Abstract
Tip60 is a histone acetyltransferase (HAT) involved in the acetyltransferase activity and the cellular response to DNA damage. Here, we show that programmed cell death 5 (PDCD5), a human apoptosis-related protein, binds to Tip60 and enhances the stability of Tip60 protein in unstressed conditions. The binding amount of PDCD5 and Tip60 is significantly increased after UV irradiation. Further, PDCD5 enhances HAT activity of Tip60 and Tip60-dependent histone acetylation in both basal and UV-induced levels. We also find that PDCD5 increases Tip60-dependent K120 acetylation of p53 and participates in the p53-dependent expression of apoptosis-related genes, such as Bax. Moreover, we demonstrate the biological significance of the PDCD5-Tip60 interaction; that is, they function in cooperation to accelerate DNA damage-induced apoptosis and knockdown of PDCD5 or Tip60 impairs their apoptosis-accelerating activity, mutually. Consistent with this, PDCD5 levels increase significantly on DNA damage in U2OS cells, as does Tip60. Together, our findings indicate that PDCD5 may play a dual role in the Tip60 pathway. Specifically, under normal growth conditions, PDCD5 contributes to maintaining a basal pool of Tip60 and its HAT activity. After DNA damage, PDCD5 functions as a Tip60 coactivator to promote apoptosis.
Introduction
Tip60 was originally identified as a protein that interacts with human immunodeficiency virus-Tat and increases Tat-dependent transcription [1]. Later studies demonstrated that Tip60 possesses histone acetyltransferase (HAT) activity in vivo and in vitro and that the role of Tip60 in transcriptional regulation has been intensively investigated [2–4]. Accumulated data suggest that Tip60 exerts diverse biological functions through mechanisms that are either dependent or independent of its intrinsic HATactivity, such as cellular signaling, DNA damage repair, cell cycle, checkpoint control, and apoptosis [5]. Tip60 is a tightly regulated transcriptional coregulator, acting in a large, multiprotein complex, with a range of transcription factors, including the androgen receptor [6], Myc [7], STAT3 [8], nuclear factor κB [9], E2F1 [10,11], and p53 [12–14]. Tip60-mediated regulation typically involves recruitment of Tip60 acetyltransferase activity to chromatin. Additionally, in response to DNA double-strand breaks, Tip60 is recruited to DNA lesions, where it participates in both the initial and the final stages of repair [15].
Programmed cell death 5 (PDCD5), formerly referred to as TF-1 cell apoptosis-related gene 19 (TFAR19), is a novel gene first cloned in our laboratory [16]. Previous studies have showed that PDCD5 can facilitate apoptosis and enhance TAJ/TROY-induced paraptosis-like cell death [17]. The PDCD5 protein is upregulated in cells undergoing apoptosis, where it translocates rapidly from the cytoplasm to the nucleus [18]. Recent studies have confirmed that two single-nucleotide polymorphisms, with linkage disequilibrium in the human PDCD5 gene 5′-regulatory region, affect promoter activity and the susceptibility of a Chinese population to develop chronic myelogenous leukemia [19]. In addition, a single-nucleotide polymorphism in the 5′-upstream region of PDCD5 is predictive of lung cancer risk and prognosis [20], suggesting that PDCD5 may represent a novel tumor suppressor gene influencing lung cancer.
The levels of both mRNA and protein of PDCD5 were analyzed in human carcinomas by different techniques. Decreased PDCD5 expression has been reported in various human tumors, such as breast cancer [21], hepatocellular carcinoma [22], lung cancer [20], gastric cancer [23], chronic myelogenous leukemia [24], and astrocytic gliomas [25]. These findings, together with our studies, suggest that decreased PDCD5 expression may be associated with carcinoma formation and malignant progression. Nevertheless, the molecular mechanism by which PDCD5 accelerates cell apoptosis remains unknown, as do the downstream events after PDCD5 nuclear accumulation in early apoptosis.
PDCD5 was recently implicated as a novel binding partner of Tip60, through a large-scale yeast two-hybrid screen [26]. However, there has yet to be no experimental evidence reported in mammalian cells or further functional investigation. In this study, we demonstrate for the first time that PDCD5 interacts with Tip60 in mammalian cells, enhances the stability of Tip60, and inhibits its proteasome-dependent degradation. After DNA damage, PDCD5 can accelerate the Tip60-mediated apoptotic cell responses. We thus conclude that PDCD5 is a positive regulator of Tip60.
Materials and Methods
Plasmids, siRNA, and Antibodies
The pcDNA3-PDCD5 and pcDNA3-PDCD5-myc plasmids used have been described previously [17]. The pCMV5-Tip60 and pCMV5 Flag-HA-Tip60 vectors were kindly provided by Dr. Amati Bruno. PcDNA3-Flag-p53 vector was a gift from Dr. Steven B. McMahon. All siRNA including PDCD5, Tip60, and the control siRNA were synthesized by GeneChem Corporation (Shanghai, China); the sequences of the various siRNA have been reported previously [13,27]. The anti-Flag, antimyc, and antiactin antibodies were purchased from Sigma (St. Louis, MO). The anti-Tip60 and anti-p53 were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-acetyl H2A (Lys5), anti-acetyl H4 (Lys8), anti-total H2A, and anti-PARP antibodies were from Cell Signaling Technology (Beverly, MA). The anti-pan-acetyl-lysine antibody, anti-acetyl H3 (Lys14), and anti-Bax were from Upstate (Waltham, MA). The anti-acetyl k120-p53 antibody was kindly provided by Dr. Steven B. McMahon. The mouse anti-PDCD5 monoclonal antibody (3A3), rabbit anti-PDCD5 polyclonal antibody, and FITC-labeled anti-PDCD5 antibody have been described previously [18]. IRDye 800-conjugated secondary antibodies against mouse, rabbit, and goat IgG were purchased from Li-Cor Bioscience (Lincoln, NE). TRITC-labeled rabbit against goat IgG was from Zhongshan Corporation (Beijing, China).
