Abstract
Background
The propensity for severe drinking is hypothesized to be regulated by differential expression of serotonin transporter gene (SLC6A4) in the human brain. The SLC6A4 promoter region 5-HTTLPR has been examined previously as a candidate polymorphic variant associated with severe drinking. In this study, we investigated whether other SLC6A4 single nucleotide polymorphisms (SNPs) are associated with drinking intensity among treatment-seeking alcoholics and whether these polymorphic variants result in differential SLC6A4 expression levels.
Methods
We analyzed associations of drinking intensity in 275 (78.5% male) treatment-seeking alcoholics of Caucasian and Hispanic origin, with 6 SLC6A4 polymorphisms. Next, to examine the functionality of the SNP that showed a significant association with drinking intensity, we transfected the two alleles of rs1042173 into HeLa cell cultures and measured serotonin transporter mRNA and protein expression levels by using qRT-PCR and western blotting techniques.
Results
One of the 6 polymorphisms we examined, rs1042173 in the 3’ untranslated region (3’-UTR) of SLC6A4, showed a significant association with drinking intensity. The G allele carriers for rs1042173 were associated with significantly lower drinking intensity (P=0.0034) compared to T allele homozygotes. In HeLa cell cultures, the cells transfected with G allele showed a significantly higher mRNA and protein levels than the T allele-transfected cells.
Conclusion
These findings suggest that the allelic variations of rs1042173 affect drinking intensity in alcoholics possibly by altering serotonin transporter expression levels. This provides additional support to the hypothesis that SLC6A4 polymorphisms play an important role in regulating propensity for severe drinking.
Keywords: SLC6A4 3’-UTR, rs1042173, intensity of drinking, gene expression
Introduction
Heavy episodic drinking is associated with numerous psychiatric and general medical conditions causing a major public health burden (Cargiulo, 2007). Several studies have reported a dose-response relationship between the extent of heavy drinking and the risk of alcohol related morbidity and mortality among heavy drinkers (Makela and Mustonen, 2007; Gastfriend et al., 2007). Consequently, reduction of heavy drinking is used as an indicator of treatment response in clinical trials aimed at treating alcohol dependence.
Of the various neurotransmitter systems through which alcohol mediates its effects, the serotonergic system has been shown to play an important role in alcohol preference and consumption (Johnson, 2004). Synaptic serotonergic neurotransmission is terminated when serotonin (5-HT) is transported back into pre-synaptic neurons by 5-HT transporters (5-HTTs) (Talvenheimo and Rudnick, 1980) and the degree of 5-HT reuptake depends on the density of 5-HTTs on presynaptic surface. The selective 5-HT reuptake inhibitors that act directly on 5-HTTs have been shown to reduce alcohol consumption in rats (Gill and Amit, 1989). However, in humans SSRIs have been effective at reducing heavy drinking only among some subtypes of alcoholics, more specifically in type A alcoholics but not in type B alcoholics (Dundon et al., 2004; Pettinati et al., 2000) who are considered to be more biologically predisposed to develop alcohol dependence. Therefore, it is reasonable to propose that allelic variations which alter expression levels of SLC6A4 gene can be expected to have an important effect on drinking intensity.
The human 5-HTT is encoded by a single gene (SLC6A4) mapped on chromosome 17q11.1–q12 (Ramamoorthy et al., 1993). The SLC6A4 gene spans ~35 kb and has 14 exons. The protein encoded by this gene, the 5-HTT, is a trans-membrane protein containing 630 amino acids (Heils et al., 1996). The expression level of SLC6A4 is regulated by at least three mechanisms: transcription regulatory elements in the promoter (Ramamoorthy et al., 1993), differential splicing (Bradley and Blakely, 1997), and the use of different 3’ polyadenylation sites (Battersby et al., 1999). Furthermore, several other polymorphisms that change amino acid sequence (Thr4Ala, Gly56Ala, Glu215Lys, Lys605Asn and Pro612Ser) of 5-HTT have been shown to affect 5-HT uptake function in cell cultures (Prasad et al., 2003).
Although the long (L) and short (S) polymorphism at 5-HTT linked polymorphic region (5-HTTLPR) of SLC6A4 has been extensively studied in the literature, the results are inconclusive. For example, in a meta-analysis of 17 studies, Feinn et al. (2005) showed that S allele was significantly associated with alcohol dependence in subjects with co-occurring serotonergic abnormalities while several other studies reported an association of alcohol dependence with the L allele (Kweon et al., 2005, Hu et al., 2005). On the other hand, numerous studies including the report by our group reveal a differential association between chronic problem-drinking and the density and function of serotonin transporters in alcoholic subjects carrying L and S variants of SLC6A4 (Little et al., 1998, Javors et al., 2005, Johnson et al., 2008).
To have a better coverage of SLC6A4, in this study, we investigate the associations of 6 genetic polymorphisms of the gene with drinking severity, in a sample of treatment-seeking alcohol dependents. In studies conducted with different psychiatric populations, all these six variants are reported to be associated with affective disorders (Mynett-Johnson et al., 2000; Ozaki et al., 2003; Hu et al., 2006). However, to our knowledge, no studies have been conducted to investigate the association of these SNPs with differences in drinking intensity among alcohol dependents.
MATERIALS AND METHODS
Subjects
A total of two hundred seventy-five alcohol-dependent subjects (78.5% male) aged between 18 and 66 years were used in this study, in which 198 of them were included in our previous study (Johnson et al., 2008). All subjects were considered to be alcohol-dependent (see below for details) and enrolled as part of a pharmacotherapy trial for the treatment of alcohol dependence at both the University of Texas Health Science Center at San Antonio and at University of Virginia. Participants were recruited by newspaper or radio advertisements, and written informed consent—approved by review boards of all participating institutes, was obtained from all participants.
