Abstract
Introduction
The expression, mechanisms of regulation, and functional impact of INHBA (activin A) in lung adenocarcinoma (AD) have not been fully elucidated.
Methods
INHBA expression was examined in 96 lung samples (86 ADs, 10 normal lung) using oligonucleotide microarrays and 187 lung samples (164 ADs, 6 bronchioalveolar carcinomas, and 17 normal lung) using immunohistochemistry. The proliferation of AD cell lines H460 and SKLU1 was examined with WST-1 assays after treatment with recombinant activin A, follistatin, and INHBA-targeting small-interfering RNA. Cells were also treated with 5-aza-2′ deoxycytidine and trichostatin A to investigate the role of epigenetic regulation in INHBA expression.
Results
Primary ADs expressed 3.1 times more INHBA mRNA than normal lung. In stage I AD patients, high levels of primary tumor INHBA transcripts were associated with worse prognosis. Immunohistochemistry confirmed higher inhibin βA protein expression in ADs (78.7%) and bronchioalveolar carcinomas (66.7%) compared with normal lung (11.8%). H460 and SKLU1 demonstrated increased proliferation when treated with exogenous activin A and reduced proliferation when treated with follistatin or INHBA-targeting small-interfering RNA. INHBA mRNA expression in H460 cells was upregulated after treatment with trichostatin A and 5-aza-2′ deoxycytidine.
Conclusions
INHBA is overexpressed in AD relative to controls. Inhibin βA may promote cell proliferation, and its overexpression is associated with worse survival in stage I AD patients. In addition, overexpression of INHBA may be affected by promoter methylation and histone acetylation in a subset of lung ADs.
Introduction
Lung cancer is currently the leading cause of cancer death in industrialized nations, with an overall 5-year survival of 15.5% [1]. Among non-small cell lung cancer variants, adenocarcinoma (AD) is the most common histologic subtype. Gene expression profiling has allowed for the simultaneous analysis of thousands of genes, and has been used to correlate lung AD expression patterns with clinical outcomes [2–4]. Elucidation of the molecular pathways that stimulate the development of lung AD will allow for improvements in prevention, diagnosis, and prognostication.
Inhibin βA (INHBA) is a ligand in the transforming growth factor β (TGF-β) superfamily [5]. Inhibin βA forms a disulfide-linked homodimer known as activin A, which was originally described in 1978 for its role in the hypothalamic-pituitary-gonadal axis [6,7]. Since that time, activin A has been implicated in multiple biologic processes, including neoplastic progression [8–13]. The inhibin βA subunit may also dimerize with the inhibin βB and inhibin α isoforms to form activin AB and inhibin A, respectively.
Overexpression of activin A in esophageal squamous cell carcinoma has been associated with advanced nodal status, clinical stage, and a worse overall prognosis [11]. In addition, chronic exposure to activin A promotes growth, tumorigenicity, invasion, and resistance to apoptosis in esophageal squamous cell carcinoma [12]. Inhibin α-knock-out mice express high levels of serum activin A and almost uniformly develop gonadal stromal tumors, supporting the role of activin A overexpression in tumorigenesis [13]. Furthermore, overexpression of activin A has been reported in colon, prostate, pancreatic, and ovarian cancers [14–17], and women with endometrial or cervical cancer exhibit elevated serum concentrations of activin [18]. However, the pleiotropic capabilities of activin A are evidenced by its growth-inhibitory effects on prostate cancer cells, vascular endothelial cells, mammary epithelial cells, plasmacytomas, and hepatocytes [19–24]. Activin A has also been shown to induce endodermal differentiation in human embryonic stem cells [25].
The role of INHBA in the pathogenesis of lung AD has not been fully elucidated. A recent study found that INHBA was one of 26 significantly mutated genes in lung AD and that INHBA mutation frequency correlated with tumor grade, suggesting a role in tumor progression [26]. Here, we demonstrate that INHBA is overexpressed in >70% of lung AD and that transcript expression inversely correlates with survival in patients with stage I disease. Our data also suggest that lung AD cell proliferation may be partially dependent on the presence of activin A. Finally, we provide evidence that an upregulated expression of INHBA in lung AD cells may be related to promoter demethylation and histone acetylation.
Materials and Methods
Patients and Tissues
The oligonucleotide microarray and tissue microarray (TMA) lung specimens were obtained from consented patients with either stage I or stage III primary lung AD who underwent surgery at the University of Michigan Health System between 1994 and 2000. Approval for this study was obtained by the local institutional review board. Tissues were transported on ice in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Inc, Carlsbad, CA), immediately frozen in liquid nitrogen, and stored at -80°C. Tissues demonstrating AD and at least 70% cellularity were identified on hematoxylin and eosin-stained frozen sections. Macrodissection was used to obtain samples 2 to 3 mm3 in size for RNA and protein isolation. The histologic diagnosis of each specimen was independently confirmed by two pathologists (T.J.G and D.G.T). Patients with stage I disease received surgery alone, whereas patients with stage III disease received surgery plus neoadjuvant chemotherapy and radiotherapy.
Cell Lines
The cell lines used in this study, H460 and SKLU1, were derived from lung AD, and have been previously described [27,28]. Both cell lines were maintained in DMEM (Invitrogen Corp., Carlsbad, CA) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin/fungizone (Life Technologies, Inc) at 37°C in 5% CO2. Cells were treated at 70% confluence.
Oligonucleotide Microarray
The RNA was isolated, cRNA was synthesized, and oligonucleotide microarrays were analyzed as previously described [29]. Results from this data set have been previously published elsewhere [2].