Cell Culture, Transfection, and Treatment
U2OS, H1299, and HeLa cell lines were cultured in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum. HeLa cells were transfected by electroporation, as described previously [27]; U2OS and H1299 cell lines were transfected using Lipofectamine 2000 (Invitrogen, Life Technologies, Inc., Carlsbad, CA) according to the manufacturer's protocol.
Proteasome inhibition was achieved by treating cells with 100 µM of N-acetyl-Leu-Leu-norleucine (ALLN; Sigma) or 10 µM of MG132 (Sigma) for 12 hours. Protein acetylation was enhanced by a 4-hour treatment of the cells with 10 mM of sodium butyrate (Sigma) before cell lysis. Protein translation inhibition was achieved by treating cells with 100 µg/ml of cycloheximide (CHX; Sigma) for different times (Figure 3C). Apoptosis was induced with various concentrations of actinomycin D (Act.D; 100 and 500 nM) or doxorubicin (Doxo; 0.25 and 1 µg/ml). UV irradiation (20 J/m2) was performed using a CX-2000 UV cross-linker (UVP, Inc., Upland, CA).
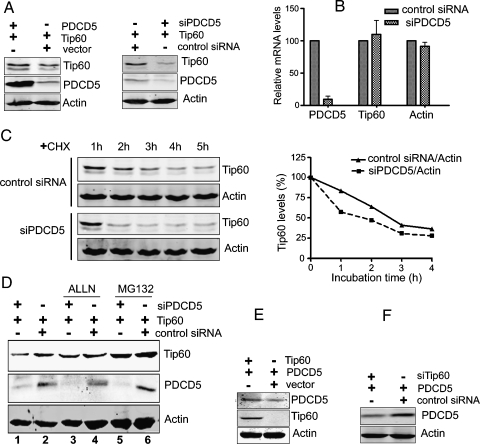
Figure 3.
PDCD5 enhances the stability of Tip60 protein. (A, left panel) Overexpression of PDCD5 increases the accumulation of Tip60. U2OS cells were transfected with the indicated plasmids for 36 hours. Total cell extracts were analyzed for the presence of Tip60, PDCD5, and actin by Western blot. (A, right panel) Knockdown of PDCD5 decreases the level of Tip60. U2OS cells were transfected with indicated plasmids and siPDCD5 for 36 hours. Other treatments just were the same as left panel. (B) Knockdown of PDCD5 fails to influence the mRNA level of Tip60. U2OS cells were transfected with siPDCD5 or control siRNA for 36 hours and then harvested and analyzed by realtime PCR. Data are presented as mean ± SD. (C) Knockdown of PDCD5 accelerates the decay of Tip60. U2OS cells were transiently cotransfected with Tip60 and either siPDCD5 or control siRNA. After 36 hours, cells were treated with 100 µg/ml of CHX for different lengths of time as indicated. Total cell lysates were analyzed for the presence of Tip60 by Western blot. Relative Tip60 protein levels were quantified using a Li-Cor Odyssey scanner (right panel). This experiment has been performed three times. The diagram presented is the statistical analysis of a single representative experiment. (D) siPDCD5-mediated decreases of Tip60 levels are attenuated by proteasome inhibition. U2OS cells were cotransfected with Tip60 and either siPDCD5 or control siRNA. After 24 hours, cells were treated with ALLN (100 µM) or MG-132 (10 µM) for 12 hours. Total cell lysates were analyzed for the presence of Tip60, PDCD5, and actin by Western blot. (E and F) Tip60 modulates PDCD5 expression. U2OS cells were transfected with the indicated plasmids (E) or siRNA (F) for 36 hours. Total cell extracts were analyzed for the presence of PDCD5, Tip60, and actin by Western blot.
Immunoprecipitation and Western Blot
For the immunoprecipitation (IP) experiment, cells were collected and lysed in lysis buffer (300 mM NaCl, 50 mM Tris pH 8.0, 0.4% NP-40, 10 mM MgCl2, and 2.5 mM CaCl2) supplemented with protease inhibitors (Complete mini EDTA-free; Roche Diagnostics, Mannheim, Germany). After centrifugation, the supernatant was measured using the BCA protein assay reagent (Pierce, Rockford, IL). Then, 1 mg of total cell extracts were diluted to 1 ml of dilution buffer (50 mM Tris, pH 8.0, 0.4% NP-40). After a precleaning step with protein G sepharose beads, cell lysates were incubated with appropriate antibodies overnight at 4°C and then with 50 µl of a 50% slurry of protein G sepharose for 2 hours. Immunoprecipitates were then washed five times in washing buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.4% NP-40, and 5 mM MgCl2) and analyzed by Western blot, as described previously [27]. The protein bands were visualized using an IRDye 800CW-conjugated secondary antibody; the infrared fluorescence image was obtained using an Odyssey infrared imaging system (Li-Cor Bioscience). In some experiments, we performed coimmunoprecipitation (CoIP) by using One-Step Complete IP-Western Kit (GeneScript Corporation, Piscataway, NJ) according to the manufacturer's protocol.
Glutathione S-Transferase Pull-down
Recombinant glutathione S-transferase (GST) or GST-PDCD5 were expressed in Escherichia coli and purified. In addition, the amounts of recombinant proteins were assessed by SDS-PAGE. Then, 1 µg of GST fusion proteins or GST was incubated with whole cell lysates extracted from Flag-HA-Tip60-transfected HeLa cells overnight at 4°C. After five washes, beads were resuspended in 2x SDS loading buffer and analyzed by SDS-PAGE followed by Western blot.