Alcohol dependence was diagnosed using the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association, 1994) Axis I Disorders, by a trained psychologist. All subjects had a score of ≥ 8 on the Alcohol Use Disorders Identification Test (AUDIT) (Babor et al., 1992) that screened for individuals with alcohol use and related problems: reported heavy drinking which was defined as drinking of ≥21 standard drinks/week for women and ≥30 standard drinks/week for men during the 90 days prior to enrollment. Absence of other substance use was confirmed by negative urine toxicological screen for narcotics, amphetamines, or sedative hypnotics at enrollment. The Subjects who met the following criteria were excluded from the study: current axis I psychiatric diagnoses other than alcohol or nicotine dependence; significant alcohol withdrawal symptoms based on the revised clinical institute withdrawal assessment for alcohol scale (Sullivan et al., 1989) score >15]; clinically significant physical abnormalities based on physical examination, electrocardiogram recording, hematological assessment, biochemistry including serum bilirubin concentration, and urinalysis; pregnant or lactating state; treatment for alcohol dependence ≤30 days prior to enrollment, and mandated incarceration or employment loss for not receiving alcohol treatment.
Drinking measurements
Self-reported drinking (measured in standard drinks) in the 90 days prior to study enrollment was quantified using the timeline follow-back method. One standard drink was defined as 0.35 L of beer, 0.15 L of wine, or 0.04 L of 80-proof liquor. The intensity of drinking was assessed by measurement of the mean drinks per drinking day and mean drinks per day. Drinks per drinking day was defined as the total number of drinks divided by the number of drinking days within the 90 days; drinks per day was defined as the total number of drinks divided by 90 days.
DNA extraction and Genotyping
Ten milliliters of blood was drawn from each subject at baseline to obtain white blood cells for the determination of 5-HTT genotypes. DNA was extracted using a Gentra Puregene® kit (QIAGEN Inc., Valencia, CA). SNPs for association analyses were selected using the National Center for Biotechnology Information (NCBI) dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) based on their functional potential and minor allele frequency (MAF) ≥ 0.05. The average SNP density is ~7 kb. Detailed information on SNP locations, chromosomal positions, allelic variants, MAF, and primer/probe sequences is summarized in Table 1. Four of the five SNPs (rs6354, rs6355 rs28914832 and rs1042173) were genotyped with TaqMan® SNP genotyping assays (Applied Biosystems, Foster City, CA). Polymerase chain reaction (PCR) conditions were 50°C for 2 min, 95°C for 10 min, 30 cycles of 95°C for 25 s, and 60°C for 1 min. Alleles of each SNP were determined with an ABI PRISM® 7900HT instrument (Applied Biosystems) and analyzed using sequence detection system (SDS) software.
Table 1.
Biological information of the 5 SNPs examined in the study
| NCBI dbSNP ID |
Physical Position |
Chromosome position |
Alleles | MAF | P values for the deviation from HWE1 | Primers and probe sequences / Context sequence ID of ABI primers and probes | |||
|---|---|---|---|---|---|---|---|---|---|
| CEUa | Caucasian1 | Hispanic1 | Caucasian | Hispanic | |||||
| 5-HTTLPR | Promoter | ~25,588,500 | L (long) | - | 0.451 | 0.430 | 0.814 | 0.624 | Forward: TCCT CCGCTTTGGCG CCTCTTCC |
| S(Short) | Reverse: TGGGGGTTGCAGGGGA GATCCTG | ||||||||
| rs25531 | Promoter | 25,588,472 | A/G | 0.100 | 0.065 | 0.079 | 0.999 | 1.000 | Forward: TCCT CCGCTTTGGCG CCTCTTCC |
| Reverse: TGGGGGTTGCAGGGGA GATCCTG The two alleles were determined using restriction enzymes Hpa11 and bcc1 |
|||||||||
| rs6354 | Exon 2 | 25,574,024 | T/G | 0.295 | 0.202 | 0.158 | 0.319 | 1.000 | C_1841706_10 |
| (5′ UTR) | |||||||||
| rs6355 | Exon 3 | 25,572,936 | C/G | 0.025 | 0.022 | 0.026 | 1.000 | 1.000 | C_11414113_20 |
| (Ala/Gly) | |||||||||
| rs28914832 | Exon 10 | 25,562,500 | A/C | 0.008 | 0.003 | 0.009 | 1.000 | 1.000 | (Custom Taqman(R) SNP Genotyping Assay) |
| (Leu/Ile) | Forward: GCAGAAGCGATAGCCAACATG | ||||||||
| Reverse: CAAGCCCAGCGTGATTAACATC | |||||||||
| Probe: CTTTCTTTGCC[C/A]TCATCT | |||||||||
| rs1042173 | Exon 15 | 25,549,137 | G/T | 0.433 | 0.419 | 0.455 | 0.138 | 1.000 | C_7473190_10 |
| (3′ UTR) | |||||||||
MAF = minor allele frequency; HWE = Hardy-Weinberg equilibrium; ABI = Applied Biosystems (Foster City, CA).
Note: European sample from HapMap project
Data from this study.
The DNA samples from the 77 subjects that were not included in our previous study were genotyped for 5’-HTTLPR L/S alleles as described previously (Johnson et al., 2008).