Immunohistochemistry of Tissue Microarray
Immunohistochemical staining was performed on the DAKO Autostainer (DAKO, Carpinteria, CA) using DAKO LSAB+ and diaminobenzadine as the chromogen, as previously described [29]. Deparaffinized sections of the TMA at 5-µm thickness were labeled with inhibin βA antibody (catalog no. MAB3381; R&D Systems, Minneapolis, MN; mouse, 1:20) after microwave antigen retrieval in 10 mM sodium citrate buffer, pH 6. Negative (no primary antibody) and positive (placenta) controls were used. The intensity and extent of immunoreactivity were scored using a three-tier (negative [-], moderate [1+], and intense [2+]) grading scheme. All tissues were examined using an Olympus BX40 microscope (Olympus, Center Valley, PA). Images were acquired using a Spot Insight model 3.2.0 camera (Diagnostic Instruments, Inc, Sterling Heights, MI) and Spot version 4.0.9 (Diagnostic Instruments, Inc) software.
Immunohistochemistry of Cell Lines
After plating on poly-l-lysine-coated glass slides, cells were air-dried and fixed in -20°C acetone for 10 minutes. Endogenous peroxidases were inactivated using 0.5% hydrogen peroxide. Cells were stained with the Vectastain ABC Kit (catalog no. PK-6102, mouse IgG; Vector Laboratories, Burlingame, CA) and Peroxidase Substrate Kit (catalog no. SK-4100; Vector Laboratories) according to the manufacturer's instructions. Inhibin βA antibody (catalog no. MAB3381; R&D Systems) at a concentration of 1:20 was used as the primary antibody. Negative controls (no primary antibody) were stained with each immunohistochemical assay.
Western Blot Analysis
Western blot analysis was performed as previously described with slight variations [30]. Briefly, 15 µl of FBS or calf serum (CS) was loaded onto a 10% SDS-polyacrylamide gel and electrophoresed. A 1:200 (2 µg/ml) concentration of inhibin βA antibody (catalog no. MAB3381; R&D Systems) and a 1:10,000 concentration of secondary antibody (catalog no. NA931V, goat antimouse; GE Healthcare UK Limited, Chalfont St. Giles, United Kingdom) were used for protein detection.
Real-time Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted using the RNeasy Mini Kit (catalog no. 74104; Qiagen, Valencia, CA), and cDNA was created using the Superscript II Kit (catalog no. 18064; Invitrogen) according to the manufacturer's instructions. Real-time reverse transcription-polymerase chain reaction (RT-PCR) was performed using the Rotor-Gene 6000 (Corbett Life Science, Sydney, Australia) with the Platinum SYBR Green kit (catalog no. 11733-038; Invitrogen). The INHBA oligo primers for real-time RT-PCR were obtained from Invitrogen and included forward INHBArt2f (5′-AAG TCG GGG AGA ACG GGT ATG TGG-3′) and reverse INHBArt2r (5′-TCT TCC TGG CTG TTC CTG ACT CG-3′). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression was used to standardize the INHBA results using GAPDH primers obtained from Invitrogen. Optimal annealing temperatures were determined, and melt curves were analyzed to ensure RT-PCR results.
WST-1 Cell Proliferation Assays
Cell viability and proliferation were assessed using Cell Proliferation Reagent WST-1 (catalog no. 11644807001; Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. The experiments were performed in 96-well plates at two different cell densities. H460 cells were plated at 1.0 x 103 and 2.0 x 103 cells per well, whereas SKLU1 cells were plated at 1.5 x 103 and 2.5 x 103 cells per well. Absorbance was measured on a Biotek EL-312e microplate reader (Biotek Instruments, Winooski, VT) at 96 hours. All activin A and follistatin treatment experiments were performed in quadruplicate.
Treatment of H460 and SKLU1 with Activin A and Follistatin
Twenty-four hours after plating, H460 and SKLU1 cells were treated with 1, 10, and 100 ng/ml recombinant activin A (catalog no. 120-14; Peprotech, Inc, Rocky Hill, NJ) in DMEM with 10% FBS or 10% CS. Similarly, H460 and SKLU1 were treated with 1, 10, and 100 ng/ml follistatin (catalog no. 120-13; Peprotech, Inc) in DMEM with 10% FBS.
Transfection of H460 Cells with INHBA-targeting Small-interfering RNA
Transient transfection of H460 cells with INHBA-targeting small-interfering RNA (siRNA) was performed as previously described [31]. Briefly, H460 cells were trypsinized at 70% confluence and plated at a density of 2.0 x 103 cells per well in DMEM with 10% FBS in a 96-well format for determination of the optimal siRNA transfection concentration. Twenty-four hours after plating, cells were treated with INHBA-targeting SMART pool siRNA (Dharmacon, Lafayette, CO) in the presence of 0.1 µl of Lipofectamine RNAiMAX (Invitrogen) in 9.9 µl of Opti-MEM I (Invitrogen). INHBA knockdown efficiency was determined by real-time RT-PCR. An optimal working concentration of 10 nM siRNA was used to perform WST-1 cell proliferation assays. Forty-eight hours after initial transfection, half of the plates were transfected with a second dose of 10 nM INHBA-targeting siRNA. Transfected cells were incubated for a total of 96 hours at 37°C before gene silencing analysis. Cells treated only with transfection reagent were used as a control. All siRNA transfection experiments were performed in quadruplicate.