Immunofluorescence Analysis
U2OS cells were plated on glass coverslips and then transfected with pCMV5-Tip60 plasmid using Lipofectamine 2000. 24 hours after transfection, cells were treated with or without UV irradiation (20 J/m2) for 5 hours. Cells were then fixed in PBS supplemented with 4% paraformaldehyde for 15 minutes at 4°C, and permeabilized in PBS supplemented with 0.2% Triton X-100 for 30 minutes. After a 30-minute incubation in blocking buffer (3% BSA in PBS), cells were incubated with goat anti-Tip60 antibodies for 1 hour at 4°C, then incubated with TRITC-conjugated rabbit antigoat IgG and FITC-labeled mouse anti-PDCD5 for 1 hour. 4′,6-Diamidino-2-phenylindole (DAPI; Sigma) was used to counterstain the nuclei. After several washing, coverslips were observed under a Leica SP2 confocal system (Germany).
Real-time Polymerase Chain Reaction Analysis
U2OS cells were transfected with either siPDCD5 or control siRNA with Lipofectamine 2000. Thirty-six hours later, total RNA were extracted from transfected U2OS cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was generated using the total RNA in a standard reverse transcriptase reaction using a poly(dT) oligonucleotide as a primer and SuperScript II reverse transcriptase (Invitrogen, Life Technologies, Inc.). The cDNA were then subjected to real-time polymerase chain reaction (PCR) analysis. The primers used in the assay are available on request.
IP-HAT Assay
This assay was performed essentially as described previously [28]. Briefly, HeLa cells were cotransfected with Flag-HA-Tip60 and PDCD5 plasmids or empty vector. Thirty-six hours after transfection, cells were harvested and IP was performed with an anti-Flag antibody. After the last wash, the supernatant was completely aspirated, and the pelleted beads were resuspended in 30 µl of HAT-reaction buffer (50 mM Tris/HCl, pH 8.0, 10% glycerol, 1 mM DTT, 1 mM PMSF, and 0.1 mMEDTA) containing 2.5 µg of histone 4 (New England BioLabs, Ipswich, MA), 10 mM sodium butyrate, and 50 nCi 3H-labeled acetyl-CoA (Amersham Pharmacia, Little Chalfont, United Kingdom). The reaction mixture was incubated at 30°C for 60 minutes. Then, the mixture was spotted onto Whatman P81 phosphocellulose squares (2 x 2 cm) and allowed to air dry. Dry filters were washed three times for 2 minutes at room temperature with 200 mM sodium bicarbonate (pH 9.2) to remove unincorporated radioactive acetyl-CoA. The filters were then placed in scintillation vials, and 2 ml of scintillation fluid was added. After incubating the vials at room temperature for 10 minutes, the amount of incorporated radioactivity was determined using a liquid scintillation counter.
Detection of Phosphatidylserine Externalization
Treated cells (5 x 105) were trypsinized, washed twice with PBS, and resuspended in 200 µl of binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 1 mM MgCl2, 5 mM KCl, and 2.5 mM CaCl2). FITC-conjugated Annexin V was added to a final concentration of 0.5 µg/ml according to the manufacturer's instructions (Beijing Biosea, China). After 20 minutes of incubation at room temperature in the dark, 400 µl of binding buffer was added, and samples were immediately analyzed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Results
PDCD5 Interacts with Tip60
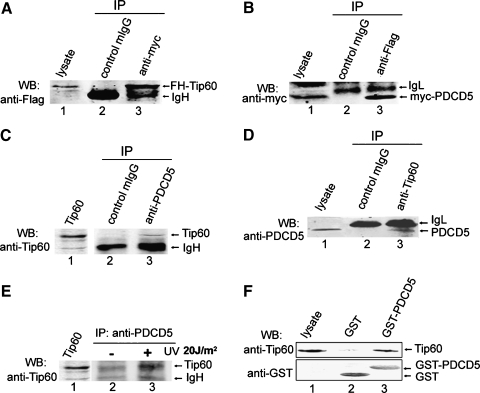
To confirm the association of Tip60 and PDCD5 in mammalian cells, Flag-HA-Tip60 and myc-PDCD5 plasmids were cotransfected into HeLa cells. Cell lysates were subjected to IP with an antimyc antibody. Western blot analysis revealed that Flag-HA-Tip60 coprecipitated with myc-PDCD5 (Figure 1A). Reciprocally, myc-PDCD5 was detected in the Flag-HA-Tip60 immunoprecipitates (Figure 1B), demonstrating that the two proteins existed in a complex in HeLa cells.
Figure 1.
PDCD5 interacts with Tip60. (A) Coimmunoprecipitation of Tip60 and PDCD5 in HeLa cells. Cells were cotransfected with Flag-HA-Tip60 and myc-PDCD5 plasmids. Total cell extracts were subjected to IP using either an antimyc or an irrelevant control IgG, as indicated. Immunoprecipitated proteins were then analyzed for the presence of FH-Tip60 by Western blot. (B) Cells were treated the same as in (A), except that cell extracts were immunoprecipitated with an anti-Flag antibody and immunoblotted with an antimyc antibody. (C) Endogenous PDCD5 interacts with Tip60 in U2OS cells. Total cell extracts were subjected to IP using either an anti-PDCD5 or an irrelevant control IgG, as indicated. Immunoprecipitated proteins were then analyzed for the presence of Tip60 by Western blot. (D) As in (C), except that cell extracts were immunoprecipitated with an anti-Tip60 antibody and immunoblotted with an anti-PDCD5 antibody. (E) Tip60 association with endogenous PDCD5 is increased after UV irradiation. U2OS cells were treated with or without UV irradiation (20 J/m2) for 5 hours. Coimmunoprecipitation was performed by using One-Step Complete IP-Western Kit. (F) Direct interaction of PDCD5 with Tip60 in vitro. The full-length GST-PDCD5 fusion protein (lane 3) and the GST protein (lane 2) were incubated with Tip60-transfected HeLa cell lysates overnight at 4°C. Washed beads were analyzed for the presence of Tip60 and GST by Western blot.