Assays for SNP rs25531 were carried out as described by Wendland et al (2006). Each assay had a total assay volume of 20 µl, and the PCR conditions were 15 min at 95°C, 35 cycles of 94°C for 30 s, 65.5°C for 90 s, and 72°C for 60 s, with a final extension step of 10 min at 72°C. Afterwards, 10 µl of PCR product was double-digested with HpaII and BccI (5 U each; New England Biolabs, Ipswich, MA) in a 20-µl reaction assay containing NEBuffer 1 and bovine serum albumin at 37°C for 5 h. Finally, 10 µl of remaining PCR product and 20 µl of restriction enzyme assay solution were electrophoresed with 3.5% UltraPure™ agarose gel (Invitrogen™, Carlsbad, CA) for 1.5–2 h at 100 V in Tris/Borate/ethylenediaminetetraacetic acid buffer and visualized by ethidium bromide staining (Sigma-Aldrich, St Louis, MO). The uncut PCR product in the lanes loaded with restriction enzyme-digested PCR products were detected as the “A” allele of rs25531, and the cut product at 402 bp were detected as the “G” allele of rs25531.
Association analyses with drinking intensity
Associations of individual SNPs with the intensity of drinking (i.e., drinks per drinking day and drinks per day) were analyzed using the analysis of variance test in SAS version 9.1 (SAS Institute Inc., Cary, NC). Three genetic models (additive, dominant, and recessive) were tested using gender and age as covariates. Pair-wise linkage disequilibrium (LD) among all 6 polymorphisms was assessed using the Haploview program (Barrett et al., 2005). All associations found to be significant were corrected for multiple testing according to Bonferroni correction by dividing the significance level by the number of polymorphisms studied.
Cloning, cell culture, and transfection
Allelic expression differences of the SNP (rs1042173) that showed a significant association with drinking intensity were studied using an in vitro system. The human 5-HTT containing the G allele of rs1042173 in pBluescript II KS (–) was a generous gift from Prof. Randy D. Blakely (Vanderbilt University School of Medicine, Nashville, TN). This 5-HTT cDNA/Bluescript construct contained the coding region as well as both 5’- and 3’-untranslated regions of the gene with a total length of 2508 bp. The human 5-HTT construct was digested with HindIII/XbaI and subcloned into pcDNA3.1(-) (InvitrogenTM) pre-digested with HindIII/XbaI as described by Qian et al (Qian et al., 1997). To produce plasmid with the T allele of rs1042173, a DNA plasmid carrying the G allele was mutated using the GeneTailor™ site-directed mutagenesis system (Invitrogen™). Both constructs were DNA sequence verified.
HeLa cells were cultured in complete medium [Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT), 10% GIBCO® fetal bovine serum (Invitrogen™), 100 U/ml penicillin, and 100 µg/ml streptomycin (Mediatech, Inc., Manassas, VA)] in 6-well plates and maintained in a humidified incubator at 37° C and 5% CO2. After the cells reached approximately 80% confluence, they were transfected with one of the two alleles (4 µg of plasmids per well) in 6-well culture plates using Lipofectamine™ 2000 (Invitrogen™) according to the manufacturer’s guidelines. RNA and proteins were extracted from HeLa cells 24 h after transfection.
RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from HeLa cells with TRIZOL® reagent (Invitrogen™). Potential DNA contaminations were removed by treating the RNA samples with RNase-free DNase I at 37°C for 30 min. Each RNA sample was reverse transcribed in vitro using SuperScript® II RT (Invitrogen™) to obtain cDNA. These cDNA samples were transcribed with TaqMan® Gene Expression Assays (Applied Biosystems) specific for 5-HTT mRNA, and the resulting 5-HTT mRNA was quantified by the ABI PRISM® 7900HT sequence detection system. TaqMan® primer/probe sets for glyceraldehyde-3-phosphate dehydrogenase (G3PDH) were used as an internal control to normalize the expression of 5-HTT. For each qRT-PCR experiment, four samples with the G allele, four samples with the T allele, and four controls with the pcDNA3.1 (-) vector only were used in cell cultures from transfections carried out on different days.
Western blotting analysis
Radioimmunoprecipitation assay buffer [Tris-HCl (pH 7.4), 1% NP-40, 150 mM NaCl, 0.25% Na-deoxycholate, and 1 mM EDTA] was added to HeLa cells after washing the cells once with ice-cold phosphate-buffered saline. The protein concentration of the cell lysates was determined using the Bio-Rad assay (Bio-Rad Laboratories, Hercules, CA). Fifteen micrograms of samples were loaded onto 10% sodium dodecyl sulfate-polyacrylamide gels (30% acrylamide) in Tris–glycine buffer containing sodium dodecyl sulfate. The separated proteins were then electrophoretically transferred to nitrocellulose membranes (PerkinElmer, Waltham, MA) overnight at 25 mA. The membranes were blocked for 1 h at room temperature with 2% non-fat dry milk diluted in Tris-buffered saline with Tween® 20 (TBST) buffer and washed three times for 10 min each in TBST buffer; then they were incubated overnight with primary antibody (1:200) at 4°C [rabbit polyclonal immunoglobulin G (IgG) corresponding to the C-terminus of a sodium-dependent 5-HTT of human origin (200 µg/ml stock solution) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA)]. Membranes then were washed three times for 10 min each in TBST buffer and incubated with secondary antibodies (1:5,000) [anti-rabbit IgG (goat), horseradish peroxidase labeled (PerkinElmer)] for 1.5 h at room temperature. The hybridized membranes were washed with TBST buffer four times for 10 min each, and the immunoreactivity of the proteins was detected using Western Lightning® Chemiluminescence Reagent Plus (PerkinElmer) and exposure to X-ray film. Tubulin protein was used as an internal control to control for discrepancies in the loading of proteins in each lane. A monoclonal antibody (mouse monoclonal antibody to α-tubulin) was used as the primary antibody (1:2,000), and an anti-mouse IgG was used as the secondary antibody in western blotting for tubulin.