Treatment of H460 and SKLU1 with Trichostatin A and 5-Aza-2′ Deoxycytidine
H460 and SKLU1 cells were serum-starved for 24 hours before treatment to synchronize cell cycles. Cell lines were then treated with trichostatin A (TSA) at a final concentration of 300 nM or with 5-aza-2′ deoxycytidine (5-AZA) at a final concentration of 5 µM in DMEM with 10% FBS. Cells receiving combination treatment were exposed to both 300 nM TSA and 5 µM 5-AZA. Trichostatin A and 5-AZA were dissolved in DMSO (catalog no. D4540; Sigma Aldrich, St. Louis, MO), and control cells were treated with equivalent amounts of DMSO. Cells treated with TSA were trypsinized and harvested at 12 and 24 hours, and those treated with 5-AZA were harvested at 48, 72, and 96 hours. Combination plates were treated with 5-AZA for 48, 72, or 96 hours and TSA for 24 hours. Cell pellets for mRNA analysis were frozen in liquid nitrogen and stored in -80°C until mRNA was extracted for real-time RT-PCR. All experiments examining the effects of TSA and 5-AZA were performed in duplicate. Cells for immunohistochemistry (IHC) analysis were immediately suspended in PBS and plated on poly-l-lysine-coated slides using a Shandon Cytospin 3 machine (Thermo Fisher Scientific, Waltham, MA).
Correlation between INHBA and Histone Deacetylase, Histone Acetyltransferase, and DNA Methyltransferase Expression
To support a role of histone acetylation and promoter methylation in the regulation of INHBA expression, we examined the oligonucleotide microarray expression of histone deacetylases (HDACs), histone acetyltransferases, and DNA methyltransferases (DNMTs). However, the Affymetrix HuFL microarray (Affymetrix, Santa Clara, CA) [2] included only one DNMT and three HDAC probe sets: DNMT1, HDAC1, HDAC2, and HDAC3. Therefore, we repeated the analysis with two publicly available expression data sets that used the Affymetrix HG-U133A oligonucleotide microarray. These data were obtained from the Gene Expression Omnibus [32]. One of the data sets used included primary lung AD and normal lung tissue [33], whereas the second included primary lung AD and lung squamous cell carcinomas [34]. For the first data set, all normal lung tissue data were excluded, and for the second data set, all squamous cell carcinoma data were excluded before analysis.
Statistical Analysis
Kaplan-Meier survival curves and the log-rank test were used to determine the association between patient survival and INHBA expression. The t-test was used to determine the significance of changes in proliferation associated with treatment. Spearman analysis was used to correlate INHBA gene expression with that of the HDACs, histone acetyltransferases, and DNMTs. P < .05 was deemed significant. All statistical analyses were performed using R version 2.7.0 (Vienna, Austria).
Results
INHBA Is Overexpressed in Lung AD and Correlates with Worse Outcome in Stage I Disease
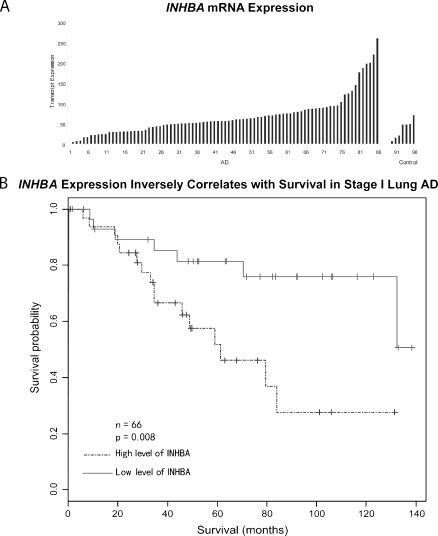
Affymetrix oligonucleotide microarrays were used to measure INHBA expression in 86 AD and 10 normal lung samples (Figure 1A). Two-fold more INHBA mRNA was detected relative to the mean of the normal lung tissue in 61 (70.9%) of 86 samples and at least four-fold more in 17 (19.7%) of 86 samples. On average, the AD samples contained 3.1-fold more INHBA transcripts than the normal lung tissue samples (67.2 vs 21.8, P = .001).
Figure 1.
(A) Affymetrix oligonucleotide microarray analysis of 86 lung AD (AD) and 10 nontumor control specimens demonstrated at least a two-fold increase in expression in 61 (70.9%) of 86 and a four-fold increase in expression in 17 (19.7%) of 86 lung AD relative to controls. The x-axis represents tumor identifier. (B) Kaplan-Meier survival analysis of 66 stage I lung AD demonstrated reduced survival in patients with high INHBA expression.
Kaplan-Meier survival analysis of the 66 stage I patients demonstrated that patients with high levels of INHBA transcripts (greater than median = 57.6; n = 33) had a significantly worse prognosis than those with low levels (less than median = 57.6; n = 33; P = .008; Figure 1B). The number of stage IA and IB patients was approximately equal between the low and high INHBA-expressing groups (23 low IA vs 20 high IA; 10 low IB vs 13 high IB). INHBA mRNA expression was not related to survival in the 20 stage III patients (P = .3).
