Abstract
Human nicotinamide phosphoribosyltransferase (NAMPT, EC 2.4.2.12) catalyses the reversible synthesis of nicotinamide mononucleotide (NMN) and inorganic pyrophosphate (PPi) from nicotinamide (NAM) and α-D-5-phosphoribosyl-1-pyrophosphate (PRPP). NAMPT, by capturing the energy provided by its facultative ATPase activity, allows the production of NMN at products/substrates ratios thermodynamically forbidden in the absence of ATP. By coupling ATP hydrolysis to NMN synthesis, the catalytic efficiency of the system is improved 1100-fold and substrate affinity dramatically increased (Km NAM from 855 nM to 5 nM) and the Keq shifted −2.1 kcal/mol toward NMN formation. ADP/ATP isotopic exchange experiments support the formation of a high-energy phosphorylated intermediate (phospho-H247) as the mechanism for altered catalysis efficiency during ATP hydrolysis. NAMPT captures only a small portion of the energy generated by ATP hydrolysis to shift the dynamic chemical equilibrium. Although the weak energetic coupling of ATP hydrolysis appears to be a non optimized enzymatic function, closer analysis of this remarkable protein reveals an enzyme designed to capture NAM with high efficiency at the expense of ATP hydrolysis. NMN is a rate-limiting precursor for recycling to the essential regulatory cofactor, nicotinamide adenine dinucleotide (NAD+). NMN synthesis by NAMPT is powerfully inhibited by both NAD+ (Ki = 0.14 µM) and NADH (Ki = 0.22 µM), an apparent regulatory feedback mechanism.
The properties of NAD+ in electron transfer are now established. As a cofactor in many metabolic pathways, NAD+ holds a key position in energy metabolism and its regulation. Over the last decade, new involvements for NAD+ were discovered. It is used by poly and mono(ADP-ribose) polymerases as a substrate for protein covalent modifications (1–5).
Sirtuins (SIRT) use NAD+ in protein deacetylation reactions with implications for metabolic control, cancer and longevity. While redox reactions do not influence the net level of NAD+ + NADH concentration, enzymes that catalyze ADP-ribosylation and deacetylation cleave the N-ribosyl bond to nicotinamide and can lead to a rapid depletion of this substrate. NAD+ levels influence all of its associated pathways and the NAD+ pool needs constant replenishment from NAM recycling since dietary sources of the vitamin may be limiting.
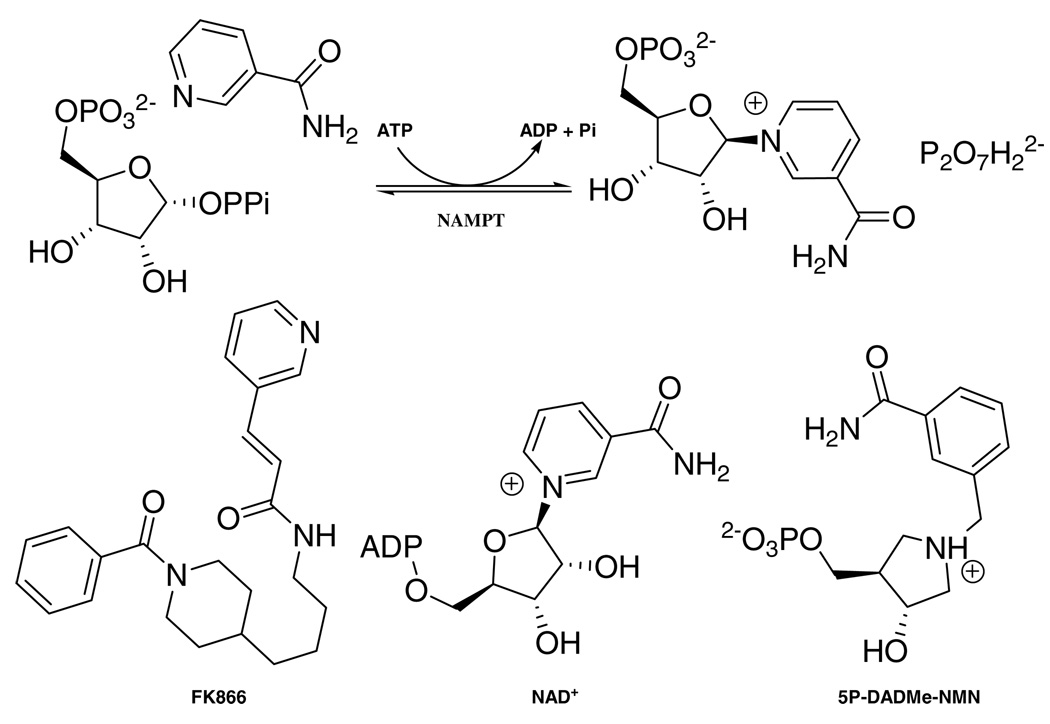
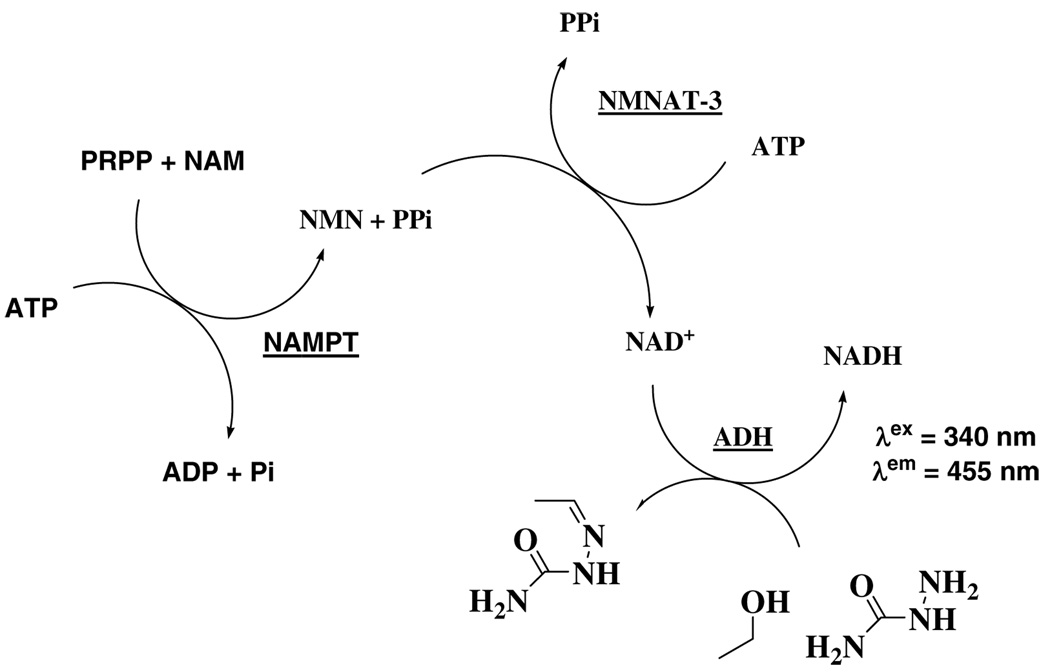
In prokaryotes and lower eukaryotes, NAD+ replenishment is achieved primarily by de novo synthesis. Quinolinic acid, derived from tryptophan, is converted into nicotinic acid mononucleotide (NAMN) by the quinolinic acid phosphoribosyltransferase (QAPT). NAMN, obtained from exogenous nicotinic acid (NA) by reaction with nicotinic acid phosphoribosyltransferase (NAPT), is converted into NAD+, via the Preiss-Handler pathway (6). The salvage pathway for those organisms (4, 7) involves the deamination of NAM into NA via the corresponding nicotinamidase (Pnc1). Mammals lack Pnc1, and directly convert NAM into nicotinamide mononucleotide (NMN) by nicotinamide phosphoribosyltransferase (NAMPT). NMN is converted to NAD+ by nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NMNAT) (scheme 1).
Scheme 1.
Reaction catalyzed by human NAMPT and inhibitors.
NAD+ is not localized uniformly in the cell, with up to 70% of the pool held in the mitochondria (8). Three isoforms of NMNAT are known, and each has a specific subcellular localization (9–13). NMNAT-3 is located mainly in the mitochondria and mitochondrial NAD+ is proposed to influence cell lifespan, together with the overexpresssion of SIRT enzymes (14, 15). In yeast, overexpression of NAPT, NMNAT and Pnc1 increased the activity of NAD+-dependant histone deacetylase (Sir2) (16–18). In mammals, NAMPT is the rate-limiting enzyme for NAD+ salvage from NAM and its overexpression lengthened cell lifespan (19). This phenomenon is apparently associated with increased catalytic activity of the mammal ortholog of Sir2, SIRT1 (20). Recently, NAMPT was identified as the enzyme regulating mitochondrial NAD+ levels (21) and increasing cell lifespan via sirtuins, SIR3 and SIR4, both located in this organelle.