Because the above experiments relied on overexpressed protein, we sought to examine whether interaction occurs between endogenous PDCD5 and Tip60. Endogenous CoIP analysis provided evidences for the endogenous PDCD5-Tip60 interaction (Figure 1, C and D). It is known that Tip60 can be induced and upregulated by UV irradiation [29]. Thus, we further tested the PDCD5-Tip60 interaction in UV-treated cells using One-Step Complete IP-Western Kit according to the manufacturer's protocol. This assay revealed that the amount of Tip60 bound to PDCD5 significantly increased after UV irradiation (Figure 1E). This interaction was specific because control IgG failed to bind either Tip60 or PDCD5 (Figure 1, A–D). The interaction between PDCD5 and Tip60 can also be observed by using a pull-down approach (Figure 1F). These findings suggest that PDCD5 directly interacts with Tip60, and the association of the two proteins is upregulated after UV irradiation. Because endogenous Tip60 protein as difficult to be detected by Western blot, the lysates of Flag-HA-Tip60 transfected HeLa cells were taken as positive controls (Figure 1, C, E, lane 1).
PDCD5 Colocalizes with Tip60
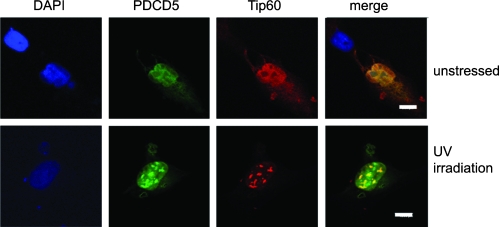
To further confirm the PDCD5-Tip60 association, we performed the colocalization assay. Owing to the very low expression level of Tip60, U2OS cells were transfected with pCMV5-Tip60 plasmids for 24 hours, then treated with or without UV irradiation (20 J/m2), and incubated for a further 5 hours before harvesting. Then, U2OS cells were incubated with goat anti-Tip60, followed by an antigoat TRITC-conjugated antibody and FITC-anti-PDCD5. Cell nucleus morphology was observed by DAPI staining. PDCD5 and Tip60 colocalized and showed a relatively diffuse nuclear distribution pattern in U2OS cells without UV irradiation (Figure 2, upper panel). Strikingly, in UV-treated U2OS cells, the localization of PDCD5 and Tip60 converted to large nuclear speckles, and the distribution of PDCD5 overlapped that of Tip60 (Figure 2, lower panel). The colocalization assay confirms the interaction between PDCD5 and Tip60.
Figure 2.
PDCD5 colocalizes with Tip60 in U2OS Cells. U2OS cells were transfected with the Tip60 plasmid and treated with or without UV irradiation (20 J/m2, 5 hours). Immunofluorescence analysis was performed as described in the Materials and Methods Tip60 expression was detected with anti-Tip60 antibody followed by an antigoat TRITC-conjugated secondary antibody. PDCD5 was detected by FITC-anti-PDCD5. Cell nuclei were stained with DAPI. They were observed using Leica SP2 confocal microscope. Scale bar, 8 µm.
The nuclear localization of Tip60 has been studied extensively and displayed large nuclear speckles [13,30,31] just as our results. Cheng et al. [32] reported that Tip60 translocated to promyelocytic leukemia (PML) bodies during DNA damage. We have even investigated the colocalization of PDCD5/Tip60 and PML. It was found that the speckles of PDCD5/Tip60 were larger than PML bodies (data not showed). The inducible formation of the Tip60-PDCD5 larger speckles may indicate that the degree of PDCD5's colocalization with Tip60 was increased under UV irradiation. This evidence supported that both proteins could act in similar regulatory pathways and play an important role in nucleus in the response to DNA damage.
PDCD5 Enhances the Stability of the Tip60 Protein
To assess the role of the PDCD5-Tip60 association, we monitored the changes in the levels of Tip60 when PDCD5 was overexpressed or depleted. U2OS cells were cotransfected for 36 hours with Tip60 and either PDCD5 or specific siRNA against PDCD5 that we had previously characterized [27]. Western blot indicated the Tip60 protein levels showed a weak increase in coexpressing PDCD5 cells compared with cells transfected with empty vector (Figure 3A, left panel). Meanwhile, Tip60 protein levels decreased dramatically 36 hours after knockdown of endogenous PDCD5, whereas actin levels were unchanged (Figure 3A, right panel). This experiment has been performed five times at least, and the results were same.
We further examined the mRNA levels of Tip60 after treatment with siRNA against PDCD5. U2OS cells were transfected for 36 hours with either siPDCD5 or control siRNA and then cells were harvested and analyzed by real-time PCR. As shown in Figure 3B, knockdown of PDCD5 failed to influence the mRNA levels of Tip60, indicating that PDCD5 might not regulate Tip60 on the transcription level. We next tested the influence of PDCD5 on Tip60 half-life. U2OS cells were transiently cotransfected with Tip60 and either siRNA against PDCD5 or control siRNA. Cells were then treated with the translation inhibitor CHX (100 µg/ml) for different periods as indicated. As shown in Figure 3C, depletion of endogenous PDCD5 was associated with a significant acceleration in Tip60 decay, which suggested that the decrease of Tip60 protein levels after PDCD5 knockdown was due to the destabilization of the Tip60 protein.
It is known that Tip60 are degraded through the proteasome pathway [29], so a series of experiments were conducted to investigate the proteasomal degradation targeting of Tip60 by PDCD5. We added the proteasome inhibitor ALLN (100 µM) or MG-132 (10 µM) to treat U2OS cells. The data obtained from Western blot showed that depletion of endogenous PDCD5 reduced Tip60 expression (Figure 3D, lane 1), which was consistent with the above observation. However, after treatment with the ALLN or MG-132 for 12 hours, the reduction in Tip60 levels was significantly blocked (Figure 3D, lanes 3 and 5) compared with lane 1. This suggested that the proteasome inhibitor (ALLN or MG-132) could rescue siPDCD5-mediated degradation of the Tip60, indicating that endogenous PDCD5 may be involved in the modulation of Tip60 levels in normal growing cells by protecting Tip60 from proteasomal degradation.
