Abstract
Self-assembled protein capsids have gained attention as a promising class of nanoparticles for biomedical applications due to their monodisperse nature and versatile genetic and chemical tailorability. To determine the plasma clearance and tissue distribution in mice of the versatile capsid of bacteriophage Qβ, the particles were decorated with gadolinium complexes using the CuI-mediated azide-alkyne cycloaddition reaction. Interior surface labeling was engineered by the introduction of an azide-containing unnatural amino acid into the coat protein for the first time. Clearance rates were conveniently monitored by quantitative detection of Gd using inductively coupled plasma-optical emission spectroscopy, and were found to be inversely proportional to the number of complexes attached to the exterior surface of the particle. This phenomenon was correlated to changes in exterior surface charge brought about by acylation of surface exposed amine groups in the initial step of the bioconjugation protocol. When primary amine groups were reintroduced by azide-alkyne coupling, the circulation time increased accordingly. These results show that nanoparticle trafficking may be tailored in predictable ways by chemical and genetic modifications that modulate surface charge.
Keywords: virus particles, phage, pharmacokinetics, surface charge
Introduction
Recent interest in the use of nanoparticles for biomedical applications is based on an expectation that such species can be imbued with multiple functions such as encapsulation of a therapeutic or diagnostic payload together with targeted trafficking to tissues or cell types of interest.1 Viruses serve as a model for such capabilities, since they act in precisely this way to infect their selected hosts. In recent years, we and others have started to use virus capsids as the basis for the creation of new functional particles by chemical and genetic means.2 The advantages of virus scaffolds include facile large-scale production and purification, robust stability, atomic-resolution structural knowledge, and tailorability of amino acid sequence by straightforward recombinant techniques.
The eventual application of these structures in vivo requires knowledge of their biodistribution, clearance, and toxicity. While the immunogenicity of virus particles is well known and has been exploited3 or combated4 as required, the bioavailability and trafficking of non-pathogenic virus particles is poorly characterized. One would expect most non-human and non-infectious virus capsids to be biocompatible, but in vivo studies are still necessary before these nanoparticles can be developed for many biomedical applications. Further, chemical modifications such as the attachment of targeting moieties and drug molecules may significantly alter the pharmacokinetics of a system. The most extensively investigated of these types of modifications has been the attachment of oligo- or poly(ethylene oxide) (PEO or PEG) chains, which often provides for increased plasma circulation time of proteins, liposomes, and polymer micelles.4b,c;5 A much smaller number of studies have addressed the effects of altering the number and polarity of charged groups on a protein's pharmacokinetic properties,6 and very few have addressed this question for virus-like particles.7 We have recently probed the trafficking in mice of cowpea mosaic virus (CPMV), showing this particle to be stable, nontoxic, and directed predominantly to the liver in the absence of targeting moieties on its surface.8
Bacteriophage Qβ is a non-enveloped icosahedral particle approximately 28 nm in diameter,9 closely related to bacteriophage MS2 but more stable due to inter-subunit crosslinking by disulfide bonds.10 We have adopted this capsid as a platform for a variety of applications,11 making genetic alterations in, and covalent attachments to, side chain residues of its 180 equivalent subunits, rather than manipulations of a C-terminal extension expressed in a fraction of the subunits per particle as has been previously reported.12 For the studies described below, we expressed two point mutant forms of the structure (K16M and T93M) in addition to the wild-type sequence in E. coli, all as non-infectious virus-like particles encapsulating random cellular RNA. The K16M chimera removes the most highly accessible lysine on the external surface, leaving two others (K2 and K12) plus the N-terminus (which is somewhat hindered) as active amine nucleophiles. The reactivity of lysines on the interior surface is strongly passivated by association with packaged polynucleotide. The K16M and T93M substitutions also allow genetic incorporation of unnatural amino acids in place of methionine on the external and internal surfaces, respectively. Following conjugation to a lanthanide-metal chelate, the plasma circulation lifetimes of surface-modified Qβ particles were investigated using inductively coupled plasma optical emission spectroscopy (ICP-OES).
Results
Labeling of Qβ Virus-Like Particles
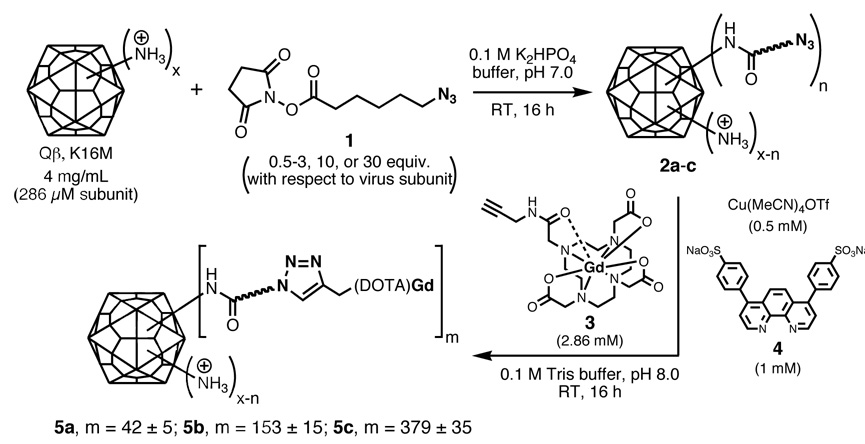
The K16M mutant form of the Qβ virus-like particle was condensed with the N-hydroxysuccinimide ester of 6-azidohexanoic acid (1) at three different concentrations to give acylated particles 2a, 2b, and 2c (Figure 1). Previous calibration experiments had established these conditions to provide a range of average “loadings” of azide functional groups on the capsid surface by acylation of side-chain lysine amino groups and the N-terminus, in analogy to the reactivity of other virus particles.13 The pendant azide groups were then addressed with Gd(DOTA)-alkyne (DOTA = 1,4,7,10-tetraazacyclododecanetetraacetic acid) derivative 311b using our standard method employing the CuI complex of ligand 4 to facilitate the process.14 Qβ particles 5a–c were obtained, bearing an average of 42, 153, and 379 Gd complexes per particle, presumably corresponding to the number of azide groups attached to 2a–c. We chose to make these attachments using the CuI-catalyzed azide-alkyne cycloaddition (CuAAC) reaction rather than other methods, such as the use of an activated ester derivative of Gd(DOTA), because of the synthetic convenience and high efficiency of the former process. Since azide and alkyne groups are unobtrusive and hydrolytically stable, they can be installed easily and stored indefinitely. The Cu-mediated cycloaddition reaction allows us to use low concentrations of the alkyne reagent, which is applicable to a variety of scaffolds and reaction conditions, and the connection is perfectly chemoselective, facilitating attachment of the Gd-alkyne complex only to azide sites. We have previously reported on the relaxivities of virus-Gd(DOTA) conjugates.11b
Figure 1.
