, Wright BD, Carpenter MK, Davison BL, Gearing DP. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene results in placental, skeletal and neural defects that result in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 25.Parfitt A, Drezner M, Glorieux F, Kanis J, Malluche H, Meunier P, Ott S, Recker R. Bone histomorphometry: Standardization of nomenclature, symbols and units. J Bone Min Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 26.Turello R, Snyder D, Hartman H, Hartman A. A modification of the osmium tetroxide post-fixation technique for the demonstration of extracellular lipid in paraffin-embedded tissue sections. J Histotech. 1984;7:75–77. [Google Scholar]
- 27.Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54:547–557. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horowitz MC, Fields A, DeMeo D, Qian H-Y, Bothwell A, Trepman E. Expression and regulation of Ly-6 differentiation antigens by murine osteoblasts. Endocrinology. 1994;135:1032–1043. doi: 10.1210/endo.135.3.7520861. [DOI] [PubMed] [Google Scholar]
- 29.Sudo H, Kodama H-A, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peister A, Melland JA, Larson BL, Hall BM, Gibson LF, Prockop DJ. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004;103:1662–1668. doi: 10.1182/blood-2003-09-3070. [DOI] [PubMed] [Google Scholar]
- 31.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 32.Kacena MA, Shivadasani RA, Wilson K, Xi Y, Troiano N, Nazarian A, Gundberg CM, Bouxsein ML, Korenzo JA, Horowitz MC. Megakaryocyte-osteoblast interaction revealed in mice deficient in transcription factors GATA-1 and NF-E2. J Bone Min Res. 2004;19:652–660. doi: 10.1359/JBMR.0301254. [DOI] [PubMed] [Google Scholar]
- 33.Gundberg CM, Clough MC, Carpenter TO. Development and validation of a radioimmunoassay for mouse osteocalcin: paradoxical response in the Hyp mouse. Endocrinology. 1992;130:1909–1915. doi: 10.1210/endo.130.4.1547718. [DOI] [PubMed] [Google Scholar]
- 34.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 35.Lagergren A, Mansson R, Zeterblad J, Smith E, Basta B, Bryder D, Akerblad P, Sigvardsson M. The CXCL12, periostin and CCL9 genes are direct targets for early B-cell factor (EBF) in OP-9 stroma cells. J Biol Chem. 2007;282:14454–144462. doi: 10.1074/jbc.M610263200. [DOI] [PubMed] [Google Scholar]
- 36.Lee SK, Kalinowski JF, Jacquin C, Adams DJ, Gronowicz G, Lorenzo JA. Interleukin-7 influences osteoclast function in vivo but is not a critical factor in ovariectomy-induced bone loss. J Bone Miner Res. 2006;21:695–702. doi: 10.1359/jbmr.060117. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Toraldo G, Li A, Yang X, Zhang H, Qian W-P, Weitzmann MN. B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo. Blood. 2007;109:3839–3848. doi: 10.1182/blood-2006-07-037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. A critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh H, Pongubala JM, Medina KL. Gene regulatory networks that orchestrate the development of B lymphocyte precursors. Adv Exp Med Biol. 2007;596:57–62. doi: 10.1007/0-387-46530-8_5. [DOI] [PubMed] [Google Scholar]
- 40.Roessler S, Grosschedl R. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin Immunol. 2006;18:12–19. doi: 10.1016/j.smim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Horowitz MC, Bothwell ALM, Hesslein DGT, Pflugh DL, Schatz DG. B cells and osteoblast and osteoclast development. Immuno Rev. 2005;208:141–153. doi: 10.1111/j.0105-2896.2005.00328.x. [DOI] [PubMed] [Google Scholar]
- 42.Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature. 1997;386:81–84. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- 43.Kieslinger M, Folberth S, Dobreva G, Dorn T, Croci L, Erben R, Consalez GG, Grosschedl R. Ebf2 regulates osteoblast-dependent differentiation of osteoclast. Dev Cell. 2005;9:757–767. doi: 10.1016/j.devcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Zhao F, McCarrick-Walmsley R, Akerblad P, Sigvardsson M, Kadesh T. Inhibition of p300/CBP by early B-cell factor. Mol Cell Biol. 2003;23:3837–3846. doi: 10.1128/MCB.23.11.3837-3846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowell P, Cooke DW. Olf-1/early B cell factor is a regulator of gult4 gene expression in 3T3-L1 adipocytes. J Biol Chem. 2002;277:1712–1718. doi: 10.1074/jbc.M108589200. [DOI] [PubMed] [Google Scholar]
- 46.Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M, Morvan F, Zhang J, Neff L, Philbrick W, Horne WC, Baron R. Overexpression of ZFP521 (MC-33), a 30 zinc finger ΔFosB-interacting protein, induces an increase in bone volume and bone formation rates in transgenic mice. J Bone Min Res. 2006;20:S47 (abs). [Google Scholar]
- 48.Ailhaud G, Grimaldi P, Negrel R. Cellular and Molecular aspects of adipose tissue development. Annu Rev Nutr. 1992;12:207–233. doi: 10.1146/annurev.nu.12.070192.001231. [DOI] [PubMed] [Google Scholar]
- 49.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 50.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gimble J, Guilak F. Adipose-derived adult stem cell: Isolation, characterization, and differemtiation potential. Cytotherapy. 2003;5:362–369. doi: 10.1080/14653240310003026. [DOI] [PubMed] [Google Scholar]
- 52.Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6:772–783. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]