Interestingly, when performing these experiments, we observed that coexpression of Tip60 caused an increase in PDCD5 accumulation (Figure 3E). Knockdown of endogenous of Tip60 consistently decreased the PDCD5 protein level (Figure 3F), indicating that these two proteins were mutually regulated in normal growing cells.
PDCD5 Promotes the HAT Activity of Tip60
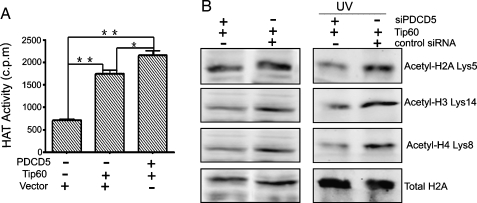
We next sought to determine whether PDCD5 binding affects Tip60 HAT activity using the IP-HAT assay. U2OS cells were cotransfected with both Flag-HA-Tip60 and PDCD5 plasmids or Flag-HA-Tip60 alone or an empty vector. Exogenous Tip60 protein was then immunoprecipitated with an anti-Flag antibody, and the HAT activity present within the immunopellets was determined using a liquid HAT assay [28]. Data from experiments showed that compared with the pCDB group, the FH-Tip60 and cotransfected groups had strong HAT activity (P < .01; Figure 4A). The difference between the FH-Tip60 and cotransfected groups was statistically significant (P < .05; Figure 4A), which indicated that PDCD5 could enhance the HAT activity of Tip60 to some extent.
Figure 4.
PDCD5 promotes the HAT activity of Tip60. (A) IP-HAT assay detects HAT activity. HeLa cells were cotransfected with Flag-HA-Tip60 and PDCD5 plasmids or empty vector. Twenty-four hours after transfection, IPs were performed using an anti-Flag antibody, and the pelleted immune complexes were tested for their ability to acetylate free histone H4. **P < .01; *P < .05. (B) Western blot analyses the acetylation of histones. U2OS cells were cotransfected with the Tip60 plasmid and either siPDCD5 or control siRNA. After 24 hours, cells were treated with or without UV irradiation for 12 hours and further incubated with 10 mM sodium butyrate for 4 hours. Then, histone fractions were extracted, and immunoblots were probed with the indicated antibodies to determine the acetylation sites of the histone.
We further evaluated the effects of PDCD5 on histone acetylation mediated by Tip60 in U2OS cells. As shown in Figure 4B (left panel), depletion of endogenous PDCD5 attenuated Tip60-mediated histone acetylation, including lysine 5 of histone H2A, lysine 14 of histone H3, and lysine 8 of histone H4. These effects remained the same under the condition of UV irradiation (Figure 4B, right panel). These findings indicate that the PDCD5-Tip60 interaction, at least in part, is necessary for the modulation of Tip60 enzymatic activity in both unstressed and stressed condition.
PDCD5 Promotes p53 Acetylation and p53-Dependent Apoptosis-Related Gene Expression Mediated by Tip60
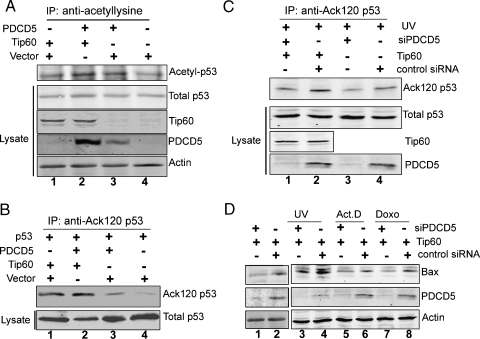
As we know, Tip60 is required for the acetylation of the endogenous p53 protein at K120 and for the p53-dependent induction of proapoptotic target genes, such as Bax, in response to DNA damage [33]. Hence, we next asked whether PDCD5 influenced either acetylation of p53 or expression of p53-dependent apoptosis-related genes mediated by Tip60. To examine this possibility, U2OS cells were cotransfected with Tip60 and PDCD5 or an empty vector. Total cell lysates were subjected to IP with a universal anti-acetyl-lysine antibody. A Western blot incubated with anti-p53 revealed that both Tip60 (Figure 5A, lane 1) and PDCD5 alone (Figure 5A, lane 3) could enhance the acetylation of p53 compared with the control vector (Figure 5A). Coexpression of Tip60 and PDCD5 resulted in a weak increase in the acetylation level of p53 (Figure 5A, lane 2).
Figure 5.
PDCD5 promotes p53 acetylation and p53-dependent apoptosis-related gene expression mediated by Tip60. (A) U2OS cells were cotransfected with Tip60 and either PDCD5 or empty vector for 36 hours. After 4 hours of treatment with 10 mM sodium butyrate, total cell lysates were subjected to IPs with a pan-acetyl-lysine antibody. Precipitates were blotted with an anti-p53 antibody to detect the p53 acetylation level. Input lysates were also blotted with anti-p53, anti-Tip60, anti-PDCD5, and antiactin antibodies. (B) H1299 cells were cotransfected as indicated for 36 hours. After 4 hours of treatment with 10 mM sodium butyrate, total cell lysates were subjected to IPs with an anti-Ack 120-p53 antibody. Precipitates were blotted with an anti-p53 antibody to detect K120 acetylation of p53. Input lysates were also blotted with anti-p53 antibodies. (C) U2OS cells were transfected as indicated. After 24 hours, cells were given UV irradiation for 12 hours. Immunoprecipitation and Western blot were performed as in (B). (D) U2OS cells were transiently cotransfected with Tip60 and either siPDCD5 or control siRNA. After 24 hours, cells were treated with or without genotoxic treatments as indicated, and then total cell lysates were analyzed for the presence of Bax, PDCD5, and actin by Western blot.