Synthesis of Gd(DOTA)-labeled Qβ particles with variable loadings; “x” represents the total number of lysine and N-termini amino groups that can affect surface charge; “n” is the number of these residues acylated in the first step; “m” is the number of pendant azide groups addressed in the second step. With the use of an excess of 3, n and m are presumed to be approximately equal.
Determination of Circulation Lifetime and Clearance Destination by ICP-OES
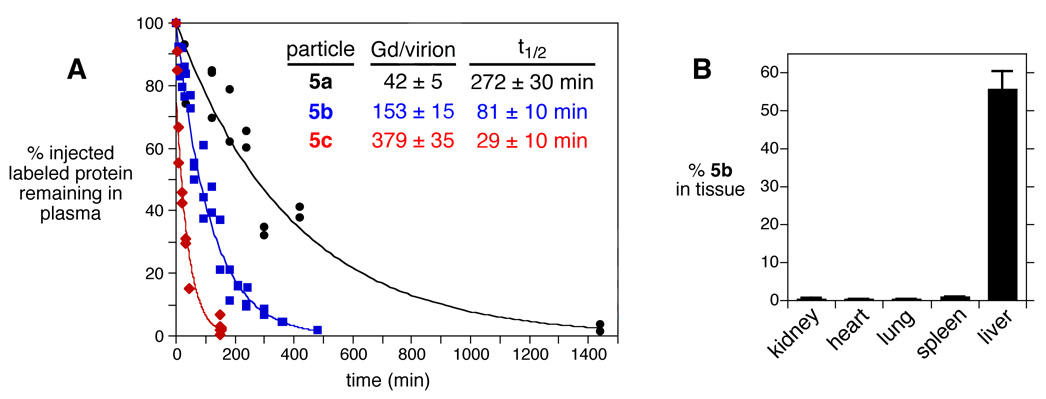
The plasma clearance rates of particles 5a–c were determined in Balb/c mice following intravenous injection of 200 µg of virus, as shown in Figure 2A. Blood from each animal was sampled at the times indicated, and the amount of Gd present in plasma was determined by ICP-OES. We have found the Qβ virus-like particle to be completely resistant to degradation in vitro in buffer and plasma for at least one week at 37 °C, as determined by size-exclusion chromatography. Extensive dialysis of Qβ-Gd(DOTA) conjugates also show no detectable leakage of Gd into the buffer, consistent with the well-known resistance of Gd(DOTA) complexes to dissociation of the metal under physiological conditions. Therefore, the amount of Gd detected reports on the presence of intact virus-like particles. A normalized clearance curve was constructed for each particle, showing half-lives inversely dependent on Gd(DOTA) loading: 5a, 272±30 min; 5b, 81±10 min; and 5c, 29±10 min.
Figure 2.
(A) Clearance of K16M Qβ particles decorated with variable loadings of Gd(DOTA) following intravenous injection. Each data point represents the result from an individual mouse. (B) Tissue distribution of Qβ-Gd(DOTA) conjugate 5b between 240 and 300 minutes after injection.
Virus particle 5b was also used to study the distribution in major organs after most of the particles were cleared from circulation (240 or 300 min after injection; >3 half-lives). The majority of detected Gd/virus was found in the liver (56 ± 5% of the injected material), whereas no Gd signal was detected in the heart, kidneys, lungs, or spleen (Figure 2B).
Effects of Surface Modifications on Clearance Rates
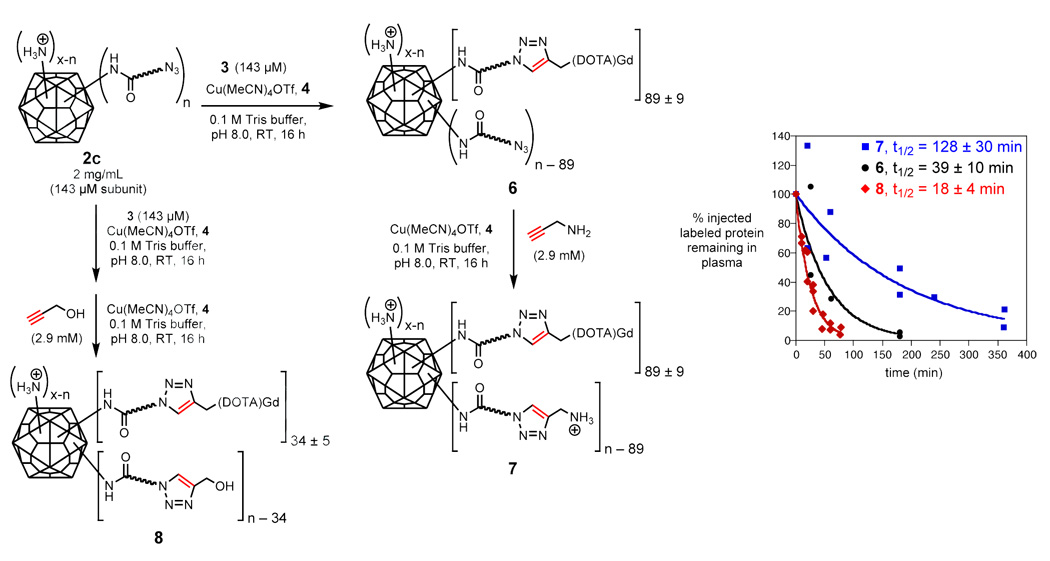
The relationship between clearance rate and Gd(DOTA) loading was further explored by varying the number of attached complexes and the number of charged surface residues, as shown in Figure 3. Qβ-azide particle 2c was addressed with only 1 equivalent of 3 and a lower concentration of Cu•4 catalyst than previously employed, providing 6 bearing an average of 89 Gd(DOTA) species per capsid and the remaining unreacted azide groups. A portion of 6 was then subjected to a second CuAAC reaction using an excess of propargyl amine. The resulting particles 7, bearing 89 Gd complexes plus primary amine groups re-installed at the remaining acylated sites, are intermediate in surface charge between the starting Qβ K16M particle and the acylated structures such as 2c, 5c, and 6. (Note that 3 is an uncharged structure, the Gd+3 ion being balanced by three coordinated carboxylates.) An analogous series of steps with 3 and propargyl alcohol gave particle 8 bearing 34 Gd complexes and the remaining acylated amine sites capped with a triazole moiety lacking added amine groups. Particle 6 was cleared at approximately the same rate as 5c (half-life ≤ 39 min; Figure 3), showing that the number of attached Gd(DOTA) complexes does not affect the circulation lifetime. The addition of triazolylamine groups (7) provided for a 3-fold increase in circulation lifetime (half-life ≈ 128 min), whereas triazolyl-alcohols (8) did not (half life ≈ 18 min).