Inhibin βA Expression Confirmed on Lung TMA
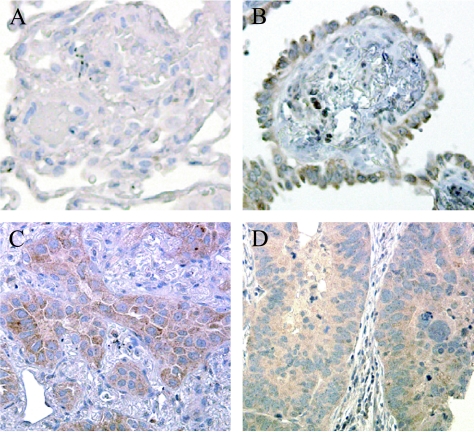
Immunohistochemical staining of a TMA containing 164 AD, 6 bronchioalveolar carcinoma (BAC), and 17 normal lung samples showed higher inhibin βA expression in AD and BAC relative to that of normal lung. All inhibin βA staining was noted to be cytoplasmic, and no correlation was noted between the intensity of staining and the differentiation or histologic subtype of AD. Immunoreactivity was found in 78.7% (moderate: 129/164, intense: 20/164) of AD (Figure 2, C and D) and 66.7% (moderate: 2/6, intense: 2/6) of BAC (Figure 2B). Only 11.8% (moderate: 2/17) of normal lung samples demonstrated inhibin βA staining, with no normal lung samples demonstrating intense staining (Figure 2A).
Figure 2.
Representative sections of TMA. Inhibin βA protein staining was not detected in normal lung tissues (A) but was seen in 66.7% (4/6) of BAC (B) and in 78.7% of lung AD by IHC (C and D). Original magnification, x40.
Activin A Induced Proliferation in H460 and SKLU1 Cells
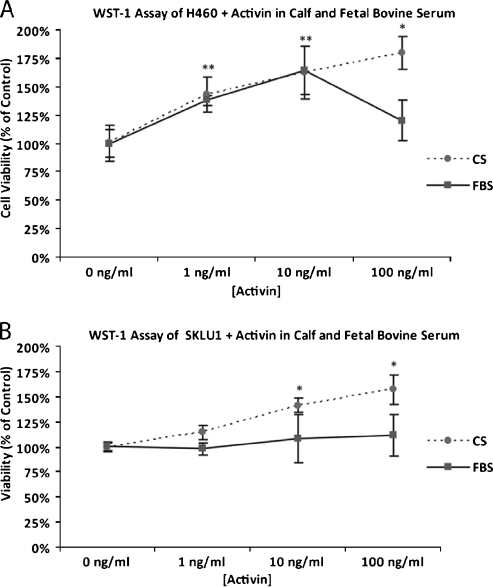
Adenocarcinoma cell lines H460 and SKLU1 treated with recombinant activin A at concentrations ranging from 1 to 100 ng/ml demonstrated increased proliferation relative to untreated controls at 96 hours. The effect of activin A treatment was greater in the presence of CS than in the presence of FBS in both cell lines. H460 cells demonstrated the greatest growth-proliferative response after treatment with 100 ng/ml activin A in CS (180%) and with 10 ng/ml activin in FBS (163%; Figure 3A). Treatment of SKLU1 cells with 100 ng/ml activin A induced the greatest growth-proliferative response in both CS and FBS, at 157% and 112% relative to controls, respectively (Figure 3B). The growth-proliferative response was inversely related to the baseline INHBA transcript expression as assessed by real-time RT-PCR. H460 cells demonstrated low INHBA transcript levels (Ct = 26.6, GAPDH = 11.8), whereas SKLU1 cells had moderate INHBA transcript expression (Ct = 18, GAPDH = 10.6). At baseline, neither H460 nor SKLU1 cells demonstrated inhibin βA immunoreactivity on Western blot or IHC.
Figure 3.
H460 (A) and SKLU1 (B) cells treated with recombinant activin A demonstrated increased proliferation at 96 hours. The effect of activin A treatment was greater in the presence of CS than in the presence of FBS. *P < .05.
Inhibin βA Is More Abundant in FBS Than CS
We suspected that the attenuated growth response seen in FBS with activin A treatment was due to a higher inhibin βA concentration in FBS than CS. On Western blot analysis, inhibin βA was seen in all four replicate FBS lanes but in none of the four CS lanes, suggesting a higher concentration of activin A in FBS than in CS. This finding was replicated and confirmed with different batches of FBS and CS (data not shown).
Inhibin βA Inhibitor Follistatin Reduces Proliferation in H460 and SKLU1 Cells
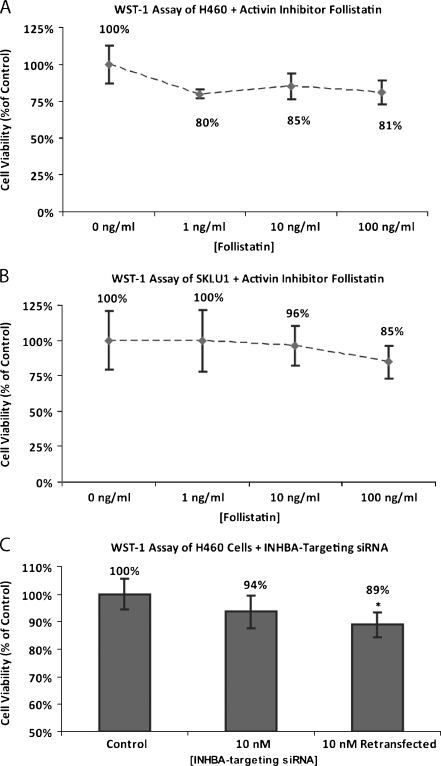
AD cell lines H460 and SKLU1 treated with activin A inhibitor follistatin at a concentration ranging from 1 to 100 ng/ml demonstrated modestly reduced cell proliferation relative to untreated controls at 96 hours. The greatest growth-inhibitory response in H460 cells was demonstrated with 1 ng/ml follistatin (80%). Higher concentrations did not further reduce cell growth (Figure 4A). Conversely, treatment of SKLU1 cells with follistatin inhibited proliferation in a dose-dependent manner, with the greatest reduction seen at 100 ng/ml (Figure 4B).
Figure 4.