We further detected the Tip60-dependent K120 acetylation of p53 in H1299 cells, which lack an endogenous p53 expression. Cells were cotransfected with Flag-p53 plasmid in the presence of PDCD5 or Tip60 or both. Total cell lysates were subjected to IP with an anti-Ack 120-p53 antibody. Western blot probed with anti-p53 revealed that the K120 acetylation level of exogenous p53 in H1299 was remarkably increased in the presence of Tip60 (Figure 5B, lane 1). PDCD5-transfected cells displayed a weak increase (Figure 5B, lane 3). Coexpression of PDCD5 enhanced K120 acetylation of p53 (Figure 5B, lane 2), indicating that the interaction of PDCD5 and Tip60 had a synergistic effect on K120 acetylation of p53. Moreover, we tested whether PDCD5 could enhance endogenous p53 acetylation at K120 in response to DNA damage. U2OS cells were treated by UV irradiation, the levels of p53 acetylation at K120 was increased in the presence of Tip60 (Figure 5C, lane 2) compared with UV irradiation alone (Figure 5C, lane 4). Knockdown of endogenous PDCD5 resulted in a striking decrease of acetyl-K120 in the presence of Tip60 overexpression (Figure 5C, lane 1). We also observed that depletion of endogenous PDCD5 could attenuate the endogenous accumulation of acetyl-K120 (Figure 5C, lane 3). These data indicate that PDCD5maybe an important regulator in K120 acetylation of p53 after genotoxic stress.
Tip60-mediated acetylation of p53 at K120 is also a key for the activation of proapoptotic molecules, such as Bax, which is a mediator of p53-dependent apoptosis. Thus, we next tested the effect of Tip60 on Bax in U2OS cells in the presence or absence of PDCD5. Knockdown of PDCD5 resulted in a decreased expression of Bax at the protein level under the unstressed condition (Figure 5D, lane 1). When cells were treated with UV irradiation, Act.D, and Doxo, the depressed Bax expression was similar to the unstressed conditions in PDCD5-depleted group (Figure 5D, lanes 3, 5, and 7). Together, these results indicate that PDCD5 plays a positive role in Tip60-mediated p53 acetylation and p53-dependent proapoptotic gene expression.
PDCD5 Is Required for Cell Apoptosis Induced by DNA Damage
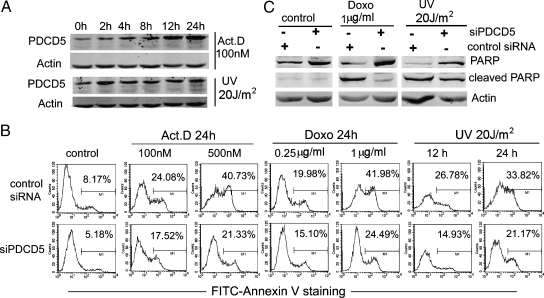
Our precious research has revealed that PDCD5 is related with apoptosis induced by DNA damage. Here, we detected endogenous PDCD5 expression in cells after DNA damage. After treatment with Act.D (100 nM) and UV irradiation (20 J/m2) for different times in U2OS cells, the PDCD5 protein exhibited a clear up-regulation, in a time-dependent manner (Figure 6A). This phenomenon was consistent with the Tip60 expression described previously [34]. These data indicate that both PDCD5 and Tip60 are upregulated after DNA damage and that they may cooperate to mediate the appropriate cellular response to DNA damage.
Figure 6.
PDCD5 is required for apoptosis induced by DNA damage. (A) PDCD5 expression was measured by Western blot in U2OS cells treated with Act.D (100 nM) and UV irradiation (20 J/m2), as indicated. (B) U2OS cells were transfected with siPDCD5 or control siRNA for 24 hours and then further incubated with different concentration of Act.D and Doxo for 24 hours or UV-irradiated for the indicated time. Then, cells were stained with FITC-Annexin V, and the percentage of cell apoptosis is shown. The data shown are from a representative experiment. (C) Cell treatment was similar to (B). Western blots analyze the presence of PARP (full-length or cleaved) and actin.
Next, we investigated whether the expression of PDCD5 is required in apoptosis induced by DNA damage. PDCD5 expression was inhibited using the specific PDCD5 siRNA, and the transfection efficiency achieved was more than 80% (data not shown). Then, U2OS cells were treated with different doses of Act.D (100 and 500 nM), Doxo (0.25 and 1 µg/ml), or 20 J/m2 of UV irradiation (12 and 24 hours). Apoptotic activity was monitored by FITC-Annexin V staining and caspase-mediated poly (ADP-ribose) polymerase (PARP) cleavage. As expected, compared with control siRNA, knockdown of PDCD5 produced a significant resistance to DNA damage-induced apoptosis in U2OS cells. This was indicated by the decrease of Annexin V-positive cells (Figure 6B, lower panel) and reduced PARP cleavage (Figure 6C). These findings provide evidences of the direct involvement of PDCD5 in the apoptotic cell response after DNA damage.
Coexpression of PDCD5 and Tip60 Accelerates DNA Damage-Induced Apoptosis
To investigate the functional significance of the targeting of Tip60 by PDCD5, a series of experiments were conducted. Our hypothesis was that the PDCD5-Tip60 association might enhance the apoptotic response of cells to genotoxic treatments, in which Tip60 has been shown to be involved [4,34,35].
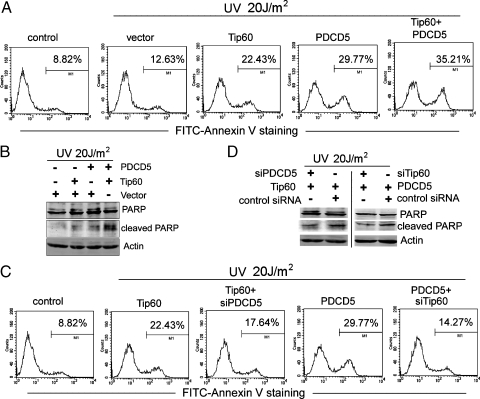
U2OS cells were transfected as indicated, and apoptosis was induced by UV irradiation. Both PDCD5 and Tip60 alone could effectively promote U2OS apoptosis compared with empty vector after UV irradiation (Figure 7A). Overexpression of both PDCD5 and Tip60 produced a synergistic effect, as indicated by the increase of Annexin V-positive cells (Figure 7A) and increased PARP cleavage (Figure 7B). Moreover, knockdown of endogenous PDCD5 caused inhibition of Tip60-dependent cell apoptosis, as indicated by the decrease in Annexin V-positive cells (Figure 7C) and reduced PARP cleavage (Figure 7D, left panel). Conversely, we also observed that knockdown of Tip60 impaired the apoptotic cell response mediated by PDCD5 overexpression (Figure 7, C and D, right panel), indicating that the apoptosis-promoting effect of PDCD5 was, at least in part, Tip60-dependent. On the basis of these data, we propose that PDCD5 and Tip60 may regulate each other. It is thus tempting to speculate that their activation and up-regulation are important events involved in DNA damage-induced apoptosis and that both are required for this process.