Figure 3.
Synthesis of Qβ particles with controlled changes in exterior surface charge. The plot shows the clearance of the resulting particles as in Figure 2.
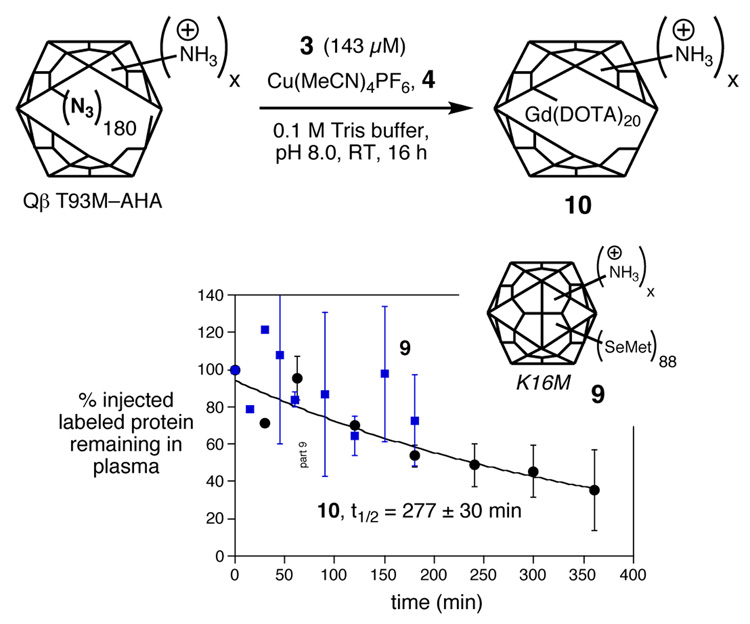
It was also of interest to determine the plasma clearance of the underivatized Qβ virion. To label the particle innocuously with an element that can be detected by ICP-OES, we wished to install selenomethionine (SelMet) in place of methionine. The wild-type Qβ coat protein sequence has no methionines other than the initiator at the N-terminus required by the bacterial translation machinery, and selenomethionine is rarely incorporated at this position. Thus, we turned to the K16M mutation, and expressed this particle in E. coli culture in the presence of SelMet. The resulting particles 9 were found to contain 88±4 Se atoms per capsid at the mutated position 16, with the other half of the 180 subunits containing methionine at this site.
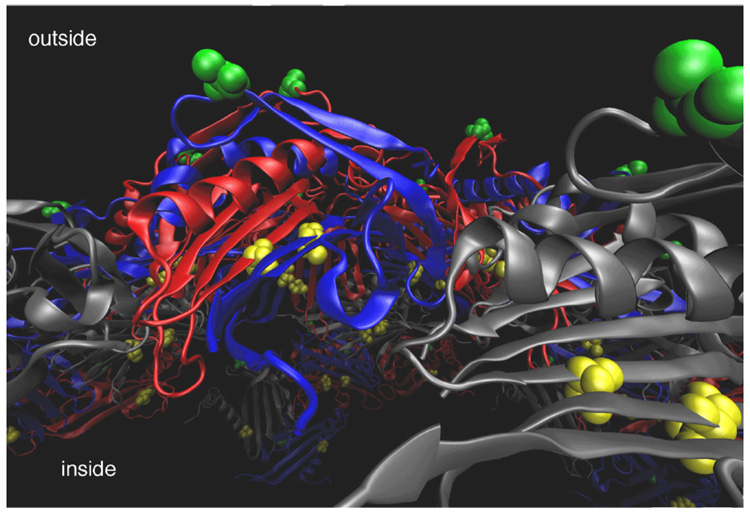
We also used a different methionine mutant to place a Gd(DOTA) label, which is more sensitively detected than selenium, at a shielded position in order to keep the exterior surface of the particle undisturbed. As shown in Figure 4, Thr93 is located on the interior capsid surface, and so the T93M mutation was created. Using the methodology of Tirrell and coworkers15 and a methionine auxotroph strain of E. coli, we incorporated the unnatural amino acid azidohomoalanine (AHA) into the Qβ structure to provide structure T93M Qβ-AHA (Figure 5). CuAAC reaction of T93M Qβ-AHA with 3 under standard conditions provided 10 containing 20±3 Gd complexes per particle. The relatively modest loading obtained in this reaction is likely a consequence of the hindered nature of the T93M site, and the reaction was not further optimized because the amount of Gd attached was sufficient for the purpose.
Figure 4.
Cross-sectional view of the structure of bacteriophage Qβ showing Lys16 on the outer surface in green and Thr93 on the inner surface in yellow. Capsid subunits are shown in ribbon representation in grey, blue, and red. Coordinates for the capsid structure were generated using the VIPER database (http://viperdb.scripps.edu/).16
Figure 5.
Qβ particles labeled with selenium (9) and interior-surface attachment of Gd(DOTA) (10), and their plasma clearance measured as in Figure 2. For clarity, averaged values are plotted from two or more independent measurements, with error bars showing the standard deviation. Data points without error bars represent single measurements.