Densitometric and statistical analysis
Western blotting films were scanned on a UMAX scanner (Techville, Inc., Dallas, TX) using Adobe Photoshop (v. 6.0; Adobe Systems Inc., San Jose, CA), and the optical densities of the G and T alleles and tubulin were measured using NIH Image software (v. 1.61). The optical densities of bands of the G and T alleles and of tubulin were quantified using densitometry. The background (the area surrounding each band) optical density values were quantified the same way as for the protein bands, and the values were subtracted from the measured optical density values for the protein bands. The ratios of the optical density values of the G and T alleles to the optical density values of tubulin in the corresponding samples were calculated to normalize the expression of the G and T alleles of the 5-HTT. Student’s t-test was used to analyze protein data to determine the significance of expression differences between the G and T alleles.
RESULTS
Genotyping and LD analysis
DNA samples from 275 alcohol-dependent subjects were genotyped in this study. Of these subjects, 165 were Caucasians (43 females and 122 males) and 110 were Hispanics (16 females and 94 males). Genotypic distributions of all 5 SNPs and 5-HTTLPR L/S alleles, conformed to the Hardy-Weinberg equilibrium (Table 1). Further, the LD analyses using Haploview revealed no haplotype blocks among the 5 SNPs and the 5’-HTTLPR L/S polymorphism according to the criteria of Gabriel et al. (2002), in Caucasian, Hispanic or pooled populations, respectively (Figure 1).
Figure 1.
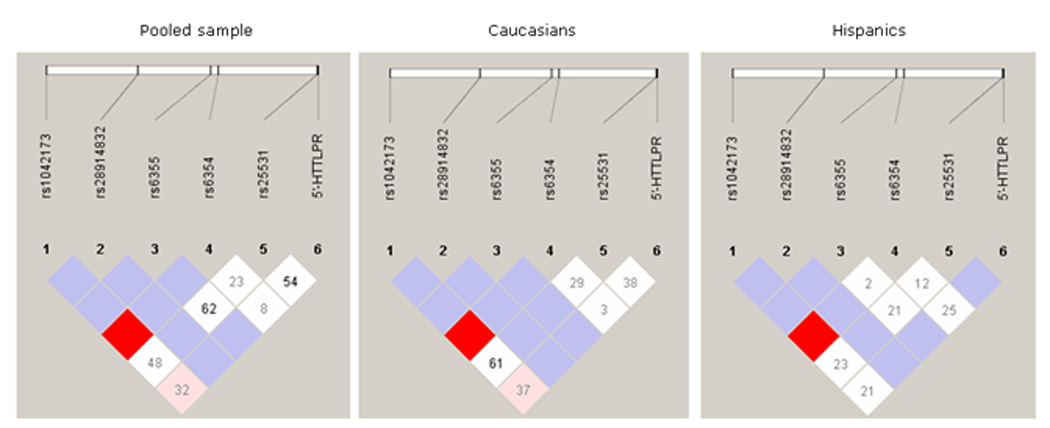
Haploview generated LD plots for the five SNPs examined in this study and 5-HTTLPR alleles of SLC6A4 gene. The pooled sample consists of subjects of Caucasian and Hispanic origin. Number in each box represents D’ values for each SNP pair.
Associations with self-reported drinking measures
To exclude potential variations caused by ethnic differences on drinking intensity, subgroups of subjects based on ethnicity were analyzed separately for all polymorphisms studied here. Among the polymorphisms analyzed individually using SAS (version 9.1) program for associations with drinking intensity, only SNP rs1042173 in the 3’ UTR of SLC6A4 showed a significant association with intensity of drinking. Table 2 shows demographic and drinking parameters of the cohort analyzed for rs1042173 association studies. No significant association was detected for other genetic polymorphisms with intensity of drinking in Caucasian, Hispanic or pooled populations (data not shown).
Table 2.
Demographics and drinking parameters in the cohort analyzed for rs1042173.
| Caucasian | Hispanic | |||||||
|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | P value | TT | TG | GG | P value | |
| Number of subjects | 47 | 77 | 41 | - | 26 | 56 | 28 | - |
| Gender (%male) | 82.97 | 64.93 | 80.49 | - | 88.46 | 85.71 | 82.14 | - |
| Age (Yr.) | 41.6±1.66 | 42.36±1.23 | 40.98±1.52 | 0.62 | 37.08±2.01 | 40.05±1.22 | 38.82±1.76 | 0.33 |
| Age of Onset of problem drinking | 29.74±1.72 | 30.61±1.25 | 28.37±1.87 | 069 | 26.44±1.67 | 26.82±1.23 | 26.36±1.87 | 0.91 |
| Baseline drinks per drinking day | 11.17±0.98 | 8.05±0.47 | 9.58±0.67 | 0.0043 | 9.99±0.71 | 10.66±0.67 | 9.76±0.58 | 0.65 |
| Baseline drinks per day | 8.99±0.96 | 6.48±0.44 | 7.72±0.58 | 0.02 | 8.23±0.75 | 7.52±0.59 | 7.92±0.55 | 0.74 |
| Years of lifetime drinking | 11.86±1.32 | 11.75±1.04 | 12.6±1.4 | 0.56 | 10.63±1.68 | 13.23±1.2 | 12.64±1.51 | 0.35 |
Notes: 1) Values are means ± S.E.M. Significant P values after correction for multiple testing are given in bold. “Years of lifetime drinking” was calculated by subtracting the age at which the subject began experiencing symptoms of alcohol dependence from their age at study enrollment.
2) The adjusted P value at the 0.05 significance level is 0.010.
