Abstract
Increasingly complex networks of small RNAs act through RNA interference pathway to regulate gene expression. Recent evidence suggests that both development and proper function of central nervous system require intricate spatiotemporal expression of a wide repertoire of small regulatory RNAs. Misregulation of these small regulatory RNAs could contribute to the abnormalities in brain development that are associated with neurodevelopmental disorders. Here, we will review recent progress made toward understanding roles of small regulatory RNAs in neurodevelopmental disorders and discuss the potential involvement of newly discovered classes of small RNAs in these disorders.
INTRODUCTION
RNAs are an integral component of chromosomes and contribute to their structural organization (1). RNAs can regulate gene expression at many levels and via an array of mechanisms. Genome projects have shown that at least 93% of human genome nucleotides analyzed are transcribed in different cells, with similar findings for the mouse and other eukaryotes, indicating that there may be a vast reservoir of biologically meaningful RNAs that could far exceed the ∼1.2% of encoding proteins (1–3). Uncovering the functions of these non-coding RNAs could significantly improve our understanding and treatment of human diseases. Recently, small non-coding RNAs were found in such abundance that they were dubbed the ‘dark matter’ of the cell (4); small non-coding RNA guides, including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs) and endogenous small interfering RNAs (esiRNAs), are 18–30 nt in length and can shape diverse cellular pathways, from chromosome architecture, development and growth control to apoptosis and stem cell maintenance (5–8).
The common trait linking the large group of neurodevelopmental disorders is that disease onset occurs during periods of ongoing maturation and development (9). These disorders are often associated with complex neuropsychiatric problems, including intellectual disability, autism, attention deficit hyperactivity disorder (ADHD) and epilepsy, among others. Neurodevelopmental disorders are caused by a wide range of genetic mutations and epigenetic and environmental factors, which lead to the changes in development, possibly via the same alterations in neurogenesis, cell migration and neuronal connectivity that are responsible for cognitive deficits in adults (9). Small regulatory RNAs, particularly miRNAs, are known to be dynamically regulated in neurogenesis and brain development (10,11). Some recent studies have suggested that the alterations in small regulatory RNAs could contribute to the pathogenesis of several neurodevelopmental disorders. In this review, we will focus on the role(s) of small regulatory RNAs in several well-defined genetic disorders, although the basic information presented here is more broadly relevant and therefore applicable to neurodevelopmental disorders in general.
BIOGENESIS OF SMALL RNAs
Given the pivotal roles of endogenous small RNAs in diverse biological pathways and the broad application of RNA interference (RNAi), understanding the mechanism of the small RNA pathway is of great importance (12). In recent years, extensive research has revealed distinct classes of small RNAs and the key protein components involved in the biogenesis of each class of small RNAs.
MicroRNAs
miRNAs are 18–25 nt, small non-coding regulatory RNAs that are known to regulate translation of target messenger RNA (mRNA) molecules in a sequence-specific manner. In mammals, the majority of endogenous miRNA genes are transcribed initially as primary transcripts (pri-miRNAs) that range from hundreds to thousands of nucleotides in length and contain one or more extended hairpin structures (13). The nuclear RNase III enzyme Drosha, working with DGCR8, cleaves both strands near the base of the primary stem-loop and yields the precursor miRNA (pre-miRNA) (Fig. 1). After being exported to the cytoplasm by exportin-5/RanGTP, pre-miRNAs are further cleaved by the RNase III Dicer, along with a double-stranded RNA (dsRNA)-binding protein, and TAR RNA-binding protein (TRBP) (13). The Dicer–TRBP complex is also required for the processing of short hairpin RNA (shRNA) into small interference RNA (siRNA) of ∼21 bp. After cleavage by Dicer and unwinding by RNA helicase, one strand of the miRNA/miRNA* or siRNA duplex (the antisense, or guide strand) is then preferentially incorporated into the RNA-induced silencing complex (RISC), whereas the other strand (the sense, or passenger strand) is degraded (Fig. 1). The RISC is a large and heterogeneous multi-protein complex. Components of the RISC that have been identified include Dicer, TRBP and Argonaute 2 protein (AGO2) (13).