Measurement of the clearance of 9 was hampered by the relatively poor sensitivity of Se detection by ICP-OES in comparison to Gd, giving rise to the widely scattered results shown in Figure 5. However, it is clear that substantial amounts of the capsid still remain in circulation after three hours. Similarly, the clearance of 10 was found to exhibit a half-life of approximately 277 min, equal to that of the lightly surface-loaded Gd species 5a. Note that 10 contains more positively charged residues on the surface than 5a because it is derived from the T93M mutant, which has the normal lysine residue at position 16; 5a is derived from K16M in which that lysine has been removed.
Summary
The results suggest that acylation of surface-exposed amine groups (and thus the neutralization of their positive charge at neutral pH), rather than the attachment of Gd(DOTA) complexes or triazole-containing linkers, affects the pharmacokinetics of Qβ. The lightly-loaded particle 5a, as well as particles bearing only an innocuous label (SelMet on 9) or a label on the particle interior (10), have the longest circulation times. Those particles that were heavily acylated were cleared approximately 10 times faster, whether all of the acyl linkages were tipped with Gd complexes (particle 5c) or not (particle 6). A capsid with an intermediate amount of acylation (5b) had an intermediate rate of clearance, and capping of a heavily-acylated structure with some primary amine groups using propargyl amine (7) caused its circulation lifetime to be increased to the intermediate range. In order to determine if the triazole linkage or the amine group was responsible for the decreased clearance rate of 7, the same heavily-acylated precursor particles were conjugated to propargyl alcohol (8). The circulation lifetime of this particle was quite short, showing that the re-introduction of surface charge by the attachment of propargylamine in 7 was the dominant factor.
Discussion
Knowledge of the plasma clearance rates and biodistribution of virus particles is required if virus-based nanoparticle platforms are to be used for targeting, imaging, or drug delivery. The subject has been addressed in several publications,7,17 in which it is shown that most particles are ultimately delivered to the liver and spleen, but clearance rates vary. The most notable examples involve adenovirus (Ad)17e and various bacteriophages (T717g, M137a, and λ7b). When administered intravenously to mice, Ad is removed from circulation within 10–15 minutes (half-life ≤ 5 min) and T7 is cleared within 60 min, while native M13 (half life = 4.5 h) and λ phage (half life = 5–6 h) have significantly longer circulation times. As shown above, Qβ is in the latter range.
The Gd(DOTA)-alkyne derivative 3 proved to be a particularly useful tag because it was readily synthesized and is stable to the conditions of the Cu+-catalyzed cycloaddition. Other lanthanide chelate-alkyne complexes designed for CuAAC coupling have recently been reported.18 Azide groups can be installed on virus capsids (2a–c) or generic proteins by a separate chemical coupling step or by incorporation of unnatural amino acids (as in T93M Qβ-AHA, Figure 5). While we will describe the technique more fully elsewhere,11a the Qβ-AHA particle represents the first report of a virus capsid incorporating unnatural amino acids for subsequent chemical derivatization.
The variation noted above in the clearance rates of Qβ particles bearing different numbers of Gd(DOTA) derivatives highlights the need to verify that the installation of labels does not alter the pharmacokinetic properties of the structure of interest. While the role of surface charge has been explored for liposomes and polymer micelles,17d,20b–d,20f,21 the subject has not received much attention for viruses. However, enough data now exist for a preliminary assessment. In the selection of long-circulating phage,17d the increase in circulation lifetime of phage λ of more than 1000-fold was traced to the mutation of a coat protein glutamic acid to lysine, thus substantially increasing the surface positive charge of the particle (although such factors as receptor binding may also be important).7b Similarly, the succinylation of lysines on M13 (converting positively charged residues to a negatively charged ones) has been reported to diminish the circulatory half-life from approximately 4.5 h to 1.5 min.7a Taken along with our Qβ results, these findings show that surface positive charge seems to inhibit clearance of three different protein particles from the bloodstream, at least in mice.
The ICP-OES method used here to measure circulation lifetime and tissue distribution has been employed several times in the past few years for small-molecule and nanoparticle magnetic resonance imaging agents.22 Atomic detection methods such as ICP-OES have several advantages relative to standard ELISA or radiolabeling methods for pharmacokinetics measurements in some applications: (1) The detection limits of ICP-OES (1–100 parts per billion, depending on the instrument and sample handling) are sufficient to allow quantitation of small amounts (100 µL) of plasma or tissue digests. (2) The metal complexes attached for the purpose of ICP-OES detection in this study are simple derivatives of molecules already approved for human use in magnetic resonance imaging at large dosages. (3) Lanthanides are not found in biological samples, and so there is no background signal; selenium background levels are also low. (4) For lanthanide labeling, only those molecules that are introduced with the label are detected, thereby distinguishing between endogenous and exogenous therapeutic proteins. (5) Up to 60 elements can be detected simultaneously; in practical terms, 2–4 different elements can be used conveniently in the same measurement, improving the accuracy and reproducibility of the results and allowing for studies to determine the co-localization of different molecules. We have taken advantage of the high stability and sensitive detection of lanthanide ions in a separate report, co-injecting virus particles labeled with Gd+3 and Eu+3 ions and observing identical clearance rates.8 (6) ICP-OES is compatible with any biological fluid or tissue, since the biological samples can be digested by strong acid prior to analysis. (7) The elements detected by ICP-OES are non-radioactive, do not otherwise decay, and the technique does not depend on the chemical or biological properties of the sample. Therefore, samples do not degrade upon storage and can be re-assayed at any time. (8) No radioactivity is used, to the benefit of both safety and convenience.
Conclusion
The well-defined and robust nature of virus capsids allows one to ask chemistry-style questions of biological systems. To probe factors influencing the plasma circulation lifetime of the Qβ nanoparticle in mice, we took advantage of the genetic and chemical tailorability of the self-assembled and highly stable protein scaffold, as well as the technique of ICP-OES to conveniently measure the concentration of labeled virions in serum and various tissues. The circulation lifetime was shown to decrease with the acylation of primary amine groups on the capsid surface, and increase upon the re-installation of amine groups by subsequent azide-alkyne conjugation. We anticipate that a combination of exterior surface charge modulation and attachment of shielding poly(ethylene oxide) chains may enable us to further limit the clearance rate of tailored Qβ capsids. Since the ultimate destination of these species is the liver, it will also be of interest to determine how the varying surface charge of Qβ particles influences interaction with cells of the reticuloendothelial system of that organ.