The crucial role of NAMPT in NAD+ biosynthesis makes it an attractive target in regulation of mammalian metabolic and regulatory pathways. Modulation of NAMPT activity through inhibition or activation, could lead to a shorter or extended lifespan, respectively. Inhibition of NAD+ salvage by specific inhibition of NAMPT has been described (22). The small molecule inhibitor, FK866, decreased the NAD+ pool via inhibition of this enzyme (Ki = 0.4 nM). Reducing the activity of NAMPT by FK866 in human vascular smooth muscular cells caused low levels of NAD+, and caused a decrease of SIRT1 activity (20). Under these conditions, SIRT1 failed to deacetylate the tumor suppressor p53 and this transcription factor was kept at levels above those which induce cell senescence.
FK866 is in clinical trials but it exhibits low bioavailability, rapid intravenous clearance and substantial drug binding to plasma proteins (23). Continuous intravenous infusions over 96 h for a period of 4 weeks showed no objective responses. Finally, inhibition by FK866 was reversible upon NAM injection. Isosteric analogues of the inhibitor have been synthesized without improved inhibition properties against NAMPT (24).
Structural analysis of mammalian NAMPTs have provided a guide to substrate-protein and inhibitor-protein interactions (25–27). Catalytic site contacts together with mechanisms from related phosphoribosyltransferases have been used to propose a molecular mechanism for NAMPT. However, human NAMPT displays weak sequence identity with other members of this family, although Thermoplasma acidophilum dimeric NAPT has been proposed as a structural homologue (28). The kinetic mechanism of Salmonella typhimurium NAPT is the most complete and is useful in analysis of NAMPT (29). This NAPT also couples ATP hydrolysis and NAMN synthesis, to shift the dynamic equilibrium toward NAMN. Its ATPase activity involves a phosphohistidine intermediate (30–32) and most rate constants fot the mechanism have been established (33, 34). NAMPT is also similar to NAPT as its catalytic efficiency is improved by ATP (35).
Unlike NAPT, the catalytic features and reaction mechanism of human NAMPT are poorly described. Crystallographic structures are useful to propose catalytic residues but offer few insights into the energetic and kinetic mechanisms. Likewise, the covalent phospho-NAMPT structure had not been revealed in the reported structures and no thermodynamic properties have been reported.
Here we describe the role of ATP in NMN synthesis catalyzed by NAMPT. The thermodynamic and kinetic properties of NAMPT demonstrate weak coupling of ATP hydrolysis to the dynamic chemical equilibrium and to the kinetic properties essential for nicotinamide salvage. The existence of a covalently phosphorylated enzyme involved in the mechanism is strongly supported by isotope exchange experiments and formation of a readily hydrolysable intermediate in the presence of ATP. The kinetic mechanism defined here provides unique insights into cellular NAD+ recycling and defines essential information required for the kinetic and thermodynamic analysis of this critical enzyme.
EXPERIMENTAL PROCEDURES
Materials
[CONH2− 14C]NAM (55 mCi mmol−1) was from American Radiolabeled Chemicals. [4-3H]NMN (1.8 Ci mmol−1) was from Moravek. [2,8-3H]ATP and ADP (respectively 27.8 and 40 Ci mmol−1) and [14C]NAD+ (253 mCi mmol−1) were from Perkin Elmer. Liquid scintillation cocktail (UltimaGold) was from Perkin Elmer. Pyruvate kinase (PK), lactate dehydrogenase (LDH), alcohol dehydrogenase (ADH) and inorganic pyrophosphatase (PPase) were from Sigma. NMNAT-3 was overexpressed as previously described (13) from the corresponding plasmid (pPROEX, generous gift from Dr. Hong Zang, Department of Biochemistry, University of Texas). Ni-NTA resin, tris(hydroxypropyl)phosphine (THP) were from Novagen, HiLoad Superdex 200GP 26/60 was from Amersham. HPLC solvents were from Fisher, other biochemicals were from Sigma.
Overexpression and Purification of human NAMPT
The enzyme was overexpressed in Escherichia coli BL21(DE3)pLys containing the expression plasmid pBAD DEST 49 with inserts of chemically synthesized DNA (DNA 2.0) encoding for human NAMPT and optimized for expression in E. coli. Cells were grown in LB broth containing 100 µg mL−1 ampicilin and 30 µg mL−1 chloramphenicol at 37 °C to an A600 of 0.4. The cultures were supplemented to 0.2% L-arabinose for induction and incubated for an additional 16 h at 37 °C preceding harvest. Resuspended cells (20 mM Tris buffer, 500 mM NaCl, 5 mM imidazole, pH 7.9, 1 mM βME, protease inhibitors cocktail) were disrupted by French Press. Debris was removed by centrifugation at 20,000 rpm for 20 min. The supernatant (150 mL from 50 g of cells) was loaded onto a Ni-NTA column (50 mL of resin pre-equilibrated with the disruption buffer). The column was washed consecutively with buffers having 10, 30, 50, 100 and 200 mM imidazole concentrations. The enzyme, eluted at 100 and 200 mM imidazole concentrations, was concentrated, desalted and further purified on a HiLoad Superdex 200GP 26/60 eluted with 100 mM HEPES pH 7.5, 100 mM NaCl, 10 mM βME at a 1 mL min−1 flow rate. The purified enzyme (10 mg mL−1) was then concentrated (50–75 mg mL−1) and frozen.
Products Purification
For the isolation of radioactive [14C]NMN formed from [14C]NAM during kinetic experiments, [3H]NAM formed from [3H]NMN during equilibrium conditions and [3H]ATP formed from [3H]ADP during isotope exchange reactions, quenched samples were injected (250 µL) onto a Luna C18 2 column (4.6 ×150 mm, 5 µm, 100 Å, Phenomenex) equilibrated in 100 mM K2HPO4, 8 mM tetrabutylammonium sulfate, pH 6.0 (Buffer A) and initially eluted with 4 mL of this buffer. Further elution was performed with a 16 mL linear gradient to 30% acetonitrile in the same buffer (Buffer B). A flow rate of 1 mL min−1 was employed. NMN elutes 2.5 mL after injection, NAM at 6.2 mL, ADP at 18.7 mL and ATP at 21.6 mL (Method 1). This purification protocol does not degrade the resolution of the column if frequent wash cycles of acetonitrile/tetrahydrofuran/acetonitrile are performed. In order to directly characterize the ATPase activity of NAMPT, quenched samples were injected (100 µL) onto a Nucleosil 4000-7 PEI strong anion exchange column (4.0 × 250 mm, 5 µm, 100 Å , Macherey-Nagel) equilibrated in 20 mM Tris pH 8.2 (Buffer C) and initially eluted with 5 mL of this buffer. Further elution was performed with a 30 mL linear gradient to 1 M KCl in the same buffer (Buffer D). A flow rate of 1 mL min−1 was employed. ADP elutes 25 mL after injection and ATP at 33 mL (Method 2).
Radioactive Samples Preparation and Reading
Each radioactive sample (HPLC) was diluted with 50 mg of cold NAM prior to freeze-drying. The resulting solid was redisolved in a mixture 1:10 (water: scintillation cocktail) prior to radioactivity determination (Wallac instrument, 20 min reading, 3 cycles).
Analysis of Data
All results from kinetic and inhibition studies were fit to the appropriate equations using KaleidaGraph (Synergy Software). The enzyme is described by its turnover rate and Michaelis constants, specific for each substrate involved (kcat and Km). The inverse of the initial velocity (Vapp A) at the corresponding inverse concentration of substrate A ([A]) follow a linear relation (Lineweaver-Burk representation) at a total enzyme concentration [E]T:
| (Equation I) |
For competitive inhibition studies, the inhibition constant Ki is described by the expression
| (Equation II) |
where VI is the initial rate at the corresponding concentration of inhibitor [I]. For low concentrations of inhibitor (<10 times [E]T) the free inhibitor concentration in solution [I]* and the inhibitor concentration added [I] are different and related by
| (Equation III) |
Where V is the initial rate of the reaction without inhibitor. Km and Ki are also related by the following relationship
| (Equation IV) |
Each experiment was carried out in duplicate.
Enzyme Activity/Stability Determination
To determinate the pH dependence of NAMPT activity, the enzyme (30 nM) was preincubated at 25 °C with 1 µM radiolabeled NAM (100 mM universal buffer with a constant ionic strength of 60 mS at various pH values, 1 mM ATP, 5 mM MgCl2, 1 mM THP) and the reaction was started by addition of α-D-5-phosphoribosyl-1-pyrophosphate (PRPP) (200 µM). Quench was performed after 1 h and samples were processed according to Method 1 to monitor NMN formation. In a similar way, to determinate its stability, 10 different mixtures of enzyme (30 nM, 1 mL at various pH) were preincubated 1 h, then concentrated and transferred to the previously described reaction mixture (at pH 7.6). The assay conditions were the same. The ATPase activity was evaluated by monitoring the steady-state formation of ADP. The reaction (0.75 µM enzyme, 100 mM universal buffer, 5 mM MgCl2, 1 mM THP) was initiated by addition of ATP (2 mM). Quenched samples were analyzed by HPLC (Method 2).