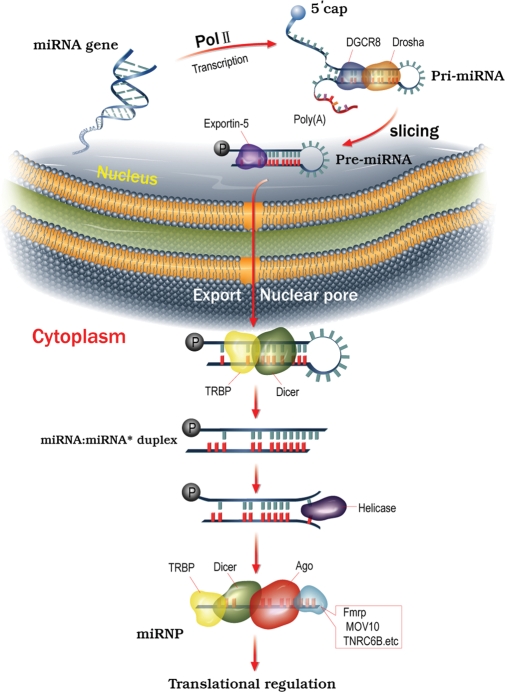
Figure 1.
miRNA biogenesis. Genes encoding miRNAs are initially transcribed by RNA polymerase II or III to generate the pri-miRNA transcripts within the nucleus. The stem-loop structure of the pri-miRNA is recognized and cleaved on both strands by a microprocessor complex, which consists of the nuclear RNase III enzyme Drosha and an RNA-binding protein, DGCR8, to yield a pre-miRNA 60–70 nt in length. The pre-miRNA is then exported from the nucleus through a nuclear pore by exportin-5 in a Ran-GTP-dependent manner and processed in the cytoplasm by the RNase III Dicer–TRBP. Sliced RNA strands are further unwound by an RNA helicase. One strand of the miRNA/miRNA* or siRNA duplex (the antisense, or guide strand) is then preferentially incorporated into the RISC (or miRNP for miRNAs) and will guide the miRNP to a target mRNA in a sequence-specific manner. Once directed to a target mRNA, the RISC can mediate translational regulation by inhibiting the initiation or elongation step or through destabilization of the target mRNA. Alternatively, miRNAs may also upregulate translation of target mRNAs under certain conditions.
The RISC uses the guide RNA to find complementary mRNA sequences via Watson–Crick base pairing, which leads to post-transcriptional gene silencing through inhibition of either translation initiation or elongation (13,14). miRNA could also negatively regulate protein expression through targeting of mRNA coding regions (15). Furthermore, miRNAs are found to upregulate the translation of target mRNAs in a cell cycle-dependent manner, switching between translational suppression in proliferating cells to translational activation in quiescent cells (16–18). Hence, a single miRNA may simultaneously regulate the expression of multiple mRNA targets and thereby act as a rheostat to fine-tune protein expression (19).
Piwi-interacting RNAs
piRNAs represent a distinct class of small RNAs that interact with Piwi proteins in both mammals and Drosophila (6–8,20). piRNAs interact with the Piwi proteins, but not Argonaute 2, the key protein in the RNAi pathway (21). Piwi protein is required for piRNA biogenesis and stability (22). piRNAs are 24–31 nt long, which differs from both siRNAs and miRNAs (21). High-throughput sequencing has revealed that the number of distinct piRNAs is much higher (more than 50,000) than miRNAs (several hundreds) (22). Most piRNAs match to the genome in clusters of 20–90 kb in a strand-specific manner, with each cluster likely representing a long single-stranded RNA precursor or, more often, two non-overlapping and divergently transcribed precursors (22). In contrast, siRNAs and miRNAs are derived from dsRNA and shRNA precursors, respectively.
Unlike siRNAs or miRNAs, the biogenesis of piRNAs is Dicer-independent (21). piRNAs are likely produced from long single-stranded precursors by yet-to-be-identified endonucleases. In Drosophila, a ‘ping-pong’ model is proposed to be involved in the generation of some transposon-derived piRNAs, although the detailed biogenesis of piRNAs in both mammals and Drosophila remains to be determined (23,24). Most piRNAs map to unique sites in the genome, including intergenic, intronic and exonic sequences. For example, only 17–20% of mammalian piRNAs map to annotated repeats, including transposons and retrotransposons (25). Thus, piRNA could have diverse functions, from epigenetic programming and repressing transposition to post-transcriptional regulation (21).