Experimental Section
General
Materials were obtained from the following commercial sources: kits for modified Lowry protein assay and SnakeSkin™ pleated dialysis tubing (10,000 MWCO) from Pierce; pQE-60 from Qiagen; IPTG (dioxane-free, high purity), ampicillin (sodium salt) and kanamycin sulfate from Calbiochem; sulfonated-bathophenanthroline ligand (4) from GFS Chemicals; Difco™ SOB medium from Becton Dickinson; and metal standards for ICP-OES from Inorganic Ventures, Inc. Linker 1 was prepared from azidohexanoic acid and N-hydroxysuccinimide using standard amide coupling methods. Complex 3 was prepared as previously reported.11b Azidohomoalanine (AHA) was synthesized based on literature procedures.15c,23 M15 methionine auxotroph (M15MA) cells were obtained from Prof. David Tirrell's laboratory. All other chemicals were purchased from Acros Organics or Sigma-Aldrich and used as received. All CuAAC reactions were performed under nitrogen in a Vacuum Atmospheres glovebox. Fermentation was done in a New Brunswick Scientific BioFlo 110 fermentor. ICP-OES measurements were obtained on a Varian VISTA AX CCD simultaneous spectrometer equipped with a Teflon nebulizer and sample uptake tubing. All standards and samples were spiked with a yttrium internal standard at 10 ppm to normalize irregularities in nebulization between samples. Selenium concentrations were determined by monitoring 2–3 wavelengths and averaging the resulting values, while Gd concentrations involved the averaging of 10 wavelengths. Mouse tissue samples were freeze-dried in a VirTis Freezemobile 25EL. Size exclusion chromatography analyses were performed with AKTA Explorer (Amersham Pharmacia Biotech) equipment, using Supersose-6™ size-exclusion columns. Ultracentrifugation was performed with a Beckman Optima L-90K Ultracentrifuge equipped with either a SW28 or 50.2 Ti rotor.
Propagation and Isolation of Virus Particles
Expression of the Qβ coat protein from a recombinant plasmid has been previously reported;24 we created our own vector to allow for more convenient genomic manipulation, as will be described in detail elsewhere.11a A 133-amino acid version of the Qβ coat protein gene was cloned into the vector pQE-60 and expressed under IPTG control in M15MA cells in SOB media. After expression, collected cells were lysed by sonication and lysozyme treatment and then centrifuged to remove insoluble cell components. Assembled particles were precipitated from the resulting supernatant using 8% PEG 8000. Following further centrifugation, the isolated pellet was resuspended in 0.1M potassium phosphate pH 7.0. The virus-like particles then underwent a final purification by ultracentrifugation through 10–40% sucrose gradients followed by ultrapelleting and resuspension in 0.1M potassium phosphate pH 7.0. Qβ concentrations were determined using the Modified Lowry Protein Assay.
Wild-type Qβ was incubated in mouse serum overnight at 37°C, after which it was recovered from solution in nearly quantitative yield with no detectable breakdown of particles (analysis by size-exclusion FPLC). The loss of material was typical for isolation by sucrose gradient ultracentrifugation, showing that the particles are stable under physiological conditions.
Incorporation of unnatural amino acids azidohomoalanine (AHA) and selenomethionine into Qβ
The T93M Qβ coat protein gene was cloned into the pQE-60 vector and expressed in minimal media under IPTG control using M15MA cells in the presence of 80 mg/L AHA, using the procedure of Tirrell and coworkers.15a,c The resulting T93M Qβ-AHA particles (10) were isolated using the above described procedure, yielding 1 mg virus/g cells.
Selenomethionine was incorporated into the structure of K16M Qβ to provide capsid 9. These particles were expressed in minimal media in a fermentor. An initial 100 mL culture in SOB was grown overnight, which was then used to inoculate a 2L culture composed of the following: 30 mM Na2HPO4·7H2O, 14 mM KH2PO4, 5.5 mM NaCl, 12 mM NH4Cl, 125 mg/L methionine, 250 mg/L serine, 125 mg/L other 18 amino acids, 4% glucose, 5 mM MgSO4, 250 µM CaCl2, 9 µM ZnSO4·7H2O, 50µM MnCl2·4H2O, 45 µM FeCl3·6H2O, 4 µM CuSO4·5H2O, 23 µM B(OH)3, 5 µM Na2MoO4·2H2O, 50 mg/L ampicillin, 100 mg/L kanamycin, and 13.6 µM propylene glycol. The culture was grown at pH 7.4 and 70% O2 for 19.5 hours and the resulting cells were collected and pelleted in several batches. The pellets were re-suspended in 125 mL of the above media, minus methionine, and grown an additional 45 min. in the fermentor. Then, solutions of selenomethionine (300 mg in 30 mL water) and IPTG (4 mL of a 1M solution) were added. The cells were then grown for an additional 7 hours, followed by cell collection and freezing of the pellets. Particles 9 were isolated as described above, providing a yield of 0.6 mg virus/g cells. The degree of incorporation of selenomethionine was determined by comparing the selenium concentration determined by ICP-OES to the virus concentration determined by modified Lowry assay.