NAMPT Auto-Phosphorylation
Although the presumed phosphorylated NAMPT was unstable to isolation, at the conditions where the assays were performed (NAMPT 46 µM, 50 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, variable ATP from 0 to 25 mM, 1 mM THP, 25 °C), its extent of phopshorylation could be monitored indirectly. Every 30 s, aliquots were quenched (EDTA 15 mM) and analyzed by HPLC (Method 1) to follow the formation of ADP. Extrapolation of the linear ATP hydrolysis profile to the original time of the experiment established the initial burst of ADP, equivalent to the phosphorylation stoichiometry for NAMPT.
Equilibrium Displacement by ATP
Equilibrium constants for the NAMPT-catalyzed reaction in the presence and absence of ATP were established by adding enzyme (0.6 µM) to a 6 mL reaction mixture (50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 100 µM PRPP, 100 µM PPi, 5 µM [4-3H]NMN) at 25 °C. At appropriate time intervals, aliquots were quenched and analyzed by HPLC (Method 1) in order to determinate the profiles of NMN and NAM. When equilibrium was established (3 h), a small volume of ATP (leading to a final concentration of 2 mM) was added and additional aliquots were processed by HPLC after quench. Variations in the equilibrium constant were evaluated by running two simultaneous reactions with similar conditions (0.6 µM enzyme, 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 100 µM PRPP, 100 µM PPi, 5 µM [14C]NAM), without and with ATP (2 mM).
NAMPT Kinetic Parameters without ATP
The kcat and Km of each substrate for the non-ATP coupled NMN synthesis were determined first in the absence, then in the presence of near-saturating phosphate concentration. To determinate the Km for PRPP, reactions were incubated at 25 °C and contained 50 to 1000 µM PRPP, 1 µM [14C]NAM, 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP. Conversion to [14C]NMN was started by enzyme addition (88 nM) and monitored by quenching three samples at 5 min intervals. Separation was achieved by HPLC (Method 1). The Km for NAM was evaluated in the same way (0.5 to 5 µM [14C]NAM, 100 µM PRPP, 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 88 nM enzyme). Phosphate influences the initial rate and was evaluated by varying [Pi] from 0.25 to 5 mM using a concentrated sodium phosphate solution prebuffered at pH 7.5. A near-saturating phosphate concentration was then used to determine Michaelis constants for PRPP and NAM in the presence of phosphate. The assays were the same except that each reaction mixture was supplemented with 2 mM phosphate.
NAMPT Kinetic Parameters with ATP
The kcat and Km of NAM for the ATP-coupled NMN synthesis were initially determined by a coupled fluorometric assay (19) slightly modified. The reactions were run at 25 °C, 2.8 nM of enzyme and varying concentrations of NAM (0.1 µM to 5 µM) reacted in a 3 mL sample containing 100 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM THP, 2.5 mM ATP, 200 µM PRPP, 100 mM ethanol, 40 mM semicarbazide (to drive the ADH reaction to the formation of acetaldehyde to be trapped as its hydrazide intermediate), 80 mU NMNAT-3 and 800 mU ADH. NADH formation was monitored over a period of 30 min by emission of fluorescence at 455 nm upon excitation at 340 nm (SlitEx = 2.5, SlitEm = 12.5). A second assay, the radio labeled assay described previously, was carried out in 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 2 mM ATP, 50 µM PRPP, 0.05 to 5 µM [14C]NAM, 1.1 nM enzyme. The corresponding Km of NAM was obtained by extrapolation to correct for the inhibition by NAD+. At a fixed 5 µM concentration of NAD+, the ATP-coupled NMN synthesis was partially inhibited (50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 2 mM ATP, 50 µM PRPP, 0.05 to 5 µM [14C]NAM, 1.1 nM enzyme). The corresponding Lineweaver-Burk plot lead to an apparent NAM Michaelis constant (Km app). By varying the NAD+ concentration from 0 to 300 µM (50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM THP, 2 mM ATP, 50 µM PRPP, 0.1 µM [14C]NAM, 1.1 nM enzyme), and measuring the initial rate of NMN formation at a single NAM concentration, several pairs of Km app/[NAD+] were obtained. Use of Equation IV provides the actual Km for NAM. The Km values for PRPP and ATP were established for the ATP-coupled reaction with, respectively, fixed substrate concentrations of 125 nM NAM, 2 mM ATP, 0.5 to 500 µM PRPP, and 100 nM NAM, 50 µM PRPP, 0.4 to 5 mM MgATP, all at 2.2 nM of enzyme.
NAMPT Inhibition
FK866, previously described as a non-competitive inhibitor of human NAMPT (22), 5P-DADMe-NMN (Scheme 1), NADH and NAD+, were evaluated as competitive inhibitors of both non-ATP and ATP-coupled NMN synthesis, versus PRPP and NAM substrates. For the reaction involving ATP, 1 mL reactions were incubated at 25 °C and contained 200 µM PRPP, 100 nM [14C]NAM, 2.5 mM ATP, appropriate inhibitor concentrations (0 to 80% inhibition), 5 mM MgCl2, and 2.2 nM enzyme in 50 mM HEPES pH 7.5 supplement with 50 mM NaCl and 1 mM THP. Three samples were quenched at 15 min intervals. Corresponding initial rates, determined by HPLC (Method 1), were fit as a function of the inhibitor concentration (Equation II and Equation III). In a similar way, but with omission of ATP, Ki for FK866 and 5P-DADMe-NMN were obtained (100 µM PRPP, 1 µM [14C]NAM, 5 mM MgCl2, 50 mM HEPES pH 7.5, 50 mM NaCl, 1 mM THP, and 88 nM enzyme).
Stimulated ATPase Activity of NAMPT
The ATPase activity was evaluated under different conditions by incubating 1.5 µM of enzyme and 1 mM of various compounds (NAM, PRPP, NMN, PPi, NAD+) in presence of 2 mM ATP (50 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM THP). Quenched samples, over 150 min, were analyzed by HPLC (Method 2) to establish the steady-state rate of ADP formation.
ADP/ATP Isotope Exchange
ADP/ATP isotope exchange reactions were carried out in 50 mM HEPES pH 7.5 with 50 mM NaCl, 5 mM free MgCl2 and 1 mM THP at 25 °C. ADP (25 to 200 µM, containing 105 cpm of [2,8-3H]ADP per 100 µM cold ADP) and ATP (0.2 to 1 mM) were present as equimolar mixtures with MgCl2. The reactions were started by addition of 0.6 µM enzyme and four samples were quenched at 15 min intervals. Nucleotides were separated by HPLC (Method 1) in order to determinate the rate of the exchange reaction. Rates of the exchange reaction were calculated as above (Equation I).
Coupling of NMN Synthesis and ATP Hydrolysis
The stoichiometry of ATP hydrolyzed per NMN formed was assayed at various ATP concentrations. The reactions (3 mL) containing 0.8 to 3 mM MgATP, 23 µM phosphoenolpyruvate (PEP), 23 µM NADH (with NAM = 400 × Km, NAD+ and NADH did not significantly inhibit the reaction), 5 mM MgCl2, 100 µM PRPP, 2 µM [14C]NAM, 50 mM HEPES pH 7.5, 50 mM NaCl, 30 mM KCl, 1 mM THP, 1 U PK and 1 U LDH were incubated at 25 °C for 15 min (to permit trace ADP to be converted to ATP) and started by the addition of 160 nM of enzyme. Decrease in the fluorescence emission from NADH permits determination of ATP hydrolysis rates with a second order polynomial calibration curve for fluorescence intensity from 0 to 25 µM NADH, using parameters: SlitEx = 2.5 and SlitEm = 2.5. The same experiments were used to measure the NMN formation rates. Seven aliquots were quenched at appropriate intervals and [14C]NMN was determined by HPLC (Method 1). The effect of PPi (0 to 75 µM) on the ratio of ADP/NMN formation was established with a similar protocol using a fixed 2.5 mM ATP concentration, where the ADP/NMN ratio is optimal at 1.1 to 1.0.