Endogenous small interfering RNAs
More recently, several groups described a rich diversity of esiRNAs in mice and Drosophila (6,8,26–28). Most of these esiRNA classes seem to be analogous between species and include those derived from transposable elements, from complementary annealed transcripts and from lone ‘fold-back’ transcripts called hairpin RNAs (29). esiRNAs in particular could be generated from mammalian pseudogene-gene pairs (26). Studies in Drosophila suggest that esiRNA biogenesis requires components involved in the siRNA/miRNA pathway; however, the mechanism of esiRNA biogenesis remains a mystery (29), as does the specific biological function(s) of esiRNAs.
miRNAs IN NEURODEVELOPMENTAL DISORDERS
Fragile X syndrome
Fragile X syndrome (FXS), one of the most common forms of inherited mental impairment, was the first genetic disorder linked to the miRNA pathway (30–32). Clinical presentations of FXS include learning disabilities and more severe cognitive or intellectual disabilities. Fragile X patients have characteristic physical and behavioral features and experience delays in speech and language development (33).
In 1991, positional cloning of the fragile X mental retardation-1 (FMR1) gene revealed the molecular basis of FXS; the syndrome is associated with a massive unstable CGG trinucleotide repeat expansion within the gene’s 5′ untranslated region (5′-UTR) (34–36). The functional FMR1 gene product, fragile X mental retardation protein (FMRP), belongs to a small and highly conserved RNA-binding protein family that has been implicated in translational control (37–41). FMRP functions as a suppressor of target mRNA translation via binding of non-coding RNA structures, including G-quartets and ‘kissing complexes’ (also known as loop-loop-pseudoknots), within the UTRs of target mRNAs (37,39–45).
FMRP interacts biochemically and genetically with known components of the miRNA pathway. Experiments in Drosophila revealed specific biochemical interactions between dFmrp and functional RISC proteins, including dAGO1, dAGO2 and Dicer (30,31,46). dFmr1 displays strong genetic interaction with dAGO1, and dAGO1 dominantly interacts with dFmr1 in both dFmr1 overexpression and loss-of-function models (32). Furthermore, dFmr1 also interacts genetically with AGO2, as exemplified by their ability of co-regulating ppk1 mRNA levels (46). Additional studies provide further evidence supporting the involvement of FMRP in miRNA-containing RISC and P body-like granules in Drosophila neurons (47). Recombinant human FMRP is able to act as an acceptor for Dicer-derived miRNAs, and importantly, endogenous miRNAs themselves are associated with FMRP in both flies and mammals (30–32). In adult mouse brain, Dicer and eIF2c2 (the mouse homolog of AGO1) interact with FMRP at postsynaptic densities (48). Presumably, this interaction works to regulate translation of target mRNAs in an activity-dependent manner. Based on these findings, it has been proposed that the RISC proteins, including Argonaute and Dicer, could interact with FMRP and use the loaded guide miRNA(s) to interact with target sequences within the 3′-UTR of mRNA bound to FMRP, and suppress translation (32,49). In this model, FMRP facilitates the interaction between miRNAs and their target mRNA sequences, ensuring proper targeting of guide miRNA-RISC within the 3′-UTRs and proper translational suppression (Fig. 2).
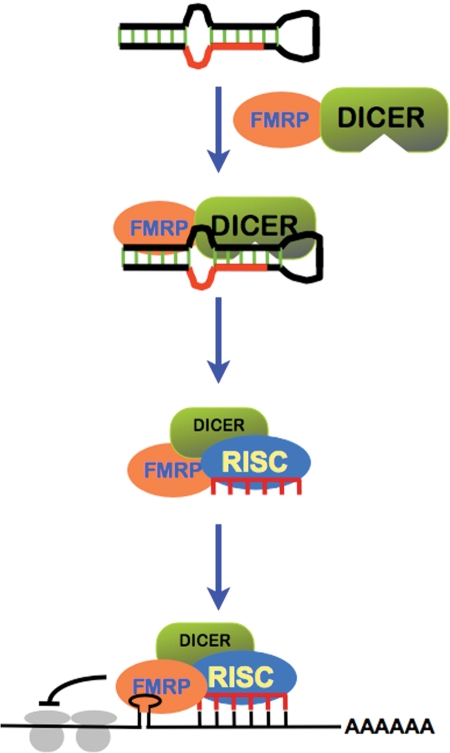
Figure 2.