Bioconjugation to Qβ Particles
The N-acylated azide particles 2a–c were prepared by reaction of K16M Qβ (final concentration 4 mg/mL, 286 µM protein subunit) with 0.5–3, 10, or 30 equivalents of N-hydroxysuccinimide ester 1, respectively, for 16 hrs at room temperature in 0.1M potassium phosphate buffer, pH 7.0. The reaction mixtures were ultracentrifuged through 10–40% sucrose gradients and then ultrapelleted. The resulting pellets were then suspended in degassed 0.1 M Tris buffer (pH 8.0) in a glove box. Virus samples were then diluted to a final concentration of 2 mg/mL (143 µM protein subunit) for the CuAAC reaction. Gd(DOTA)-alkyne 3 in degassed buffer was added to a final concentration of 2.86 mM, followed by a mixture of Cu(CH3CN)4OTf and ligand 4 (final concentrations of 0.72 and 1.43 mM, respectively, with the mixture being made from 100 mM stock solutions of Cu(CH3CN)4OTf in degassed CH3CN and 4 in degassed buffer). The reactions were sealed, brought out of the glove box, and agitated gently on a rotisserie mixer overnight (16–24 hrs). Each reaction was conducted on 1 mL of total solution in 2 mL eppendorf tubes; larger-scale reactions were performed by multiplying the number of these tubes, rather than by using larger reaction volumes. The individual reaction mixtures were combined and the resulting particles (5a–c) were purified on 10–40% sucrose gradients followed by ultrapelleting and resuspension in 10 mM potassium phosphate pH 7.4. The number of attached complexes per virion was determined by measurement of Gd concentration by ICP-OES and protein concentration by modified Lowry assay. Previous control experiments established that the purification procedure removes all traces of free Gd complex.
Particles 6 and 7 were prepared and purified as described above for 5c, with the following minor modifications. The CuAAC reaction was performed with 143 µM 3, 143 µM Cu(MeCN)4OTf, and 286 µM 4, yielding 6. A portion of the resulting 6 underwent a second CuAAC reaction using 2.86 mM propargylamine, 715 µM CuI and 1.43 mM 4, to give 7. Particle 8 was prepared and purified as described for 7, except the initial CuAAC reaction used the following conditions: 2 mg/mL virus particles, 429 µM Gd(DOTA)-alkyne, 143 µM CuI, and 286 µM sulfonated-bathophenanthroline; and the second CuAAC reaction used the following conditions: 2 mg/mL virus particles, 2.86 mM propargyl alcohol, 715 µM CuI, and 1.43 mM sulfonated-bathophenanthroline. Particle 10 was prepared from T93M Qβ-AHA by the CuAAC reaction using the procedure described above for 5c. The integrity of all particles was verified by size exclusion chromatography. All virus conjugates described in this work are overwhelmingly composed of intact, non-aggregated particles.
Pharmacokinetics and biodistribution measurements
Adult Balb/c mice approximately 8 to 10 weeks of age, with an average of 25 grams body weight, were obtained from the TSRI rodent breeding colony. All experiments were performed according to protocols approved by the TSRI Institutional Animal Care and Use Committee (IACUC). To measure the plasma circulation time, animals were injected intravenously with the species of interest (200 µg 5a, 5b, 5c, 6, 7, and 8; 900 µg 9; 400 µg 10). Heparinized blood samples were collected by cardiac puncture at the indicated time points. Cells were removed by centrifugation at 500 × g for 10 minutes and the resulting plasma isolated. For tissue biodistribution studies, mouse organs (liver, spleen, kidneys, heart, and lungs) were collected between 240 and 300 minutes after injection. Each organ was lyophilized and then digested with nitric acid (70%, purified by double-distillation, Sigma-Aldrich) for 18–24 h. A small amount of insoluble material was occasionally observed following acid digestion and was removed by filtering through a plug of glass wool. The plasma and tissue samples were diluted, when necessary, using 0.1 M potassium phosphate pH 7.0 and examined by ICP-OES for the concentration of Gd or Se. The amount of protein and, thus, normalized % virus detected was calculated from the Gd or Se concentration and the predetermined loadings of Gd(DOTA) complexes or selenomethionine for each virus. For calculations, it was estimated that the average mouse contains 0.07 mL blood per gram body weight.25
Acknowledgment
This work was supported by the National Institutes of Health (CA112075 and EB000432), The Skaggs Institute for Chemical Biology, The David & Lucille Packard Foundation Interdisciplinary Science Program, and the W.M. Keck Foundation. We are grateful to Prof. David Tirrell and Mr. A. James Link for auxotroph cells and helpful discussions.
References
- 1.Singh P, Gonzalez MJ, Manchester M. Drug Devel. Res. 2006 67;:23–41. [Google Scholar]