RESULTS
NAMPT Stability
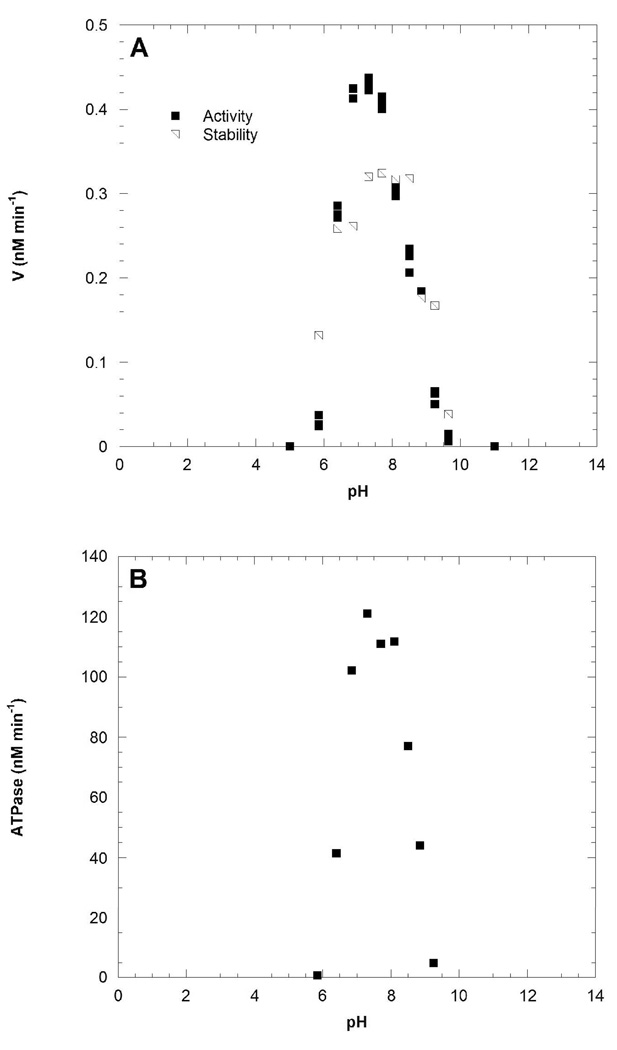
NAMPT was found to be catalytically stable from pH 6.5 to 8.5 for the experimental time ranges. NMN synthesis was catalyzed with or without ATP. Optimal activity is restricted to a narrow pH range for both NMN synthesis and ATPase activities. The apparent pH optimus is 7.5 and this pH was used for the remainder of the experiments (Figure 1A).
Figure 1.
NAMPT activity as a function of pH for the ATP-coupled NMN synthesis reaction (A). Concentrations fixed at 1 µM [14C]NAM, 200 µM PRPP, 1 mM ATP and 5 mM MgCl2 with 30 nM enzyme. NAMPT ATPase activity as a function of pH (B). Concentrations were fixed at 2 mM ATP and 5 mM MgCl2 with 0.75 µM enzyme.
Auto-Phosphorylation of NAMPT
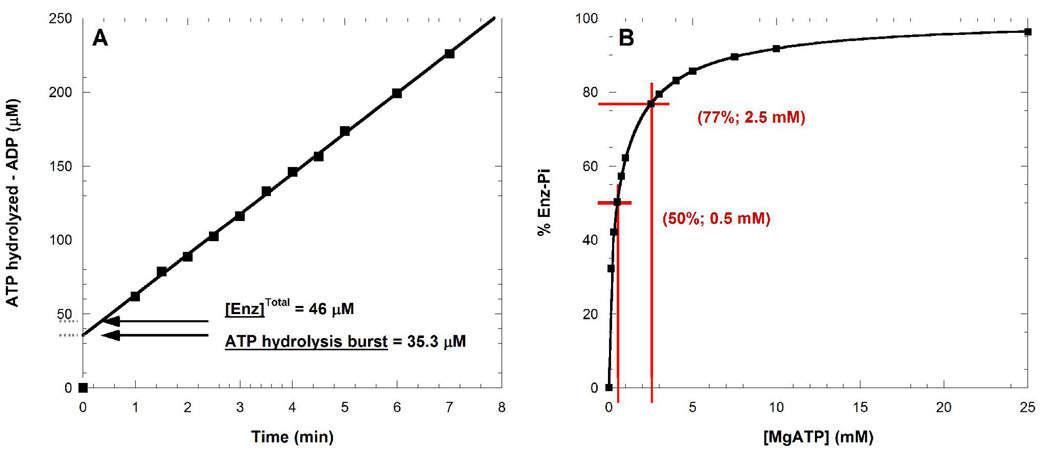
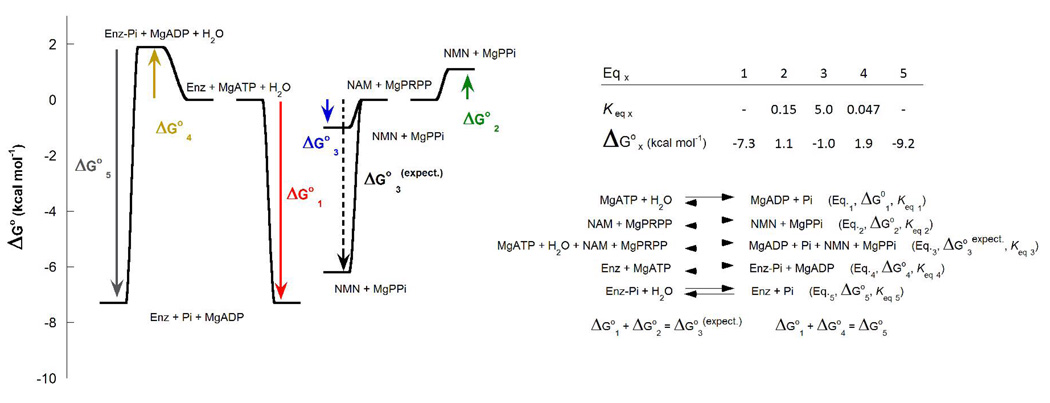
The slow steady-state ATP hydrolysis catalyzed by NAMPT was preceded by an initial burst of ADP (Figure 2A). The burst of ADP is saturable and represents the auto-phosphorylation of the enzyme. At 2.5 mM MgATP concentration, 77% of NAMPT can be found phosphorylated (Figure 2B). With an estimated value of 0.047 for its Keq (ΔG° = 1.9 kcal mol−1), the NAMPT phopshorylation is an unfavorable reaction that leads to a high-energy species that hydrolyzes at 0.8 min−1 under these experimental conditions (Figure 3; Eq.4 and Eq.5).
Figure 2.
Auto-phopshorylation of NAMPT by ATP. Assay performed by HPLC measuring ADP formation. In (A) NAMPT was present at 46 µM with 2.5 mM MgATP. The initial burst of ADP characterized the extent of enzyme phosphorylation. Estimated fraction of NAMPT phosphorylated at various MgATP concentrations (B) from experiment shown in (A).
Figure 3.
Thermodynamic model for NAMPT. Determination of Keq and ΔG° for the NAMPT auto-phosphorylation, the non-ATP and ATP-coupled NMN synthesis reactions at pH 7.5 (HEPES 50 mM) in presence of 100 mM KCl, and 1 mM THP at 25 °C.
The Effect of ATP on the Dynamic Chemical Equilibrium of NAMPT
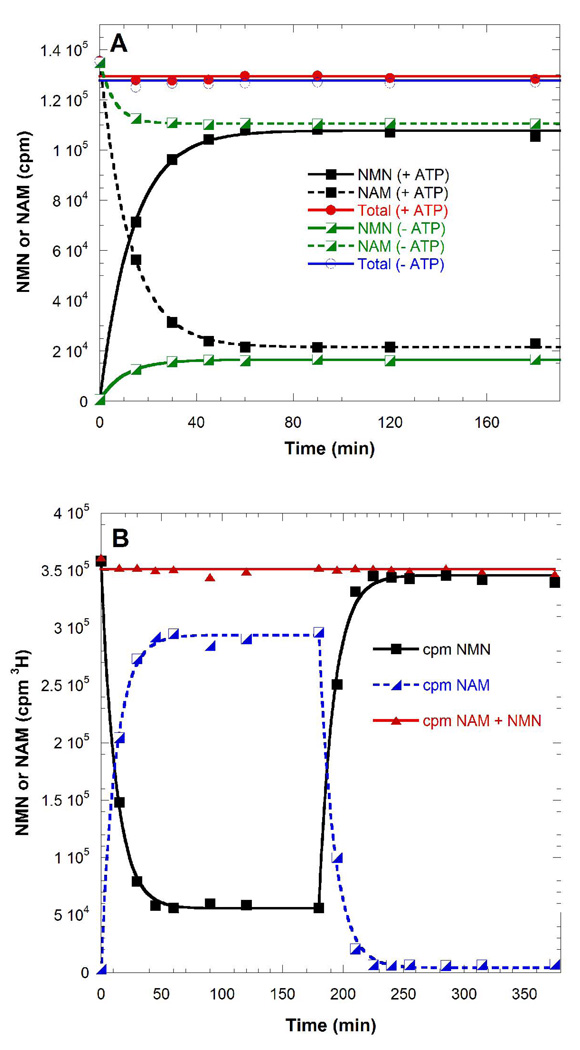
The NAM transferase activity of NAMPT gave different equilibrium positions in the presence of ATP, consistent with coupling to the transferase reaction (Figure 4A). Adding ATP to a preestablished equilibrium of NAMPT in a non ATP-coupled reaction, the position of the chemical equilibrium was reversed (Figure 4B). A value of 35 was obtained for the ratio KATP/K ([NMN]ATP[NAM]/[NMN][NAM]ATP), which is the ratio of equilibrium constants for the NAMPT reaction without including ATP, ADP and Pi concentrations. This effect represents an energy difference of −2.1 kcal mol−1, a small energy compared to the −7.3 kcal mol−1 available from ATP hydrolysis at physiological conditions, in the presence of Mg2+ (Figure 3; Eq.1, Eq.2 and Eq.3). Thus, hydrolytic coupling of ATP energy into the NAMPT reaction is weakly coupled. Nevertheless, the energy generated by ATP hydrolysis serves to thermodynamically switch the unfavorable products/substrates ratio toward production of NMN.