Figure 7.
Coexpression of PDCD5 and Tip60 accelerate DNA damage-induced apoptosis. (A) U2OS cells were transfected with PDCD5 and Tip60 plasmids alone or together. After 24 hours, they were treated with UV irradiation for 12 hours. Then, cells were stained with FITC-Annexin V, and the percentage of apoptosis was determined. The data shown are from a representative experiment. (B) Cell treatment was the same as in (A). Western blots analyze the presence of PARP (full-length or cleaved) and actin. (C) U2OS cells were transfected with the indicated siRNA and plasmids. After 24 hours, they were treated and analyzed as in (A). The data shown are from a representative experiment. (D) As in (C), except that total cell extracts were analyzed by Western blot for the presence of PARP (full-length or cleaved) and actin.
Discussion
PDCD5 has been described as an apoptosis-promoting molecule. Here, we provide evidences for the first time that PDCD5 is a novel partner of the Tip60 protein, confirmed by CoIP, GST pull-down, and colocalization assay. Data from our consistent experiments suggest that PDCD5 can stabilize Tip60 levels and inhibit its proteasomal degradation. PDCD5 may play an important role in the modulation of Tip60 levels in normal growing cells by protecting Tip60 from proteasomal degradation. This analysis of the underlying mechanism has revealed new aspects of the regulation of cellular Tip60.
The activity of HATs is important for the proper control of gene expression. Accordingly, many mechanisms regulating their activity have been described. One mechanism is through protein-protein interactions. We showed that, in normal growing conditions and stressed condition, suppressed endogenous PDCD5 could decrease Tip60-mediated histone acetylation, including histone H2A, histone H3, and histone H4. Consistent with this, an in vitro HAT assay further confirmed that the interaction of PDCD5 with Tip60 enhanced the HAT activity of Tip60. We have even performed the HAT assay with purified recombinant PDCD5 protein; unfortunately, PDCD5 alone has no obvious HAT activity (data not shown). Therefore, there was a good relationship between the ability of PDCD5 to interact with Tip60 and the promotion of its HAT activity. It is thus tempting to speculate that PDCD5, through its capacity to interact with Tip60, may act as an additional stability factor and further enhance the enzyme-substrate interaction. This may also be one of the mechanisms by which PDCD5 promotes programmed cell death.
Tip60 involvement in DNA repair and apoptosis in response to DNA damage is complex and elaborately regulated by multiple factors in mammalian cells. Our results indicated that PDCD5 expression was upregulated and required for apoptosis induced by DNA damage (Figure 6). Because the association between PDCD5 and Tip60 was increased under stressed conditions, we further detected an apoptosis-accelerating effect of PDCD5 on Tip60. Overexpression of PDCD5 can give cells a remarkable sensitivity to genotoxic treatments, and there was a synergistic effect when PDCD5 and Tip60 were co-overexpressed. Knockdown of endogenous PDCD5 could impair the Tip60-mediated apoptotic response including phosphatidylserine externalization and caspase-mediated PARP cleavage. These findings suggest that nuclear accumulation of endogenous PDCD5 in the preapoptotic or early apoptotic stage can enhance the PDCD5-Tip60 interaction and function as a cofactor for the Tip60-dependent apoptotic response.
Published data [33,36] demonstrate that Tip60 interacts physically with p53 and increases the stability and acetylation level of p53. Notably, Tip60-dependent acetylation of p53 at the K120 site is a key for p53-dependent apoptosis, through regulating expression of apoptosis gene, such as Bax. Our results, together with others, indeed demonstrate that Tip60 strongly induced K120 acetylation of exogenous p53 in H1299 cells. Moreover, PDCD5 overexpression can enhance this process, compared with Tip60 alone. Additionally, knockdown of PDCD5 in U2OS cells reduced the expression level of K120 acetylation with or without Tip60 overexpression (Figure 5C). At same time, depletion of PDCD5 also reduced the expression level of Bax protein in the presence of Tip60. Thus, we suggest that the interaction of PDCD5 and Tip60 in the nucleus may increase the acetyl-K120 form of p53 specifically accumulating at proapoptotic target genes, promote the ability of p53 to activate Bax expression, and subsequently increase apoptosis. Interestingly, we also found that PDCD5 has a potential ability to interact with p53 identified by GST pull-down assay (Figure W1). Our results raise the possibility that PDCD5, Tip60, and p53 should form a ternary complex, and there is an interregulatory network among them. PDCD5 plays a positive regulatory role in the Tip60-p53-apoptosis pathway. This needs more investigation and evidence in vitro and in vivo, especially evidences from PDCD5 transgenic mice or PDCD5 knockout mice.
Tip60 may influence tumorigenesis in multiple ways [37]. A recent report showed that Tip60, in both mice and humans, has haploinsufficient tumor suppressor activity and is required for the Myc-induced DNA damage response [38]. It has also been found that the human Tip60 gene is a frequent target for monoallelic loss in human lymphomas and head-and-neck and mammary carcinomas, with concomitant reduction in mRNA levels. In the present study, immunohistochemical analysis demonstrated loss of nuclear Tip60 staining in mammary carcinomas. These events correlated with disease grade and frequently concurred with mutations in p53. Regarding PDCD5, its expression level, at both the mRNA and protein levels, was lower in many carcinomas. We propose that decreased PDCD5 may impair Tip60 stability and the Tip60-mediated DNA damage response, thereby counteracting the Tip60-p53 apoptosis pathway or a tumor suppressor pathway. Key levels of Tip60, manipulated by PDCD5, are required to control incipient tumor cell development, the failure of which may synergize with Tip60 mutations, toward tumor progression.