H460 (A) and SKLU1 (B) cells treated with activin inhibitor follistatin demonstrated modestly reduced proliferation at 96 hours. (C) H460 cells treated with 10 nM INHBA-targeting siRNA demonstrated reduced cell proliferation at 96 hours relative to mock controls. Repeat transfection 48 hours after initial transfection resulted in further growth inhibition relative to controls.
Transfection of H40 with INHBA-Targeting siRNA Reduces INHBA Expression and Cell Proliferation
As assessed using real-time RT-PCR, an INHBA-silencing efficiency of 93% relative to mock controls was achieved by transfecting H460 cells with INHBA-targeting siRNA at a concentration of 10 nM. This concentration of INHBA-targeting siRNA produced a 6% reduction in proliferation at 96 hours. When cells were retransfected 48 hours after the initial transfection, an 11% reduction in proliferation was observed at 96 hours (Figure 4C).
Epigenetically Mechanisms May Influence INHBA Expression in Lung AD Cell Lines
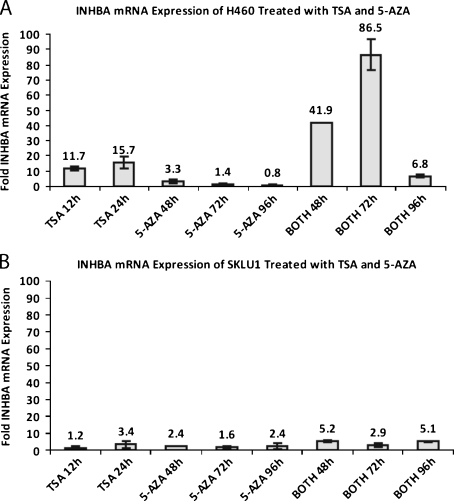
We treated H460 and SKLU1 cells with 5-AZA and TSA to determine whether epigenetic regulation influenced INHBA expression. H460, a cell line with low baseline INHBA expression, demonstrated a 15.7-fold (range, 13.4- to 18-fold) increase in mRNA production relative to the mean of the controls with 24 hours of TSA exposure (Figure 5A). When cells were treated with a combination of TSA for 24 hours and 5-AZA for 72 hours, an 86.5-fold (range, 79.3- to 93.7-fold) increase in INHBA mRNA expression was observed. SKLU1 cells, which demonstrated higher baseline INHBA expression, showed a blunted INHBA up-regulation on treatment with TSA and 5-AZA (Figure 5B). Treatment with TSA and 5-AZA, alone or in combination, induced a 1.2- to 5.2-fold increase in SKLU1 INHBA mRNA production. The INHBA mRNA up-regulation observed in the H460 and SKLU1 cells were not able to be demonstrated on Western blot or IHC.
Figure 5.
INHBA mRNA expression in H460 (A) and SKLU1 (B) cells was upregulated on treatment with HDAC inhibitor TSA and demethylating agent 5-AZA. “Both” refers to the combination of TSA and 5-AZA treatment.
Correlation between INHBA and DNMT1 and HDAC6 Expression
We compared the expression of DNMT1, HDAC1, HDAC2, and HDAC3 on the original oligonucleotide microarray with INHBA expression using Spearman correlation analysis. DNMT1 was found to have a strong negative correlation with INHBA expression (P = .003). However, no HDACs included on the microarray correlated with INHBA expression. We then performed the analysis using two publicly available Affymetrix HG-U133A lung cancer data sets. DNMT1 was confirmed to have a significant negative correlation with INHBA expression in the first lung AD data set [32] (P = .026, n = 58). In addition, significantly negative correlations were found between INHBA and HDAC6 expression in both data sets [32,33] (P = .014 and P = .029 [n = 60], respectively).
Discussion
We have demonstrated that INHBA mRNA and protein are overexpressed in primary lung AD relative to normal lung tissue. On Kaplan-Meier analysis, the level of INHBA mRNA expressed by primary ADs inversely correlates with patient survival in stage I disease. The prognostic capability of INHBA may be related to the proliferative effect activin A exposure has on AD cells. Furthermore, AD activin dependency is suggested by the decrease in proliferation seen with follistatin and INHBA-targeting siRNA treatment in AD cell lines.
Interestingly, exposure of H460 cells to high doses of exogenous activin A in the presence of FBS resulted in a blunted proliferative response. This effect was not seen when the cells were treated in the presence of CS, which, unlike FBS, contains no detectable activin A by Western blot. These findings suggest that at high concentrations, activin A might exert growth-inhibitory effects on some cell lines. Activin A's complex function is characteristic of members of the TGF-β superfamily, which are known to have both tumor suppressive and oncogenic effects. In the current paradigm, the tumor suppressor activities of the TGF-β cascade dominate in normal tissues, but during tumorigenesis, changes in cellular response may promote oncogenesis [35,36]. Activin A seems to exert a similar dose-dependent dual effect on H460 cells, which do not express large amounts of activin A. The moderate INHBA-expressing cell line, SKLU1, demonstrated a less pronounced proliferative response when treated with activin A. However, the growth-proliferative effects of activin are seen in a dose-dependent manner, suggesting that SKLU1 cells tolerate higher concentrations of activin without growth inhibition.
Although the levels of specific growth factors are not available for most commercially obtained serums, multiple studies have demonstrated that the concentration of activin A rises during pregnancy in maternal serum and is detectable at high levels in amniotic fluid and fetal serum [37–43]. In addition, studies have shown that deletion of INHBA in mice results in severe craniofacial abnormalities, including cleft palate and lack of lower incisors. Furthermore, lack of type IIA activin receptors in fetal mice results in impaired reproductive function [44,45]. Such evidence implicates the role of activin A in fetal development and is consistent with our Western blots demonstrating the presence of inhibin βA in FBS.