Figure 4.
Reaction equilibrium of non-ATP and ATP-coupled NMN synthesis (A). NAM was originally present at 5 µM, PRPP and PPi at 100 µM and ATP at 2 mM, therefore there is no significant depletion of ATP during formation of NMN and PPi. Reversible chemical equilibrium of NAMPT as a function of ATP hydrolysis (B). NMN was originally present at 5 µM, PRPP and PPi at 100 mM with no ATP (0 to 180 min). At 180 min the solution was made to 2 mM with ATP which altered the equilibrium as shown.
ATP Effects on Kinetic Parameters
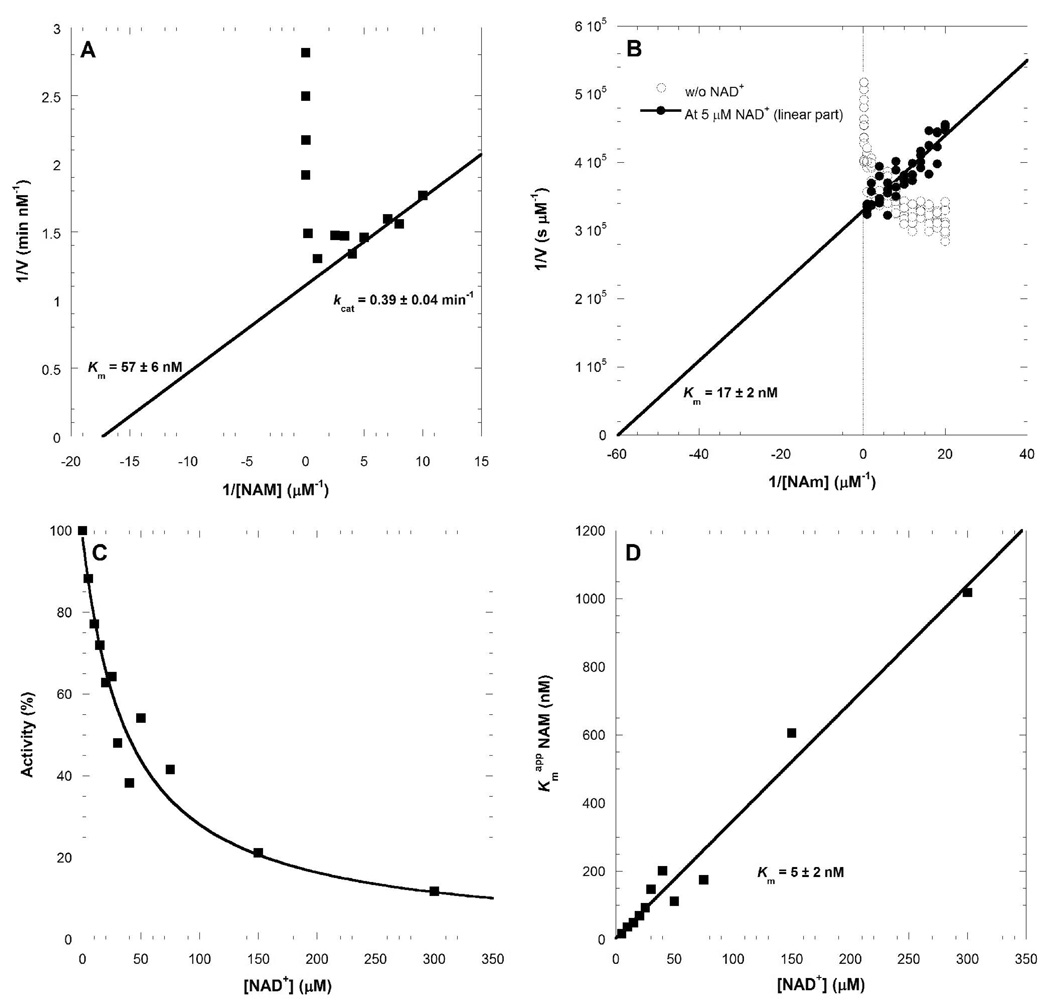
To evaluate the effects of ATP, the kinetic parameters for NAM and PRPP were determined without ATP, in the presence of Pi, and with ATP. The uncoupled reaction was studied with a radio-labeled assay, and two independent methods were used to determine the kinetic parameters on the ATP-coupled reaction. The continuous assay (Scheme 2) revealed a lower Km value for NAM (Figure 5A) in the presence of ATP (Km of 57 ± 6 nM with ATP compared to 855 ± 22 nM without ATP). The direct assay with [14C]NAM revealed an even tighter affinity of 17 ± 2 nM for nicotinamide (Figure 5B). The inverse of the initial rate (1/V) as a function of reciprocal NAM concentration indicated substrate-inhibition by nicotinamide concentrations as low as 250 nM. The addition of NAD+ (5 µM) altered the kinetic properties to give linear 1/V versus 1/[NAM] plots. Assays with variable NAD+ permitted the determination of an apparent Km (17 ± 2 nM). Even in the presence of NAD+, nicotinamide causes substrate inhibition but with an inhibition constant near 3 µM.
Scheme 2.
Continuous fluorometric assay for the Km determination of NAM (ATP-coupled reaction)
Figure 5.
Determination of kinetic parameters for NAMPT. Lineweaver-Burk representation for the Km determination of NAM using the fluorometric coupled assay and fitting only the linear portion of the curve (A). Lineweaver-Burk representation for the Km determination of NAM using the direct radiolabeled assay, without and with the presence of NAD+ (B). Profile of the NAD+ inhibition of the ATP-coupled NMN synthesis reaction (C). Km determination of NAM for the ATP-coupled NMN synthesis. The value of the apparent Km app is plotted as function of concentration with the value for the inhibition-free Km value for NAM taken from the ordinate intercept to give 5 ± 2 nM (D).
Using NAD+ as a competitive inhibitor of NMN synthesis (Figure 5C, D), several sets of Km app:[NAD+] pairs were obtained and reploted, leading to a linear relationship. This representation (Equation IV), extrapolated to its ordinate intercept, allowed the determination of the actual Km for NAM (5 ± 2 nM). This unusually small Km value has significance for the cellular function of NAMPT in salvage of NAM for NAD+ synthesis.
While NAMPT exhibits high affinity for NAM, especially in the presence of ATP, the Km for PRPP was less affected by ATP (Table 1). The Km value for MgPRPP was 7 to 12 µM in the absence of ATP but showed increased affinity (Km = 0.63 µM) in the presence of ATP.
Table 1.
Kinetic parameters of human NAMPT for both coupled and non ATP-coupled reactions.
| Non ATP-coupled NMN synthesis |
ATP-coupled NMN synthesis |
||
|---|---|---|---|
| No Pi | Pi | ||
| Km | |||
| NAM (nM) | 855 ± 22 | 235 ± 17 | 5 ± 2 |
| PRPP (µM) | 7.2 ± 0.6 | 11.7 ± 0.8 | 0.63 ± 0.03 |
| ATP (Pi)*(mM) | 0.18 ± 0.02* | 7.4 ± 1.5 | |
| kcat (min−1) | 0.082 ± 0.001 | 0.085 ± 0.001 | 0.46 ± 0.09 |
| kcat/Km (M−1 s−1)† | 1.60 ± 0.06 × 103 | 6.1 ± 0.5 × 103 | 1.8 ± 0.9 × 106 |
Km of Pi for the non ATP-coupled NMN synthesis.
kcat/Km regarding to NAM.
The weak affinity between ATP and the protein is consistent with physiological conditions with cellular levels of ATP in mM range. High ATP concentrations were required in order to saturate NAMPT (Km = 7.4 ± 1.5 mM) and are complicated by inhibition at concentrations above 4 mM.
Despite the low catalytic turnover rate of NAMPT, the catalytical efficiency (kcat/Km) of 1.8 × 106 M−1 s−1 is high because of the nanomolar Km for NAM (Table 1). The ATP-coupled reaction is efficient for NAM salvage since the enzyme will catalyze formation of NMN for most collisions of a nicotinamide molecule at the active site.