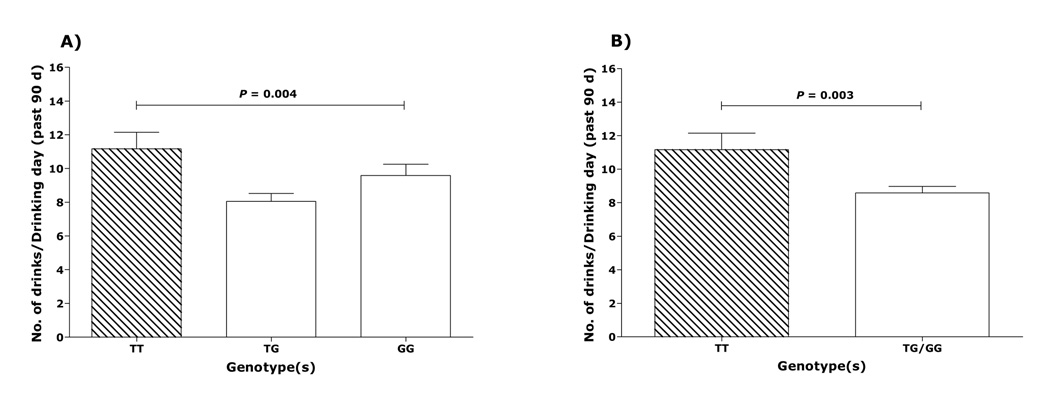
Among Caucasian subjects, mean drinks per drinking day differed significantly among TT, TG, and GG genotypes (F=5.625; p=0.004). Using Tukey’s post-hoc multiple comparison test, the differences between TG heterozygotes and TT homozygotes were statistically significant (d=4.721; p=0.002); however, the differences between TG heterozygotes and GG homozygotes were not (d=2.175; p=0.20). When TT and TG were combined and compared with GG using Student’s t-test, the means did not differ significantly (t=0.32; p=0.75). The combined means of TG and GG (Figure 2A) were significantly lower than the mean of TT (t=2.97; p=0.003). This suggests a dominant effect of the G allele over the T allele. The difference between the means of drinks per drinking day in G-carriers and TT genotypic group was 2.59±0.87 (95% CI - 0.879 to 4.297).
Figure 2.
Amounts of drinking in 165 Caucasian male and female alcoholics. (A) Amounts of drinking as a function of TT, TG and GG genotypes of rs1042173 (N of subjects in each group is: 47 TT, 77 TG, and 41 GG). Mean drinks per drinking day (± SEM) for the TT, TG and GG subjects were 11.17 ± 0.98 vs. 8.05 ± 0.47 and 9.58 ± 0.67, respectively (F=5.63; p=0.004).(B) Drinks per drinking day variance as a function of the TT and G carriers (N of subjects in each genotype is: 47 TT, 118 G carriers). Mean drinks per drinking day (± SEM) for the T homozygotes and G carriers were 11.17±0.98 vs. 8.58±0.39 respectively (t=2.97; p= 0.003).
Estrogen has been shown to modulate the synthesis, release, and metabolism of 5-HT (Bethea et al., 2002; Frackiewicz et al., 2000; Pivac et al., 2004). Thus, to examine the impact of gender on these associations, we repeated the analyses on male subjects only. In Caucasian males, the mean difference of standard drinks per drinking day between G-carriers and TT genotypic group, was 2.89±1.07 (95% CI - 0.771 to 5.009), which was similar to the mean difference in combined Caucasian male and female subjects. Therefore, we did not find a significant effect of gender on the associations between rs1042173 genotypes and drinks per drinking day.
However, among Hispanic subjects, we did not detect a significant effect of rs1042173 genotypes on both measures of drinking intensity, drinks per drinking day (F=0.935; p=0.397) and drinks per day (F = 0.299; p = 0.74).
Considering that 5’-HTTLPR L/S polymorphism has implicated as functional in many reported studies, we examined potential interactive effect of 5’-HTTLPR L/S and rs1042173 alleles on drinking intensity by using a newly developed algorithm for detecting gene-gene interaction, called generalized multifactor dimensionality reduction (GMDR) method (Lou et al. 2007). Our GMDR analyses revealed no significant interaction between these two functional SNPs (P=0.623).
5-HTT mRNA expression in cells transfected with plasmid carrying either T or G alleles
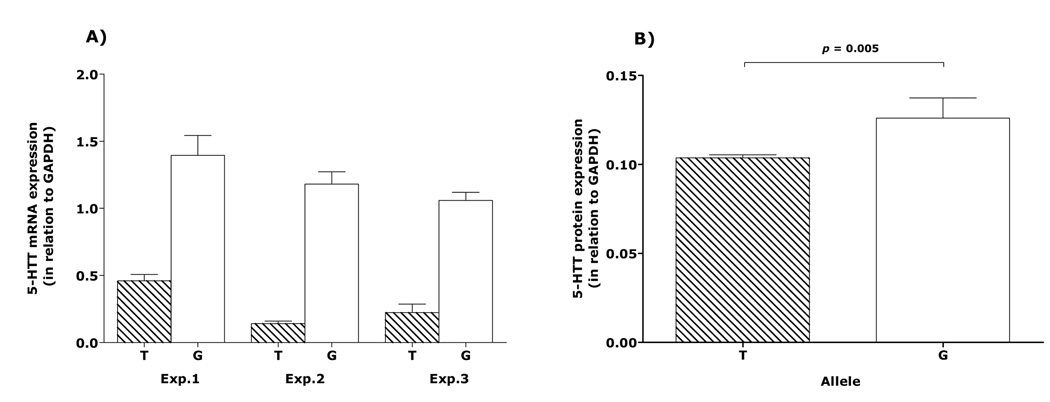
To study whether the T and G alleles of rs1042173 leads to differential expression levels of 5-HTTs, we transfected plasmids carrying the T and G alleles of rs1042173 into HeLa cells and quantified mRNA levels by using the qRT-PCR assay. Results were analyzed for allelic differences using the ΔΔCt method described by Winer et al (1999). Figure 3A depicts the mean 5-HTT mRNA expression levels for the T and G alleles from three independent transfection experiments. The G allele yielded significantly higher mRNA expression level compared with the T allele. In the three independent experiments, the G allele-transfected HeLa cells, compared with their T allele-transfected counterparts, always produced a >50% higher 5-HTT mRNA level, an effect that was significant statistically (p<0.0001).