- 2.(a) Whaley SR, English dS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]; (b) Wang Q, Lin T, Tang L, Johnson JE, Finn MG. Angew. Chem. Int. Ed. 2002;41:459–462. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]; (c) Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Science. 2004;303:213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]; (d) Chatterji A, Ochoa W, Shamieh L, Salakian SP, Wong SM, Clinton G, Ghosh P, Lin T, Johnson JE. Bioconj. Chem. 2004;15:807–813. doi: 10.1021/bc0402888. [DOI] [PubMed] [Google Scholar]; (e) Flenniken ML, Liepold LO, Crowley BE, Willits DA, Young MJ, Douglas T. Chem. Commun. 2005:447–449. doi: 10.1039/b413435d. [DOI] [PubMed] [Google Scholar]; (f) Schlick TL, Ding Z, Kovacs EW, Francis MB. J. Am. Chem. Soc. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]; (g) Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Bioconj. Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- 3.(a) Lomonossoff GP, Johnson JE. Curr. Opin. Struct. Biol. 1996;6:176–182. doi: 10.1016/S0959-440X(96)80072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Porta C, Lomonossoff G. Rev. Med. Virol. 1998;8:25–41. doi: 10.1002/(sici)1099-1654(199801/03)8:1<25::aid-rmv212>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]; (c) Ulrich R, Nassal M, Meisel H, Kruger DH. Adv. Virus Res. 1998;50:141–182. doi: 10.1016/s0065-3527(08)60808-8. [DOI] [PubMed] [Google Scholar]; (d) Kruger DH, Ulrich R, Gerlich WH. Biol. Chem. 1999;380:275–276. doi: 10.1515/BC.1999.037. [DOI] [PubMed] [Google Scholar]; (e) Langeveld JPM, Brennan FR, Martínez-Torrecuadrada JL, Jones TD, Boshuizen RS, Vela C, Casal JI, Kamstrup S, Dalsgaard K, Meloen RH, Bendig MM, Hamilton WDO. Vaccine. 2001;19:3661–3670. doi: 10.1016/s0264-410x(01)00083-4. [DOI] [PubMed] [Google Scholar]; (f) Pardoll DM. Nat. Rev. Immun. 2002;2:227–238. doi: 10.1038/nri774. [DOI] [PubMed] [Google Scholar]
- 4.(a) Paillard F. Hum. Gene Ther. 1999;10:2575–2576. doi: 10.1089/10430349950016618. [DOI] [PubMed] [Google Scholar]; (b) O'Riordan CR, Lachapelle A, Delgado C, Parkes V, Wadsworth SC, Smith AE, Francis GE. Hum. Gene Ther. 1999;10:1349–1358. doi: 10.1089/10430349950018021. [DOI] [PubMed] [Google Scholar]; (c) Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Biomacromolecules. 2003;4:472–476. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 5.(a) Katre K. Adv. Drug Del. Rev. 1993;10:91–114. [Google Scholar]; (b) Caliceti P, Veronese FM. Adv. Drug Del. Rev. 2003;55:1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]; (c) Owens DEI, Peppas NA. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]; (d) Panagi Z, Beletsi A, Evangelatos G, Livaniou E, Ithakissios DS, Avgoustakis K. Int. J. Pharm. 2001;221:143–152. doi: 10.1016/s0378-5173(01)00676-7. [DOI] [PubMed] [Google Scholar]
- 6.(a) Moore AT, Williams KE, Lloyd JB. Biochem. J. 1977;164:607–616. doi: 10.1042/bj1640607. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wachsmuth ED, Klingmüller D. J. Reticuloendothel. Soc. 1978;24:227–241. [PubMed] [Google Scholar]; (c) Roser M, Fischer D, Kissel T. Eur. J. Pharm. Biopharm. 1998;46:255–263. doi: 10.1016/s0939-6411(98)00038-1. [DOI] [PubMed] [Google Scholar]; (d) Yamasaki Y, Sumimoto K, Nishikawa M, Yamashita F, Yamaoka K, Hashida M, Takakura Y. J. Pharmacol. Exp. Ther. 2002;301:467–477. doi: 10.1124/jpet.301.2.467. [DOI] [PubMed] [Google Scholar]
- 7.(a) Molenaar TJM, Michon I, de Haas SAM, van Berkel TJC, Kuiper J, Biessen EAL. Virology. 2002;293:182–191. doi: 10.1006/viro.2001.1254. [DOI] [PubMed] [Google Scholar]; (b) Vitiello CL, Merril CR, Adhya S. Virus Res. 2005;114:101–103. doi: 10.1016/j.virusres.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Singh P, Prasuhn J, D E, Yeh RM, Destito G, Rae C, Osborn K, Finn MG, Manchester M. J. Control. Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liljas L, Golmohammadi R. Structure. 1996;4:543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 10.Ashcroft AE, Lago H, Macedo JMB, Horn WT, Stonehouse NJ, Stockley PG. J. Nanosci. Nanotech. 2005;5:1–8. doi: 10.1166/jnn.2005.507. [DOI] [PubMed] [Google Scholar]
- 11.(a) Strable E, Prasuhn D, Udit A, Brown S, Finn MG. Virus-Like Particles Incorporating Unnatural Amino Acids. manuscript in preparation. [Google Scholar]; (b) Prasuhn J, D E, Yeh RM, Obenaus A, Manchester M, Finn MG. Chem. Commun. 2007:1269–1271. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 12.(a) Kozlovska TM, Cielens I, Vasiljeva I, Strelnikova A, Kazaks A, Dislers A, Dreilina D, Ose V, Gusars I, Pumpens P. Intervirology. 1996;39:9–15. doi: 10.1159/000150469. [DOI] [PubMed] [Google Scholar]; (b) Vasiljeva I, Kozlovska T, Cielens I, Strelnikova A, Kazaks A, Ose V, Pumpens P. FEBS Lett. 1998;431:7–11. doi: 10.1016/s0014-5793(98)00716-9. [DOI] [PubMed] [Google Scholar]
- 13.(a) Wang Q, Kaltgrad E, Lin T, Johnson J, Finn MG. Chem. Biol. 2002;9:805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]; (b) Taylor DJ, Wang Q, Bothner B, Natarajan P, Finn MG, Johnson JE. Chem. Commun. 2003:2770–2771. doi: 10.1039/b310533d. [DOI] [PubMed] [Google Scholar]