Follistatin is a 32- to 39-kDa monomeric protein that was first identified as a potent inhibitor of pituitary follicle-stimulating hormone secretion [46] and subsequently found to serve as an activin A-binding compound [47]. Follistatin binds activin A with high affinity (50–500 pM) to form inactive complexes, thereby regulating the function of activin A [48]. When we treated H460 and SKLU1 cells with follistatin, a moderate decrease in proliferation was seen in both cell lines, which was consistent with activin A inhibition. We confirmed the growth-inhibitory effects of activin A inhibition by treatment of H460 with INHBA-targeting siRNA. Although follistatin does bind activin B and some members of the bone morphogenetic protein family with very low affinity and siRNA transfection has the potential to exert off-target effects, our data consistently demonstrated a reduction in cell growth with differing methods of activin A inhibition. The growth reduction consistently seen with activin A inhibition suggests that agents targeting INHBA may be therapeutically useful.
The inhibin βA subunit may also dimerize with inhibin α and inhibin βB subunits to form inhibin A and activin AB, respectively. Within the reproductive axis, inhibin A and activin A oppose each other in the control of steroid, androgen, and placental hormone production [49]. Interestingly, on oligonucleotide microarray analysis, our ADs expressed approximately half the a subunit mRNA transcripts as the normal tissue samples. In addition, a direct correlation was identified between survival and α subunit expression when the 66 stage I ADs (P = .017) and all 86 ADs (P = .008) were examined. Assuming inhibin A and activin A are similarly antagonistic in the regulation of AD growth, these correlations may represent inhibition of activin A's proliferative effect on AD cells.
INHBA mRNA expression has prognostic significance in patients with stage I AD, but not stage III AD, most likely because mediastinal lymph node spread is a stronger predictor of survival than primary tumor growth rate. Perhaps, by the time that AD gain metastatic potential, the proliferative rate of the primary tumor, which INHBA expression may be associated with, does not alter outcome. The prognostic significance of INHBA mRNA expression in stage I AD, combined with its growth-proliferative effects in two AD cell lines, prompted us to explore potential mechanisms for overexpression. Previous reports have established that epigenetic mechanisms affect patterns of gene expression in normal and neoplastic tissues [50,51]. Our cell line work and microarray correlation data suggest that the overexpression seen in a subset of primary tumors might be due to histone acetylation and/or promoter demethylation. The greatest increase in INHBA mRNA expression occurred in the low-INHBA expressing cell line, H460, after a combination of 72 hours of 5-AZA and 24 hours of TSA exposure. Treatment of H460 with 5-AZA beyond 72 hours, alone or in combination with TSA, led to a smaller increase in INHBA expression. Because 5-AZA is a global demethylating agent, prolonged exposure may also affect promoters of other genes that result in the down-regulation of INHBA transcription. The overall blunted up-regulation of INHBA transcription seen in SKLU1 compared with H460 may be a reflection of its higher baseline expression, thus resulting in a smaller fold increase.
Although 70% to 80% of primary AD overexpressed INHBA mRNA and protein, inhibin βA immunoreactivity was not observed on Western blot or IHC of H460 and SKLU1 cells treated with TSA and 5-AZA. The basis for this difference is not clear; however, the low-to-moderate baseline production of INHBA in both cell lines may allow for a significant relative up-regulation of mRNA but an absolute increase that does not translate into a detectable difference in protein production. Our inability to detect the up-regulation of inhibin βA protein in the cell lines may also be related to the soluble nature of activin A and its known secretion from the cell. Differences between the in vivo and in vitro microenvironment may also play a role and posttranscriptional interference. Forty-nine microRNA are predicted to bind to the INHBA mRNA, suggesting an area for future investigation [52].
INHBA may represent a novel target in the treatment of lung AD. It is clear that INHBA overexpression is not the sole mechanism for the uncontrolled growth of lung AD because some ADs do not express high levels of INHBA and inhibition of INHBA does not fully arrest AD growth. However, our data suggest that activin A exposure does promote proliferation and that lung AD cells are partially dependent on activin A exposure. Activin-binding agents, such as follistatin, monoclonal antibodies, and small molecule inhibitors, all hold therapeutic potential, especially in combination with other treatments. However, because INHBA is expressed in a wide array of human tissues and has diverse biologic functions, the feasibility of systemic therapy needs further investigation before in vivo application. This study provides data to support continued investigation into the role of INHBA in the progression of lung AD.
Abbreviations
- 5-AZA
5-aza-2′ deoxycytidine
- AD
lung adenocarcinoma
- BAC
bronchioalveolar carcinoma
- CS
calf serum
- DMEM
Dulbecco's modified Eagle's medium
- DNMT
DNA methyltransferase
- FBS
fetal bovine serum
- HDAC
histone deacetylase
- IHC
immunohistochemistry
- RT-PCR
reverse transcription-polymerase chain reaction
- TGF-β
transforming growth factor β
- TMA
tissue microarray
- TSA
trichostatin A
Footnotes
Grant support: National Cancer Institute Early Detection Research Network (EDRN) Biomarker Development Laboratory grant EDRN 051717 (D.G.B.); National Institutes of Health through the University of Michigan's Cancer Center support grant 5 P30 CA46592. The authors have no potential conflict of interest relevant to this article.