Figure 3.
(A) Serotonin transporter (5-HTT) mRNA expression levels in HeLa cell cultures quantified by quantitative real-time PCR assay. The data shown here are mean ± SEM of four replicates for 5-HTT mRNA expressed by the T and G alleles in three separate experiments (Exp.) conducted on separate times. GAPDH = glyceraldehyde-3-phosphate dehydrogenase. (B) Average differences in optical densities of bands seen on immunoblots for serotonin transporter (5-HTT) protein expression in HeLa cell cultures for the T and G allele specific expression in three cell cultures (G: 1.23 + 0.07; T: 0.28 + 0.05; N=4)
5-HTT protein expression in the T and G alleles of the rs1042173 SNP
To determine if the allele-associated RNA difference can be translated into protein, we measured allele-specific differences in 5-HTT protein levels between the two alleles. After normalization with tubulin for the loading difference, we found that the 5-HTT protein level with G allele (0.137 ± 0.006) is significantly higher than that of T allele (0.104 ± 0.002) (t=5.53; p=0.005; Figure 3B). These results were reproduced in western blotting experiments from several independent replications. Notably, the expression of both mRNA and 5-HTT proteins was in the same direction—the G allele being associated with higher mRNA and protein expression levels than the T allele.
DISCUSSION
Our findings provide evidence that rs1042173, a SNP in the 3’ UTR of the SLC6A4 gene, was associated with intensity of drinking among Caucasians dependent on alcohol. By using a site-directed mutagenesis approach, we further showed that rs1042173 is a functional polymorphism that resulted in a difference in 5-HTT expression levels in HeLa cell cultures, with G allele associated with higher 5-HTT mRNA and protein expression levels than the T allele. Of multiple approaches used to determine whether a polymorphism is a function one, a direct comparison of expression level between two alleles through an in vitro expression system as used in this study represents one of the most convenient molecular techniques in the field. We expect such an approach will become more popular as more and more potential causative polymorphisms are being identified through association study.
Alcohol-dependent individuals who were G-allele carriers for rs1042173 showed less intensity of drinking compared with those who were homozygous for the T allele. Importantly, the average intensity of drinking for both of these allelic groups exceeded the threshold for heavy drinking (i.e., ≥5 and ≥4 standard drinks/day for men and women, respectively), and all were dependent on alcohol. At the time of entry, subjects in both allelic groups were not statistically significantly different in average chronological age and duration of alcohol dependency. It is, therefore, reasonable to propose that alcohol-dependent individuals with the TT genotype might constitute a subtype of more intense drinkers among heavy-drinking alcoholics of European descent.
To our knowledge, this is the first study to investigate the function of the rs1042173 SNP in an alcohol-dependent population. The rs1042173 polymorphism is not only located at a putative polyadenylation signal site in the 3’ UTR of the 5-HTT gene but also near a potential binding site for microRNA miRNA-135 according to a bioinformatics prediction with PicTar program (Chen et al., 2006). It has been hypothesized that a variant at this location may change expression levels by affecting the stability of mRNA (Battersby et al., 1999; Beaudoing et al., 2000; Chen et al., 2006). Therefore, our finding that those homozygous for the T allele, compared with their G-allelic counterparts, have lower mRNA and protein expression levels, providing support evidence for this hypothesis although the mechanism(s) by which these mRNA and protein expression levels change remains to be further explored in the future. Our findings have been further supported by two recent reports. The first study reported by Vallender et al. (2008) revealed that a functional haplotype containing T allele of rs1042173 was associated with higher mRNA expression in HEK293 cells compared to the haplotype consisting of G allele. Another study reported by Lim et al. (2006) showed that G-allele had increased allelic expression imbalance (AEI) in Epstein-Barr virus transformed lymphoblast cells while human pons tissue showed a decreased AEI for G-allele. Although the expression levels associated with each allele of rs1042173 are inconsistent among these studies (likely due to different reporter genes and/or cell lines used among them), they all reveal that rs1042173 is a functional one.
It is, therefore, tempting to speculate as to the mechanism whereby alcohol-dependent individuals who are homozygous for the T allele drink more severely than carriers of the G allele. Plausibly, those homozygous for the T allele compared with their Gallele-carrying counterparts will have lower expression of 5-HTT mRNA and protein levels and, therefore, a relatively higher intrasynaptic 5-HT level. Because 5-HTTs in the raphe nuclei are somatodendritic (Little et al., 1998), a reduction in their numbers will be associated with reduced 5-HT firing rates due to increased self-inhibition and, consequently, an upregulation of postsynaptic 5-HT receptors mediating the reinforcing effects of alcohol. Hence, chronic alcohol intake, which generally increases 5-HT turnover (LeMarquand et al., 1994), might be serving as a neuroadaptive response to “normalize” serotonergic function in the raphe nuclei.
We considered the possibility that a potential confound to our results was that the associations of the T and G alleles at the 3’ UTR SNP rs1042173 with intensity of drinking were being influenced by allelic differences in the promoter region of the 5-HTT gene; hence, these associations could be spurious. However, a lack of significant linkage disequilibrium and interaction between 3’ UTR SNP rs1042173 and 5-HTTLPR region L/S alleles excluded this possibility.