- 14.Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Bioconj. Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- 15.(a) Van Hest JCM, Kiick KL, Tirrell DA. J. Am. Chem. Soc. 2000;122:1282–1288. [Google Scholar]; (b) Link AJ, Mock ML, Tirrell DA. Curr. Opin. Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]; (c) Link AJ, Tirrell DA. J. Am. Chem. Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 16.Reddy V, Natarajan P, Okerberg B, Li K, Damodaran K, Morton R, Brooks CI, Johnson J. J. Virol. 2001;75:11943–11947. doi: 10.1128/JVI.75.24.11943-11947.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Brunner KT, Hurez D, McCluskey RT, Benacerraf B. J. Immunol. 1960;85:99–105. [PubMed] [Google Scholar]; (b) Schultz I. J. Immunol. 1966;97:629–633. [PubMed] [Google Scholar]; (c) Geier MR, Trigg ME, Merril CR. Nature. 1973;246:221–223. doi: 10.1038/246221a0. [DOI] [PubMed] [Google Scholar]; (d) Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. Proc. Natl. Acad. Sci. USA. 1996;93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Alemany R, Suzuki K, Curiel DT. J. Gen. Virol. 2000;81:2605–2609. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]; (f) Schellinger D, Rainov NG, Breakefield XO, Weissleder R. Gene Ther. 2000;7:1648–1655. doi: 10.1038/sj.gt.3301272. [DOI] [PubMed] [Google Scholar]; (g) Srivastava AS, Kaido T, Carrier E. J. Virol. Methods. 2004;115:99–104. doi: 10.1016/j.jviromet.2003.09.009. [DOI] [PubMed] [Google Scholar]; (h) Zou J, Dickerson MT, Owen NK, Landon LA, Deutscher SL. Mol. Biol. Rep. 2004;31:121–129. doi: 10.1023/b:mole.0000031459.14448.af. [DOI] [PubMed] [Google Scholar]
- 18.(a) Viguier RFH, Hulme AN. J. Am. Chem. Soc. 2006;128:11370–11371. doi: 10.1021/ja064232v. [DOI] [PubMed] [Google Scholar]; (b) Dijkgraaf I, Rijnders AY, Soede A, Dechesne A, Van Esse GW, Brouwer AJ, Corstens FHM, Boerman OC, Rijkers DTS, Liskamp RMJ. Org. Biomol. Chem. 2007;5:935–944. doi: 10.1039/b615940k. [DOI] [PubMed] [Google Scholar]
- 19.(a) Croyle MA, Yu Q-C, Wilson JM. Hum. Gene Ther. 2000;11:1713–1722. doi: 10.1089/10430340050111368. [DOI] [PubMed] [Google Scholar]; (b) Lecolley F, Tao L, Mantovani G, Durkin I, Lautru S, Haddleton DM. Chem. Commun. 2004:2026–2027. doi: 10.1039/b407712a. [DOI] [PubMed] [Google Scholar]; (c) Kreppel F, Gackowski J, Schmidt E, Kochanek S. Mol. Ther. 2005;12:107–117. doi: 10.1016/j.ymthe.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 20.(a) Abraham I, Goundalkar A, Mezei M. Biopharm. Drug Disposition. 1984;5:387–398. doi: 10.1002/bdd.2510050409. [DOI] [PubMed] [Google Scholar]; (b) Aoki H, Tottori T, Sakurai F, Fuji K, Miyajima K. Int. J. Pharmaceutics. 1997;156:163–174. [Google Scholar]; (c) Nomura T, Koreeda N, Yamashita F, Takakura Y, Hashida M. Pharm. Res. 1998;15:128–132. doi: 10.1023/a:1011921324952. [DOI] [PubMed] [Google Scholar]; (d) Allen C, Dos Santos N, Gallagher R, Chiu GNC, Shu Y, Li WM, Johnstone SA, Janoff AS, Mayer LD, Webb MS, Bally MB. Biosci. Rep. 2002;22:225–250. doi: 10.1023/a:1020186505848. [DOI] [PubMed] [Google Scholar]; (e) Shi N, Pardridge WM. Proc. Natl. Acad. Sci. USA. 2004;97:7567–7572. doi: 10.1073/pnas.130187497. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Simoes S, Moreira JN, Fonseca C, Duzgunes N, De Lima MCP. Adv. Drug Del. Rev. 2004;56:947–965. doi: 10.1016/j.addr.2003.10.038. [DOI] [PubMed] [Google Scholar]; (g) Gabizon A, Shmeeda H, Horowitz AT, Zalipsky S. Adv. Drug Del. Rev. 2004;56:1177–1192. doi: 10.1016/j.addr.2004.01.011. [DOI] [PubMed] [Google Scholar]; (h) Visser CC, Stevanovic S, Voorwinden LH, Van Bloois L, Gaillard PJ, Danhof M, Crommelin DJA, De Boer AG. Eur. J. Pharm. Sci. 2005;25:299–305. doi: 10.1016/j.ejps.2005.03.008. [DOI] [PubMed] [Google Scholar]; (i) Torchilin VP. Adv. Drug Del. Rev. 2005;57:95–109. doi: 10.1016/j.addr.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 21.(a) Gabizon A, Papahadjopoulos D. Biochem. Biophys. Acta - Biomembranes. 1992;1103:94–100. doi: 10.1016/0005-2736(92)90061-p. [DOI] [PubMed] [Google Scholar]; (b) Yamamoto Y, Nagasaki Y, Kato Y, Sugiyama Y, Kataoka K. J. Contr. Rel. 2001;77:27–38. doi: 10.1016/s0168-3659(01)00451-5. [DOI] [PubMed] [Google Scholar]; (c) Levchenko TS, Rammohan R, Lukyanov AN, Whiteman KR, Torchilin VP. Int. J. Pharm. 2002;240:95–102. doi: 10.1016/s0378-5173(02)00129-1. [DOI] [PubMed] [Google Scholar]; (d) Nicolazzi C, Mignet N, De la Figuera N, Cadet M, Ibad RT, Seguin J, Scherman D, Bessodes M. J. Contr. Rel. 2003;88:429–443. doi: 10.1016/s0168-3659(03)00067-1. [DOI] [PubMed] [Google Scholar]; (e) Reddy LH, Sharma RK, Chuttani K, Mishra AK, Murthy RR. AAPS Journal. 2004;6:1–10. [Google Scholar]
- 22.(a) Frame EMS, Uzgiris EE. Analyst. 1998;123:675–679. doi: 10.1039/a708905h. [DOI] [PubMed] [Google Scholar]; (b) Dafni H, Gilead A, Nevo N, Eilam R, Harmelin A, Neeman M. Magnetic Resonance in Medicine. 2003;50:904–914. doi: 10.1002/mrm.10638. [DOI] [PubMed] [Google Scholar]; (c) Allen MJ, MacRenaris KW, Venkatasubramanian PN, Meade TJ. Chem. Biol. 2004;11:301–307. doi: 10.1016/j.chembiol.2004.03.003. [DOI] [PubMed] [Google Scholar]; (d) Parac-Vogt TN, Kimpe K, Laurent S, Vander Elst L, Burtea C, Chen F, Muller RN, Ni Y, Verbruggen A, Binnemans K. Chem. Eur. J. 2005;11:3077–3086. doi: 10.1002/chem.200401207. [DOI] [PubMed] [Google Scholar]
- 23.Mangold JB, Mischke MR, LaVelle JM. Mutation Research. 1989;308:33–42. doi: 10.1016/0027-5107(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 24.Kozlovska TM, Cielens I, Dreilinna D, Dislers A, Baumanis V, Ose V, Pumpens P. Gene. 1993;137:133–137. doi: 10.1016/0378-1119(93)90261-z. [DOI] [PubMed] [Google Scholar]
- 25.(a) Bannerman R. Hematology. Vol. III. New York: Academic Press; 1983. [Google Scholar]; (b) Hoff J. Lab. Animal. 2000;29:47–53. [Google Scholar]