References
- 1.Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, Howlader N, Homer MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, et al. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD: National Cancer Institute; [April 1, 2008]. Available at: http://seer.cancer.gov/csr/1975_2005/. [Based on November 2007 SEER data submission, posted to the SEER Web site] [Google Scholar]
- 2.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaddy-Kurten D, Tsuchida K, Vale W. Activins and the receptor serine kinase superfamily. Recent Prog Horm Res. 1995;50:109–129. doi: 10.1016/b978-0-12-571150-0.50010-x. [DOI] [PubMed] [Google Scholar]
- 6.Vale W, Rivier C, Hsueh A, Campen C, Meunier H, Bicsak T, Vaughan J, Corrigan A, Bardin W, Sawchenko P, et al. Chemical and biological characterization of the inhibin family of protein hormones. Recent Prog Horm Res. 1988;44:1–34. doi: 10.1016/b978-0-12-571144-9.50005-3. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzen JR, Channing CP, Schwartz NB. Partial characterization of FSH suppressing activity (follicostatin) in porcine follicular fluid using the metestrous rat as an in vivo bioassay model. Biol Reprod. 1978;19:635–640. doi: 10.1095/biolreprod19.3.635. [DOI] [PubMed] [Google Scholar]
- 8.Boitani C, Stefanini M, Fragale A, Morena AR. Activin stimulates Seroli cell proliferation in a defined period of rat testis development. Endocrinology. 1995;136:5438–5444. doi: 10.1210/endo.136.12.7588293. [DOI] [PubMed] [Google Scholar]
- 9.Green JB, New HV, Smith JC. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 10.Smith JC, Price JMB, van Nimmen K, Huylebroeck D. Identification of a potent Xenopus mesoderm-inducing factor as a homologue of activin A. Nature. 1990;345:729–731. doi: 10.1038/345729a0. [DOI] [PubMed] [Google Scholar]
- 11.Yoshinaga K, Mimori K, Yamashita K, Utsunomiya T, Inoue H, Mori M. Clinical significance of the expression of activin A in esophageal carcinoma. Int J Oncol. 2003;22:75–80. [PubMed] [Google Scholar]
- 12.Yoshinaga K, Yamashita K, Mimori K, Tanaka F, Inoue H, Mori M. Activin A causes cancer cell aggressiveness in esophageal squamous cell carcinoma cells. Ann Surg Onc. 2008;15:96–103. doi: 10.1245/s10434-007-9631-1. [DOI] [PubMed] [Google Scholar]
- 13.Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumor suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 14.Thomas TZ, Wang H, Niclasen P, O'Bryan MK, Evans LW, Groome NP, Pedersen J, Risbridger GP. Expression and localization of activin subunits and follistatins in tissues from men with high grade prostate cancer. J Clin Endocrinol Metab. 1997;82:3851–3858. doi: 10.1210/jcem.82.11.4374. [DOI] [PubMed] [Google Scholar]
- 15.Kleeff J, Ishiwata T, Friess H, Büchlor MW, Koro M. Concomitant over-expression of activin/inhibin beta subunits and their receptors in human pancreatic cancer. Int J Cancer. 1998;77:860–868. doi: 10.1002/(sici)1097-0215(19980911)77:6<860::aid-ijc11>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Zheng W, Luo MP, Welt C, Lambert-Messerlian G, Sung CJ, Zhang Z, Ying SY, Schneyer AL, Lauchlan SC, Felix JC. Imbalanced expression of inhibin and activin subunits in primary epithelial ovarian cancer. Gynecol Oncol. 1998;69:23–31. doi: 10.1006/gyno.1998.4958. [DOI] [PubMed] [Google Scholar]
- 17.Wildi S, Kleeff J, Maruyama H, Maurer CA, Buchler MW, Korc M. Overexpression of activin A in stage IV colorectal cancer. Gut. 2001;49:409–417. doi: 10.1136/gut.49.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petraglia F, Florio P, Luisi S, Gallo R, Gadducci A, Viganò P, Di Blasio AM, Genazzani AR, Vale W. Expression and secretion of inhibin and activin in normal and neoplastic uterine tissues. High levels of serum activin A in women with endometrial and cervical carcinoma. J Clin Endocrinol Metab. 1998;33:1194–1200. doi: 10.1210/jcem.83.4.4689. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Zhao Y, Batres Y, Lin MF, Ying SY. Regulation of growth and prostatic marker expression by activin A in a androgen sensitive prostate cancer cell line LNCaP. Biochem Biophys Res Comm. 1997;234:362–365. doi: 10.1006/bbrc.1997.6649. [DOI] [PubMed] [Google Scholar]
- 20.Dalkin AC, Gilrain JT, Bradshaw D, Myers CE. Activin inhibition of prostate cancer cell growth: selective actions on androgen-responsive LNCaP cells. Endocrinology. 1996;137:5230–5235. doi: 10.1210/endo.137.12.8940339. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy SA, Bicknell R. Inhibition of vascular endothelial cell growth by activin A. J Biol Chem. 1993;268:23066–23071. [PubMed] [Google Scholar]
- 22.Liu QY, Niranjan B, Gomes P, Gomm JJ, Davies D, Coombes RC, Buluwela L. Inhibitory effects of activin on the growth and morphogenesis of primary transformed mammary epithelial cells. Cancer Res. 1996;56:1155–1163. [PubMed] [Google Scholar]
- 23.Sternberg D, Honigwachs-Sha'anani J, Brosh N, Malik Z, Burstein Y, Zipori D. Restrictin-P/stromal activin A, kills its target cells via an apoptotic mechanism. Growth Factors. 1995;12:277–287. doi: 10.3109/08977199509028966. [DOI] [PubMed] [Google Scholar]