We took into consideration the potential for ethnicity and gender to confound our results by performing independent statistical tests to show that these factors did not influence our findings; thus, our cohort was relatively homogenous. The finding of no association between rs1042173 genotype and intensity of drinking in Hispanics, which differed from that of an association among Caucasians, while the allelic frequencies for T and G alleles in Caucasians and Hispanics were not significantly different, does suggest the possibility of differential regulation of gene expression by ethnic group. Due to the relatively small sample size of the cohort, such a premise needs to be treated as preliminary and confirmed by larger studies.
Another caveat to our results is that our cohort was a population of treatment-seeking, alcohol-dependent individuals who were motivated to decrease or cease their alcohol consumption. Therefore, this cohort may be more motivated and perhaps less dense in drinking pathophysiology compared with alcohol-dependent individuals derived from community samples (Wrase et al., 2006).
We also considered the possibility that the specified time point of 90 days prior to enrollment for measuring the level of drinking intensity might be an arbitrary point at which to perform comparative analyses with the genotype. We were, however, able to show that the association between the intensity of drinking and the genotype remained significant in the same manner even if we varied the drinking period prior to enrollment within a range of 14 to 90 days (data not shown). The consistency of these results strengthened our findings.
In summary, we detected that a subgroup of treatment-seeking adult Caucasian male and female alcohol-dependent individuals who were homozygous for the T allele of the 3’ UTR SNP rs1042173 in SLC6A4 gene were more severe drinkers than their G-allele-carrying counterparts. Using an in vitro approach, we demonstrated that the T allele, which is associated with greater intensity of drinking, expressed lower mRNA and 5-HTT protein levels compared with the G-allele carriers of rs1042173 SNP. Taken together, these findings suggest the possibility that two different subgroups of treatment-seeking alcoholics with allelic differences at the 3’ UTR SNP rs1042173 can differ in their intensity of drinking, an effect that might be associated with underlying differences in expression of 5-HTT. Further studies with a large sample size are needed to confirm our findings, and to determine whether alcohol-dependent individuals with these allelic differences at the 3’ UTR SNP rs1042173 would vary in response to different types of specific serotonergic medication.
ACKNOWLEDGMENTS
This study was funded by grants from the National Institute on Alcohol Abuse and Alcoholism to B.A.J. (Grants 7 U10 AA011776-10, 1 N01 AA001016-000, 7 R01 AA010522-12, and 5 R01 AA012964-06) and N.A.-D. (Grant 5 K23 AA000329-06) and from the National Institute on Drug Abuse to M.D.L. (Grants R01 DA012844 and 5 R01 DA013783). We are grateful to Prof. Randy D. Blakely, Director, Center for Molecular Neuroscience, Vanderbilt University School of Medicine, Nashville, Tennessee, for his generous gift of human serotonin transporter plasmid construct.
We appreciate the skilled technical assistance of the staff at the South Texas Addiction Research and Technology Center, Department of Psychiatry, University of Texas Health Science Center at San Antonio and at the Department of Psychiatry and Neurobehavioral Sciences, University of Virginia. We also are grateful to Drs. Jinxue Wei, Qing Xu, Xiang-Yang Lou, and Guobo Chen for their expert advice, Eva Jenkins-Mendoza, BS, for her service as project coordinator, and Robert H. Cormier, Jr., BA, for his assistance with manuscript preparation.
REFERENCES
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th ed. Washington, D.C: American Psychiatric Association; 1994. [Google Scholar]
- Babor TF, de la Fuente JR, Saunders J, Grant M. Geneva, Switzerland: World health organization; AUDIT: The alcohol use disorders identification test. 1992
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM, Harmar AJ. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3' untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72:1384–1388. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea CL, Lu NZ, Gundlah C, Streicher JM. Diverse actions of ovarian steroids in the serotonin neural system. Front Neuroendocrinol. 2002;23:41–100. doi: 10.1006/frne.2001.0225. [DOI] [PubMed] [Google Scholar]
- Bradley CC, Blakely RD. Alternative splicing of the human serotonin transporter gene. J Neurochem. 1997;69:1356–1367. doi: 10.1046/j.1471-4159.1997.69041356.x. [DOI] [PubMed] [Google Scholar]
- Cargiulo T. Understanding the health impact of alcohol dependence. Am J Health Syst Pharm. 2007;64(5 Suppl 3):S5–S11. doi: 10.2146/ajhp060647. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet. 2006;120:1–21. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Dundon W, Lynch KG, Pettinati HM, Lipkin C. Treatment outcomes in type A and B alcohol dependence 6 months after serotonergic pharmacotherapy. Alcohol Clin Exp Res. 2004;28(7):1065–1073. doi: 10.1097/01.alc.0000130974.50563.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinn R, Nellissery M, Kranzler HR. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2005;133B(1):79–84. doi: 10.1002/ajmg.b.30132. [DOI] [PubMed] [Google Scholar]
- Frackiewicz EJ, Sramek JJ, Cutler NR. Gender differences in depression and antidepressant pharmacokinetics and adverse events. Ann Pharmacother. 2000;34:80–88. doi: 10.1345/aph.18465. [DOI] [PubMed] [Google Scholar]
