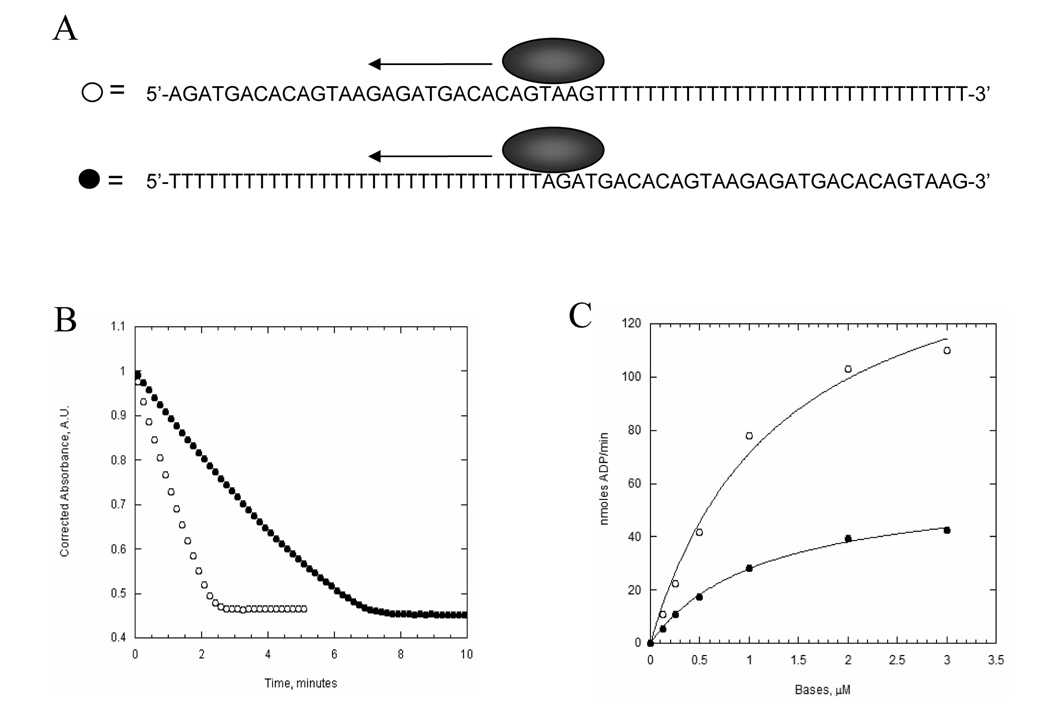
Figure 4. The poly-dT assay for translocation directionality.
A. An illustration depicting the 3’ to 5’ translocation of UvsW helicase on ssDNA containing d(T)30 on either the 5’ or 3’ end of d(N)30. UvsW will bind at random positions throughout the ssDNA with an average position in the center of the substrate. Because activation of ATP hydrolysis is minor with d(T)30 as compared to d(N)30, translocation away from the d(T)30 segment leads to higher ATPase activity than translocation into the d(T)30 segment. B. Time courses of ATP hydrolysis by UvsW (20 nM) in the presence of 2 µM 5’-d(T)30-d(N)30-3’ (closed circles) or 5’-d(N)30-d(T)30-3’ (open circles). C. Rate of ATP hydrolysis by UvsW as a function of 5’-d(T)30-d(N)30-3’ (closed circles) or 5’-d(N)30-d(T)30-3’ (open circles) concentration. The solid lines are fits to the data using the Michaelis-Menten equation and these values are found in Table II.