- 24.Yasuda H, Mine T, Shibata H, Eto Y, Hasegawa Y, Takeuchi T, Asano S, Kojima I. Activin-A: an autocrine inhibitor of initiation of DNA synthesis in rat hepatocytes. J Clin Invest. 1993;92:1491–1496. doi: 10.1172/JCI116727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, Zweig MH, Minna JD. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 28.Chen G, Wang H, Gharib TG, Huang CC, Thomas DG, Shedden KA, Kuick R, Taylor JM, Kardia SL, Misek DE, et al. Overexpression of oncoprotein 18 correlates with poor differentiation in lung adenocarcinomas. Mol Cell Prot. 2003;2:107–116. doi: 10.1074/mcp.M200055-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JM, Lee N, Misek DE, Greenson JK, Kardia SL, Beer DG, et al. Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol. 2001;159:1231–1238. doi: 10.1016/S0002-9440(10)62509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes SJ, Nambu Y, Soldes OS, Hamstra D, Rehemtulla A, Iannettoni MD, Orringer MB, Beer DG. Fas/APO-1 (CD95) is not translocated to the cell membrane in esophageal adenocarcinoma. Cancer Res. 1997;57:5571–5578. [PubMed] [Google Scholar]
- 31.Lin J, Raoof DA, Wang Z, Lin MY, Thomas DG, Greenson JK, Giordano TJ, Orringer MB, Chang AC, Beer DG, et al. Expression and effect of inhibition of the ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia. 2006;8:1062–1071. doi: 10.1593/neo.05832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. [June 24, 2008.];Gene Expression Omnibus Web site. Available at: http://www.ncbi.nlm.nih.gov/geo/
- 33.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son D, Kim S, Jo J, Kim H, Choi YH, Jung Y, Park M, Lim Y, Lee J, Lee E, et al. [June 24, 2008];Prediction of Recurrence-Free Survival in Postoperative NSCLC Patients—a Useful Prospective Clinical Practice. Available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE8894.
- 35.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 36.Massagué J, Blain SW, Lo RS. TGF-beta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 37.Petraglia F, Garg S, Florio P, Sadick M, Gallinelli A, Wong WL, Krummen L, Comitini G, Mather J, Woodruff TK. Activin A and activin B measured in maternal serum, cord blood serum and amniotic fluid during human pregnancy. Endocr J. 1993;1:323–327. [Google Scholar]
- 38.Riley SC, Balfour C, Wathen NC, Chard T, Evans LW, Groome NP, Wallace EM. Follistatin and activin A in extra-embryonic coelomic and amniotic fluids and maternal serum in early pregnancy. Hum Reprod. 1998;13:2624–2628. doi: 10.1093/humrep/13.9.2624. [DOI] [PubMed] [Google Scholar]
- 39.Luisi S, Battaglia C, Florio P, D'Ambrogio G, Taponeco F, Santuz M, Genazzani AR, Petraglia F. Activin A and inhibin B in extra-embryonic coelomic and amniotic fluids, and maternal serum in early pregnancy. Placenta. 1998;19:435–438. doi: 10.1016/s0143-4004(98)90085-6. [DOI] [PubMed] [Google Scholar]
- 40.Fowler PA, Evans LW, Groome NP, Templeton A, Knight PG. A longitudinal study of maternal serum inhibin-A, inhibin-B, activin-A, activin-AB, pro-C and follistatin during pregnancy. Hum Reprod. 1998;13:3530–3536. doi: 10.1093/humrep/13.12.3530. [DOI] [PubMed] [Google Scholar]
- 41.Wallace EM, D'Antona D, Shearing C, Evans LW, Thirunavukarasu P, Ashby JP, Shade M, Groome NP. Amniotic fluid levels of dimeric inhibins, pro-C inhibin, activin A and follistatin in Down's syndrome. Clin Endocrinol. 1999;50:669–673. doi: 10.1046/j.1365-2265.1999.00716.x. [DOI] [PubMed] [Google Scholar]
- 42.Muttukrishna S, Chamberlain P, Evans LW, Asselin J, Groome NP, Ledger WL. Amniotic fluid concentrations of dimeric inhibins, activin A and follistatin in pregnancy. Eur J Endocrinol. 1999;140:420–424. doi: 10.1530/eje.0.1400420. [DOI] [PubMed] [Google Scholar]
- 43.Debieve F, Beerlandt S, Hubinont C, Thomas K. Gonadotropins, prolactin, inhibin A, inhibin B, and activin A in human fetal serum from mid-pregnancy and term pregnancy. J Clin Endocrinol Metab. 2000;85:270–274. doi: 10.1210/jcem.85.1.6249. [DOI] [PubMed] [Google Scholar]
- 44.Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, Bradley A. Functional analysis of activins during mammalian development. Nature. 1995;374:354–356. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- 45.Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 46.Ueno N, Ling N, Ying S-Y, Esch F, Shimasaki S, Guillemin R. Isolation and partial characterization of follistatin: a single-chain 35,000 monomeric protein that inhibits the release of follicle stimulating hormone. Proc Natl Acad Sci USA. 1987;84:8282–8286. doi: 10.1073/pnas.84.23.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura T, Takio K, Eto Y, Shibai H, Titani K, Sugino H. Activin binding protein from rat ovary is follistatin. Science. 1990;247:836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- 48.Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 49.Woodruff TK, Mather JP. Inhibin, activin and female reproductive axis. Annu Rev Physiol. 1995;57:219–244. doi: 10.1146/annurev.ph.57.030195.001251. [DOI] [PubMed] [Google Scholar]
- 50.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 52. [May 29, 2008];microRNA.org Web site. Available at: www.microrna.org.