- Gastfriend DR, Garbutt JC, Pettinati HM, Forman RF. Reduction in heavy drinking as a treatment outcome in alcohol dependence. J Subst Abuse Treat. 2007;33(1):71–80. doi: 10.1016/j.jsat.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nature Reviews/Genetics. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Gill K, Amit Z. Serotonin uptake blockers and voluntary alcohol consumption. A review of recent studies. Recent Dev Alcohol. 1989;7:225–248. doi: 10.1007/978-1-4899-1678-5_12. [DOI] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, Lesch KP. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An Expanded Evaluation of the Relationship of Four Alleles to the Level of Response to Alcohol and the Alcoholism Risk. Alcohol Clin Exp Res. 2005;29(1):8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78(5):815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Swift RM, Ait-Daoud N, DiClemente CC, Javors M, Malcolm RJ., Jr Development of novel pharmacotherepies for the treatment of alcohol dependence: focus on antiepileptics. Alcohol Clin Exp Res. 2004;28(2):295–301. doi: 10.1097/01.alc.0000113409.47937.6c. [DOI] [PubMed] [Google Scholar]
- Javors MA, Seneviratne C, Roache JD, Ait-Daoud N, Bergeson SE, Walss-Bass MC, Akhtar FZ, Johnson BA. Platelet serotonin uptake and paroxetine binding among allelic genotypes of the serotonin transporter in alcoholics. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29(1):7–13. doi: 10.1016/j.pnpbp.2004.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Javors MA, Roache JD, Seneviratne C, Bergeson SE, Ait-Daoud N, Dawes MA, Ma JZ. Can serotonin transporter genotype predict serotonergic function, chronicity, and severity of drinking? Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(1):209–216. doi: 10.1016/j.pnpbp.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon YS, Lee HK, Lee CT, Lee KU, Pae CU. Association of the serotonin transporter gene polymorphism with Korean male alcoholics. Journal of Psychiatric Research. 2005;39:371–376. doi: 10.1016/j.jpsychires.2004.10.005. [DOI] [PubMed] [Google Scholar]
- LeMarquand D, Pihl RO, Benkelfat C. Serotonin and alcohol intake, abuse, and dependence: clinical evidence. Biol Psychiatry. 1994;36:326–337. doi: 10.1016/0006-3223(94)90630-0. [DOI] [PubMed] [Google Scholar]
- Lim JE, Papp A, Pinsonneault J, Sadee W, Saffen D. Allelic expression of serotonin transporter (SERT) mRNA in human pons: lack of correlation with the polymorphism SERTLPR. Mol Psychiatry. 2006;11(7):649–662. doi: 10.1038/sj.mp.4001797. [DOI] [PubMed] [Google Scholar]
- Little KY, McLaughlin DP, Zhang L, Livermore CS, Dalack GW, McFinton PR, DelProposto ZS, Hill E, Cassin BJ, Watson SJ, Cook EH. Cocaine, ethanol, and genotype effects on human midbrain serotonin transporter binding sites and mRNA levels. Am J Psychiatry. 1998;155:207–213. doi: 10.1176/ajp.155.2.207. [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela P, Mustonen H. How do quantities drunk per drinking day and the frequencies of drinking those quantities contribute to self-reported harm and positive consequences? Alcohol Alcohol. 2007;42(6):610–617. doi: 10.1093/alcalc/agm066. [DOI] [PubMed] [Google Scholar]
- Mynett-Johnson L, Kealey C, Claffey E, Curtis D, Bouchier-Hayes L, Powell C, McKeon P. Multimarkerhaplotypes within the serotonin transporter gene suggest evidence of an association with bipolar disorder. Am J Med Genet. 2000;96(6):845–849. doi: 10.1002/1096-8628(20001204)96:6<845::aid-ajmg30>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, Rudnick G, Murphy DL. Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry. 2003;8(11):933–936. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, Rudnick G, Murphy DL. Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry. 2003;8(11):933–936. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- Pivac N, Muck-Seler D, Mustapic M, Nenadic-Sviglin K, Kozaric-Kovacic D. Platelet serotonin concentration in alcoholic subjects. Life Sci. 2004;76:521–531. doi: 10.1016/j.lfs.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Prasad HC, Zhu CB, McCauley JL, Samuvel DJ, Ramamoorthy S, Shelton RC, Hewlett WA, Sutcliffe JS, Blakely R. Human serotonin transporter variants display altered sensitivity to protein kinase G and p38 mitogen-activated protein kinase. PNAS. 2005;102(32):11545–11550. doi: 10.1073/pnas.0501432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Galli A, Ramamoorthy S, Risso S, DeFelice LJ, Blakely RD. Protein kinase C activation regulates human serotonin transporters in HEK-293 cells via altered cell surface expression. J Neurosci. 1997;17:45–57. doi: 10.1523/JNEUROSCI.17-01-00045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Bauman AL, Moore KR, Han H, Yang-Feng T, Chang AS, Ganapathy V, Blakely RD. Antidepressant- and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc Natl Acad Sci U S A. 1993;90:2542–2546. doi: 10.1073/pnas.90.6.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Sykora K, Schneiderman J, Naranjo CA, Sellers EM. Assessment of alcohol withdrawal: the revised clinical institute withdrawal assessment for alcohol scale (CIWA-Ar) Br J Addict. 1989;84:1353–1357. doi: 10.1111/j.1360-0443.1989.tb00737.x. [DOI] [PubMed] [Google Scholar]
- Talvenheimo J, Rudnick G. Solubilization of the platelet plasma membrane serotonin transporter in an active form. J Biol Chem. 1980;255:8606–8611. [PubMed] [Google Scholar]
- Vallender EJ, Priddy CM, Hakim S, Yang GL, Miller GM. Functional variation in the 3' untranslated region of the serotonin transporter in human and rhesus macaque. Genes, Brain Behav. 2008 doi: 10.1111/j.1601-183X.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11:224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Wrase J, Reimold M, Puls I, Kienast T, Heinz A. Serotonergic dysfunction: brain imaging and behavioral correlates. Cogn Affect Behav Neurosci. 2006;6:53–61. doi: 10.3758/cabn.6.1.53. [DOI] [PubMed] [Google Scholar]