Inhibition Studies
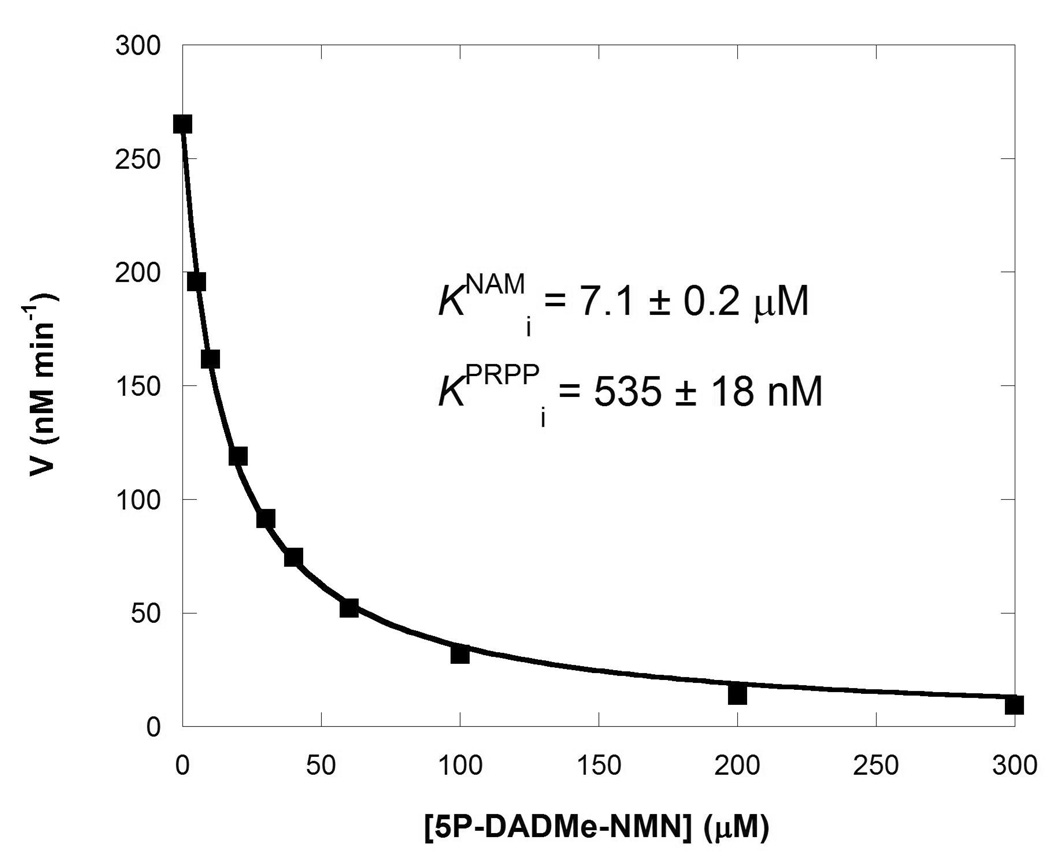
Four inhibitors (FK866, 5P-DADMe-NMN, NAD+ and NADH; scheme 1) were assayed against the ATP-coupled and non ATP-coupled reactions. In the presence of ATP, NAD+ and NADH were competitive inhibitors versus PRPP. The relatively high affinities of 0.14 and 0.22 µM, respectively for these molecules are significant for the biological pathway as regulators of NAM recycling. Exhibiting a picomolar Ki, and high ratios Km/Ki (with or without ATP) FK866 is a powerful inhibitor of NAMPT (Table 2). In contrast, 5P-DADMe-NMN (Scheme 1) was a relatively poor inhibitor of the transfer reaction (Figure 6) with Ki values of 0.5 and 7 µM with respect to PRPP and NAM. In the presence of ATP, its binding increased 3-fold (Table 2).
Table 2.
Inhibition results for both coupled and non ATP-coupled reactions.
| Non ATP-coupled NMN synthesis |
ATP-coupled NMN synthesis* |
|||
|---|---|---|---|---|
| PRPP | NAM | PRPP | NAM | |
| Ki | ||||
| FK866 (pM) | 730 ± 75 (9863)† | 5000 ± 500 (171) | 10 ± 1 (63000) | 150 ± 18 (33) |
| 5P-DADMe-NMN (µM) | 0.53 ± 0.02 (14) | 7.1 ± 0.2 (<1) | 0.160 ± 0.015 (4) | 2.5 ± 0.2 (<1) |
| NAD+ (µM) | > 100 | 0.140 ± 0.014 (5) | 2.1 ± 0.2 (<1) | |
| NADH (µM) | > 100 | 0.22 ± 0.02 (3) | 3.2 ± 0.2 (<1) | |
Ki determined at 2.5 mM ATP.
Ratio Km/Ki corresponding.
Figure 6.
Inhibition of the non ATP-coupled reaction by 5P-DADMe-NMN.
Stimulated ATPase Activity
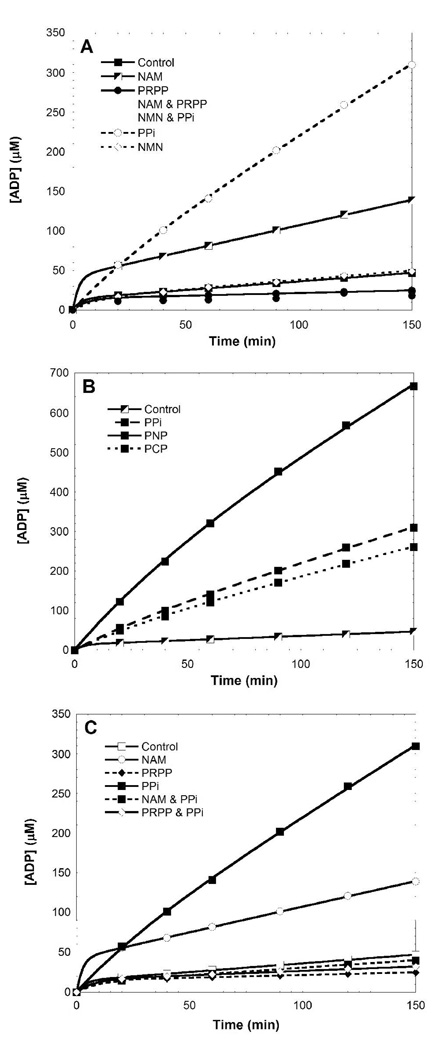
ATPase activity is not tightly coupled to NMN synthesis and the enzyme had the ability to hydrolyze ATP in absence of other substrates (Figure 2A, Figure 7A). Upon addition of single substrates of the transferase reaction (or mixtures of those), the ATPase function could be modulated. As described in the experimental section, the free-enzyme hydrolyzed the nucleotide at 0.11 min−1 and became as slow as 0.02 min−1 when both substrates (forward and reverse reactions) were bound. Nicotinamide and pyrophosphate enhanced ATP hydrolysis (0.37 and 1.1 min−1 respectively), while PRPP inhibited ATP hydrolysis (Figure 7A).
Figure 7.
NAMPT catalyzed ATP hydrolysis and the effect of substrates and products on ATPase reaction (A). ATPase activity of NAMPT and activation by PPi and its analogues (B). The presence of PRPP and/or NAM reverses the PPi-stimulation of the NAMPT ATPase reaction (C).
The PPi-stimulated ATPase activity was further explored with the methylene and imido analogues of PPi (PCP and PNP), assayed in the same conditions (Figure 7B). PNP had the greatest effect on ATP hydrolysis (2.6 min−1), greater than the effect of pyrophosphate. However, this effect was reversible upon addition of NAM or PRPP (Figure 7C).