Our findings are the first to demonstrate a role for PDCD5 in interfering with the expression of cellular genes, through modulation of the enzymatic activity of Tip60. PDCD5 may play a dual role in the Tip60 pathway: under normal growth conditions, PDCD5 contributes to maintaining a basal pool of Tip60 and HAT activity; after DNA damage, PDCD5 functions as Tip60 coactivator to promote apoptosis through the Tip60-p53 pathway. Further clarifying the relationship between the ectopic expression of PDCD5-Tip60 and carcinomas will be interesting.
Supplementary Material
Acknowledgments
The authors thank Steven B. McMahon for the generous gift of anti-acetyl k120-p53 antibody and Bruno Amati and Steven B. McMahon for the gifts of various plasmids used in this work. The authors thank Qihua He for the help with confocal imaging.
Footnotes
This work was supported by grants from the National Natural Sciences Foundation of China (30871263) and the National High Technology Research and Development Program of China (2006AA02A305).
This article refers to supplementary material, which is designated by Figure W1 and is available online at www.neoplasia.com.
References
- 1.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 2.Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 4.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 5.Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 7.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 9.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 10.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeRan M, Pulvino M, Greene E, Su C, Zhao J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol Cell Biol. 2008;28:435–447. doi: 10.1128/MCB.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 13.Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M, Trouche D. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 2004;279:44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 14.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Wang Y, Zhang Y, Song Q, Di C, Chen G, Tang J, Ma D. TFAR19, a novel apoptosis-related gene cloned from human leukemia cell line TF-1, could enhance apoptosis of some tumor cells induced by growth factor withdrawal. Biochem Biophys Res Commun. 1999;254:203–210. doi: 10.1006/bbrc.1998.9893. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, Ma D. An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci. 2004;117:1525–1532. doi: 10.1242/jcs.00994. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Sun R, Han W, Zhang Y, Song Q, Di C, Ma D. Nuclear translocation of PDCD5 (TFAR19): an early signal for apoptosis? FEBS Lett. 2001;509:191–196. doi: 10.1016/s0014-5793(01)03062-9. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Ruan G, Wang Y, Li Q, Zhu P, Qin YZ, Li JL, Liu YR, Ma D, Zhao H. Two single-nucleotide polymorphisms with linkage disequilibrium in the human programmed cell death 5 gene 5′ regulatory region affect promoter activity and the susceptibility of chronic myelogenous leukemia in Chinese population. Clin Cancer Res. 2005;11:8592–8599. doi: 10.1158/1078-0432.CCR-05-0039. [DOI] [PubMed] [Google Scholar]
- 20.Spinola M, Meyer P, Kammerer S, Falvella FS, Boettger MB, Hoyal CR, Pignatiello C, Fischer R, Roth RB, Pastorino U, et al. Association of the PDCD5 locus with lung cancer risk and prognosis in smokers. J Clin Oncol. 2006;24:1672–1678. doi: 10.1200/JCO.2005.04.4339. [DOI] [PubMed] [Google Scholar]
- 21.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 22.Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, Cai T, Zhang X, Xiao HS, Qu J, et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci USA. 2001;98:15089–15094. doi: 10.1073/pnas.241522398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang YH, Zhao M, Li WM, Lu YY, Chen YY, Kang B, Lu YY. Expression of programmed cell death 5 gene involves in regulation of apoptosis in gastric tumor cells. Apoptosis. 2006;11:993–1001. doi: 10.1007/s10495-006-6714-6. [DOI] [PubMed] [Google Scholar]
- 24.Ruan GR, Qin YZ, Chen SS, Li JL, Ma X, Chang Y, Wang YZ, Fu JY, Liu YR. Abnormal expression of the programmed cell death 5 gene in acute and chronic myeloid leukemia. Leuk Res. 2006;30:1159–1165. doi: 10.1016/j.leukres.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Wang Q, Gao F, Zhu F, Wang X, Zhou C, Liu C, Chen Y, Ma C, Sun W, et al. Reduced expression of PDCD5 is associated with high-grade astrocytic gliomas. Oncol Rep. 2008;20:573–579. [PubMed] [Google Scholar]
- 26.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Chen LN, Wang Y, Ma DL, Chen YY. Short interfering RNA against the PDCD5 attenuates cell apoptosis and caspase-3 activity induced by Bax overexpression. Apoptosis. 2006;11:101–111. doi: 10.1007/s10495-005-3134-y. [DOI] [PubMed] [Google Scholar]
- 28.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 29.Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002;21:1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logan IR, Sapountzi V, Gaughan L, Neal DE, Robson CN. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J Biol Chem. 2004;12:11696–11704. doi: 10.1074/jbc.M312712200. [DOI] [PubMed] [Google Scholar]
- 31.Halkidou K, Logan IR, Cook S, Neal DE, Robson CN. Putative involvement of the histone acetyltransferase Tip60 in ribosomal gene transcription. Nucleic Acids Res. 2004;32:1654–1665. doi: 10.1093/nar/gkh296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Z, Ke Y, Ding X, Wang F, Wang H, Wang W, Ahmed K, Liu Z, Xu Y, Aikhionbare F, et al. Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene. 2008;27(7):931–941. doi: 10.1038/sj.onc.1210710. [DOI] [PubMed] [Google Scholar]
- 33.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 35.Tyteca S, Vandromme M, Legube G, Chevillard-Briet M, Trouche D. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 2006;25:1680–1689. doi: 10.1038/sj.emboj.7601066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.