ADP/ATP Exchange Catalyzed by NAMPT
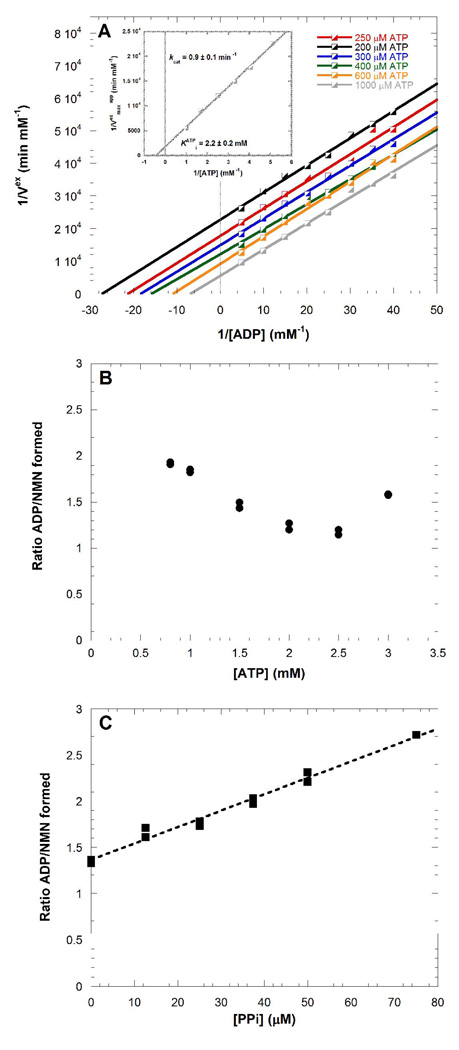
Phosphate is a product of the ATPase reaction of NAMPT. Comparing the catalytic efficiencies of the enzyme in the presence of inorganic phosphate for the non ATP-coupled NMN synthesis, and the ATP-coupled reaction, it is clear that Pi effects are smaller than those with ATP (Table 1). A catalytically altered His247 phosphoenzyme is proposed to exist in NAMPT. ADP/ATP isotope exchange explores conservation of a chemically-exchangeable high-energy phosphate on the enzyme. ADP/ATP exchange in the absence of other reactants can only occur through the action of a high-energy phosphorylated enzyme. Figure 8A shows the results of the kinetic study for the exchange reaction. The kcat for exchange was 0.9 ± 0.1 min−1 and the ATP concentration required for half-maximal exchange activity was 2.2 ± 0.2 mM and was 0.44 ± 0.05 mM for ADP. ATP binds weakly to the phosphorylated enzyme and exhibited inhibition of NMN synthesis at concentrations higher than 4 mM.
Figure 8.
ADP/ATP isotope exchange under conditions where no NAM to NMN catalysis occurred. NAMPT at a 0.6 mM concentration, with 4 samples quenched every 15 minutes (A). Stoichiometry between ATP hydrolysis and NMN synthesis (B). Comparison of ATP hydrolysis and NMN synthesis at increasing concentration of PPi (C).
ADP/ATP exchange occurs without NMN formation, however the similar rates for ADP/ATP exchange (0.9 ± 0.1 min−1) and NMN synthesis (0.46 ± 0.09 min−1) in optimized reaction conditions suggests that formation or hydrolysis of the phosphoenzyme may be rate-limitating for the overall reaction.
ADP/ATP exchange was also carried out in presence of 10 µM P1,P5-di(adenosine-5’-) pentaphosphate (Ap5A), a potent inhibitor of adenylate kinase, and the NAMPT retained the same exchange rate. Thus, adenylate kinase activity is insignificant and the catalytic exchange site for NAMPT does not accommodate Ap5A. Another control was performed by measuring the ADP/ATP exchange in the presence of 100 µM PRPP. This gave near-complete inhibition in the exchange, establishing structural or conformational linkage between these sites.
Coupling of NMN Synthesis and ATP Hydrolysis
Since only 2.1 kcal mol−1 of energy from the ATP hydrolysis is coupled to product formation (Figure 3), chemical stoichiometry by tight coupling can be eliminated. By comparing the rates of ATPase and NMN synthesis in the coupled transfer reaction, this stoichiometry (ratio R) was found to be ATP dependent (Figure 8B). Only at ATP concentrations of 2.0 to 2.5 mM was this ratio near 1. Low concentrations of ATP result in 2 moles of ATP hydrolysis per NMN formed and likewise, high concentrations of ATP also increased the ratio to > 1. When the ATPase-transferase system was near optimal conditions (2 µM NAM, 100 µM PRPP, 2.5 mM MgAT, 5 mM MgCl2), the ratio was increased in a linear fashion with increasing PPi concentration (Figure 8C).
DISCUSSION
The use of ATP hydrolysis to drive enzymatic reactions requires a mechanism to couple hydrolytic bond energy to the linked process. This distinction allows distribution of ATPases into two groups: in the first, enzymes chemically couple ATP hydrolysis by phosphorylation of a substrate-derived intermediate, and in the second, the coupling does not involve substrate phosphorylation but linked conformational changes. By similarity with bacterial NAPT (34), NAMPT is not catalytically dependent on the presence of ATP but has the ability to couple ATP hydrolysis and NMN synthesis. During ATP hydrolysis, NAMPT catalysis is more catalytically efficient, forward turnover is accelerated, and the unfavorable thermodynamic equilibrium for NMN formation is overcome. As ATP is consumed, the reaction returns to the non-ATP equilibrium position.
NAMPT uses ATP hydrolysis for thermodynamic drive, allowing the enzymatic system to build up 35-fold higher concentrations of NMN than observed in the absence of ATP. Variations in the substrates/enzyme affinities are also influenced by ATP. The Km value for PRPP is lowered 10-fold to a submicromolar value, ensuring adequate saturation at cellular levels of PRPP. The presence of ATP dramatically changed the interaction between NAMPT and NAM, lowering its Km 150-fold. Moreover, with a 1.8×106 M−1 s−1 kcat/Km value, the enzymatic efficiency of the ATP-coupled system, is improved 1100-fold. These values are similar to the Salmonella typhimurium NAPT (34). The Km values for NA (NAPT) and NAM (NAMPT) are both lowered by > 100-fold and the catalytical efficiency increased 1000 to 2000-fold (Table 3). However, NAMPT is unique in its unprecedented affinity for NAM, allowing the enzyme to recycle the cofactor at low nanomolar concentrations of the vitamin.
Table 3.
Comparison of kinetic parameters for human NAMPT and Salmonella typhimurium NAPT.
| Non ATP-coupled NMN/NAMN synthesis |
ATP-coupled NMN/NAMN synthesis |
|||
|---|---|---|---|---|
| NAMPT | NAPT | NAMPT | NAPT | |
| Km | ||||
| NAM (nM) | 855 ± 22 | 290000 ± 50000 | 5 ± 2 | 1500 ± 300 |
| PRPP (µM) | 7.2 ± 0.6 | 4500 ± 2200 | 0.63 ± 0.03 | 22 ± 3.3 |
| kcat (min−1) | 0.082 ± 0.001 | 17 ± 3 | 0.46 ± 0.0.09 | 174 ± 4 |
| kcat/Km (M−1 s−1)† | 1.60 ± 0.06 × 103 | 1.0 ± 0.4 × 103 | 1.8 ± 0.9 × 106 | 1.9 ± 0.6 × 106 |
kcat/Km regarding to NAM/NA.
Unlike ATP, inorganic phosphate does not induce large differences in substrate affinity or catalysis. Previous studies on NAPT, the bacterial counterpart of NAMPT, demonstrated conformational modifications induced by ATP binding to the enzyme (31, 32) and from chemical modification (30) via phosphorylation of His219, the structural equivalent of His247 in human NAMPT. With NAMPT, positive ADP/ATP isotope exchange experiments establish the existence of an active phosphorylated enzyme intermediate although attempts to isolate and characterize this species were not successful. Energetics of the NAMPT auto-phosphorylation describe a high-energy unstable species with a ΔG° of hydrolysis = −9.2 kcal mol−1, consistent with a phospho-histidine intermediate (36, 37). Similarities between NAPT and NAMPT allow us to propose His247 as a candidate for the phosphorylation. This assignment is also supported by mutagenesis studies of His247 on the mouse and human enzymes (25, 27). The His247Glu and His247Ala mutants exhibit a decrease of substrates affinity and a lower enzymatic efficiency. Recent efforts to structurally characterize complexes of human NAMPT have provided us with X-Ray data sets (38) containing a BeF3 − ion in an apparent covalent interaction to His247 (in preparation).
His247 overlaps the catalytic site (27) and explains the competitive and substrate inhibitions seen in the kinetic data but structural reasons for His247 phosphorylation causing NAMPT activation are not clear. Having an additional negative charge in the active site has been proposed to influence the formation of an oxacarbenium ion transition state (27), but this change would influence catalysis in both directions, and is an unlikely explanation for forward activation. Considering the small change in kcat between the ATP-coupled and uncoupled reactions (6-fold), and assuming that NMN synthesis proceeds through a single transition state, the presence of phospho-His247 would only lower the transition state barrier by 1 kcal mol−1. Such considerations suggest that other parameters involving conformational changes favorable for NAM binding, are important to explain the mechanism for ATP-coupled activation.
Hydrolysis of the phospho-enzyme is required for each catalytic turnover in NMN synthesis in the presence of ATP. The ability of exogenous PPi to enhance ATPase indicates that hydrolysis might be triggered by this product, signaling that the reaction is complete. In stoichiometry studies with NAMPT there is always one or more ATPs hydrolyzed per NMN formed, supporting this mechanism. It is also possible to imagine phospho-enzyme hydrolysis and rephosphorylation without full clearance of the catalytic site, consistent with more than one ATP hydrolysis event per NMN formed.
With only 2.1 kcal mol−1 transferred to the enzyme in the form of chemical equilibrium shifting, one could criticize the thermodynamic imperfection in this machinery. Only a few conditions result in a 1 to 1 stoichiometry between the ATP hydrolysis and NMN formation and slight variations of ATP or PPi concentrations destabilized ratio. Linkage between His247 phosphorylation and the catalytic site is demonstrated by PPi-stimulated ATPase inhibition with either PRPP or NAM, and PRPP also inhibited the rate of ADP/ATP isotope exchange. Those observations support an overlap between the ATP/ADP binding site and those for PRPP/NAM.
Comparing the thermodynamic and kinetic aspects of NAMPT establish that the phospho-enzyme is poised to capture NAM and PRPP and in turn, both of these stabilize the phospho-enzyme from hydrolysis. NMN and PPi can then be formed at concentrations that art thermodynamically prohibited without ATP coupling. The apparent energy leak in this weakly coupled reaction is transferred into a NAMPT conformation favoring the forward steps in the reaction. PPi formation signals completion of the reaction and stimulates hydrolysis of the phosphoenzyme. The affinity of NAMPT for ATP is consistent with cellular ATP levels. Although the enzyme uses ATP energy to recycle nicotinamide, the expenditure ensures cellular NAD+ levels with feedback regulation from NAD+/NADH levels.
An unknown feature of phosphoribosyltransferase reactions that use ATP to shift their equilibrium is the need for ATP hydrolysis, when product PPi hydrolysis would be more efficient in shifting the equilibrium toward products. However, coupled PPi hydrolysis would not provide the altered kinetic properties found for NAM binding as a consequence of ATP activation.
NAMPT is essential for NMN synthesis and highly adapted to replenish the NAD+ pool from NAM and PRPP. When NAD+ is used in regulatory biological processes (proteins ADP-ribosylation, deacetylation), free NAM can be recycled efficiently by NAMPT using available energy in the form of ATP. Metabolic and cell developmental studies have revealed links from glucose restriction through AMP-activated protein kinase to altered transcription levels of NAMPT and altered NAD+ levels (39). Finally, the inhibitions by NAD+ and NADH are sufficiently powerful to balance NMN synthesis with needs as a function of the NAD+/NADH pool.
ACKNOWLEDGMENT
We thank Dr. Hong Zang (Department of Biochemistry, University of Texas) for the gift of NMNAT-3 (pPROEX plasmid).
ABBREVIATIONS
- NAMPT
nicotinamide phosphoribosyltransferase
- NMN
nicotinamide mononucleotide
- PPi
pyrophosphate
- PCP
methylenediphosphate
- PNP
imidodiphosphate
- NAM
nicotinamide
- PRPP
α-D-5-phosphoribosyl-1-pyrophosphate
- NAD+
nicotinamide adenine dinucleotide
- SIRT
sirtuins
- NAPT
nicotinic acid phosphoribosyltransferase
- NAMN
nicotinic acid mononucleotide
- NA
nicotinic acid
- QAPT
quinolinic acid phosphoribosyltransferase
- NMNAT
nicotinamide/nicotinic acid mononucleotide adenylyltransferase
- FK866
(E)-N-[4-(1-benzoylpiperidin-4-yl) butyl]-3-(pyridin-3-yl) acrylamide
- THP
tris(hydroxypropyl)phosphine
- Ap5A
P1,P5-di(adenosine-5’-) pentaphosphate
Footnotes
This work was supported by research grant GM41916 from the NIH.
REFERENCES
- 1.Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267(6):1550–1564. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 2.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. (4) 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32(Pt 6):904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- 4.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61(1):19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305(5686):1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 6.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958;233(2):488–492. [PubMed] [Google Scholar]
- 7.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 8.Di Lisa F, Ziegler M. Pathophysiological relevance of mitochondria in NAD(+) metabolism. FEBS Lett. 2001;492(1–2):4–8. doi: 10.1016/s0014-5793(01)02198-6. [DOI] [PubMed] [Google Scholar]
- 9.Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem. 2001;276(1):406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 10.Raffaelli N, Sorci L, Amici A, Emanuelli M, Mazzola F, Magni G. Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem Biophys Res Commun. 2002;297(4):835–840. doi: 10.1016/s0006-291x(02)02285-4. [DOI] [PubMed] [Google Scholar]
- 11.Rizzi M, Schindelin H. Structural biology of enzymes involved in NAD and molybdenum cofactor biosynthesis. Curr Opin Struct Biol. 2002;12(6):709–720. doi: 10.1016/s0959-440x(02)00385-8. [DOI] [PubMed] [Google Scholar]
- 12.Schweiger M, Hennig K, Lerner F, Niere M, Hirsch-Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M. Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett. 2001;492(1–2):95–100. doi: 10.1016/s0014-5793(01)02180-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Kurnasov OV, Karthikeyan S, Grishin NV, Osterman AL, Zhang H. Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis. J Biol Chem. 2003;278(15):13503–13511. doi: 10.1074/jbc.M300073200. [DOI] [PubMed] [Google Scholar]
- 14.Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28(1):41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 15.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299(5611):1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277(21):18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 18.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 19.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 20.van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282(15):10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63(21):7436–7442. [PubMed] [Google Scholar]
- 23.Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drugs. 2008;26(1):45–51. doi: 10.1007/s10637-007-9083-2. [DOI] [PubMed] [Google Scholar]
- 24.Galli U, Ercolano E, Carraro L, Blasi Roman CR, Sorba G, Canonico PL, Genazzani AA, Tron GC, Billington RA. Synthesis and Biological Evaluation of Isosteric Analogues of FK866, an Inhibitor of NAD Salvage. ChemMedChem. 2008;3(5):771–779. doi: 10.1002/cmdc.200700311. [DOI] [PubMed] [Google Scholar]
- 25.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13(7):582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 26.Kim MK, Lee JH, Kim H, Park SJ, Kim SH, Kang GB, Lee YS, Kim JB, Kim KK, Suh SW, Eom SH. Crystal structure of visfatin/pre-B cell colony-enhancing factor 1/nicotinamide phosphoribosyltransferase, free and in complex with the anti-cancer agent FK-866. J Mol Biol. 2006;362(1):66–77. doi: 10.1016/j.jmb.2006.06.082. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Zhang X, Bheda P, Revollo JR, Imai S, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol. 2006;13(7):661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 28.Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J Biol Chem. 2005;280(18):18326–18335. doi: 10.1074/jbc.M501622200. [DOI] [PubMed] [Google Scholar]
- 29.Vinitsky A, Grubmeyer C. A new paradigm for biochemical energy coupling. Salmonella typhimurium nicotinate phosphoribosyltransferase. J Biol Chem. 1993;268(34):26004–26010. [PubMed] [Google Scholar]
- 30.Gross J, Rajavel M, Segura E, Grubmeyer C. Energy coupling in Salmonella typhimurium nicotinic acid phosphoribosyltransferase: identification of His-219 as site of phosphorylation. Biochemistry. 1996;35(13):3917–3924. doi: 10.1021/bi9517906. [DOI] [PubMed] [Google Scholar]
- 31.Rajavel M, Gross J, Segura E, Moore WT, Grubmeyer C. Limited proteolysis of Salmonella typhimurium nicotinic acid phosphoribosyltransferase reveals ATP-linked conformational change. Biochemistry. 1996;35(13):3909–3916. doi: 10.1021/bi951791y. [DOI] [PubMed] [Google Scholar]
- 32.Rajavel M, Lalo D, Gross JW, Grubmeyer C. Conversion of a cosubstrate to an inhibitor: phosphorylation mutants of nicotinic acid phosphoribosyltransferase. Biochemistry. 1998;37(12):4181–4188. doi: 10.1021/bi9720134. [DOI] [PubMed] [Google Scholar]
- 33.Gross JW, Rajavel M, Grubmeyer C. Kinetic mechanism of nicotinic acid phosphoribosyltransferase: implications for energy coupling. Biochemistry. 1998;37(12):4189–4199. doi: 10.1021/bi972014w. [DOI] [PubMed] [Google Scholar]
- 34.Grubmeyer CT, Gross JW, Rajavel M. Energy coupling through molecular discrimination: nicotinate phosphoribosyltransferase. Methods Enzymol. 1999;308:28–48. doi: 10.1016/s0076-6879(99)08004-0. [DOI] [PubMed] [Google Scholar]
- 35.Elliott GC, Rechsteiner MC. Evidence for a physiologically active nicotinamide phosphoribosyltransferase in cultured human fibroblasts. Biochem Biophys Res Commun. 1982;104(3):996–1002. doi: 10.1016/0006-291x(82)91348-1. [DOI] [PubMed] [Google Scholar]
- 36.Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino Acids. 2007;32(1):145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 37.Stock JB, Stock AM, Mottonen JM. Signal transduction in bacteria. Nature. 1990;344(6265):395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 38.Data sets are deposited on the Protein Data Bank. Accession codes 3DKL for the PRPP/Benzamide/BeF3 − complexed to NAMPT, and 3DHF for the NMN/PPi/BeF3 − complexed to NAMPT
- 39.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14(5):661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]