miRNA pathway in FMRP-mediated translational control. FMRP interacts with Dicer and RISC, and could participate the processing of miRNA precursors into mature miRNAs (i.e. miR-124a in Drosophila). FMRP could bind to mRNA through either G-quartet/stem structure or ‘kissing complexes’. Once FMRP binds to its mRNA ligands, it could recruit RISC along with specific miRNAs (i.e. bantam in Drosophila) to its mRNA ligands and facilitate the recognition between miRNAs and mRNAs, which could modulate the translation of the bound mRNA ligands.
The fact that FMRP is associated with Dicer, miRNAs and specific mRNA targets raised the question of whether FMRP is associated with specific miRNAs and modulates their processing. To address this question, the expression and processing of miRNAs were examined in Drosophila dfmr1 mutants. In fly brain, dFmrp was found to be specifically associated with miR-124a, a nervous-system-specific miRNA (50). The proper processing of pre-miR-124a requires dFmrp, whereas the loss of dFmr1 leads to a reduced level of mature miR-124a and an increased level of pre-miR-124a. These results suggest a modulatory role for dFmrp to maintain proper levels of miRNAs during neuronal development (50). In our own studies, we have shown that dFmr1 plays a role in the proper maintenance of germline stem cells in Drosophila ovary, potentially through the miRNA pathway (51). To further test this hypothesis we used an immunoprecipitation assay and revealed that specific miRNAs, particularly the bantam miRNA (bantam), are physically associated with dFmrp in ovary (52). We found that like dFmr1, bantam is not only required for repressing primordial germ cell differentiation, but also functions as an extrinsic factor for germline stem cell maintenance (52). Furthermore, we showed that bantam genetically interacts with dFmr1 to regulate the fate of germline stem cells (52). Collectively, our results support the notion that the FMRP-mediated translational pathway functions through specific miRNAs to control stem cell regulation; however, we saw no effect of dFmrp on the biogenesis of the bantam miRNA. Whether FMRP is associated with specific miRNAs in mammalian cells remains to be determined.
Intriguingly, another member of the fragile X-related (FXR) protein family, FXR1, has also been implicated in miRNA-mediated translational upregulation through an association with AGO2 on AU-rich 3′-UTRs in quiescent cells (16–18). Nonetheless, the relevance of these observations to FMRP-mediated translational regulation requires further exploration.
Rett syndrome
De novo mutations in MECP2 are known to cause the X-linked dominant neurodevelopmental disorder Rett syndrome (RTT) (53). MECP2 encodes the DNA methyl-CpG-binding protein, MeCP2 (54). The general association of methyl CpG dinucleotides with heterochromatic or transcriptionally silent regions of the genome led to the hypothesis that MeCP2 normally functions as a component of transcriptional repressor complexes (55,56). More recently, MeCP2 has also been shown to function as a transcriptional activator at certain loci (57). MeCP2 null and MeCP2 transgenic mouse models, which, respectively, mimic loss-of-function MECP2 mutations and MECP2 gene duplications, also display RTT-like phenotypes (55). Furthermore, recent clinical observations correlated duplications of MECP2 with Rett-like phenotypes, although overall such duplications result in clinically distinct phenotypes (55). Together, these observations are consistent with a dose-dependent mechanism for MeCP2-mediated regulation of target transcripts whose misexpression during development is pathogenic. To date, most concerted efforts to identify MeCP2 target transcripts have focused on protein-coding mRNA transcripts. These approaches have revealed a number of direct MeCP2 target genes in specific cell and tissue types; however, there are several recent observations surrounding the involvement of small regulatory RNAs in MeCP2 function.
Examination of an imprinted locus on mouse chromosome 9, in which genes are known to be imprinted and expressed specifically in brain, revealed that MeCP2 binds upstream and regulates the paternal expression of an miRNA (miR-184) located within 55 kb of the imprinted locus. Moreover, the induction of miR-184 expression in depolarized cultured neurons is concomitant with a loss of MeCP2-binding upstream of the miR-184 locus. These data suggest that the regulation of miR-184 expression by MeCP2 is activity-dependent; however, the expression of miR-184 was not significantly changed in whole brain tissue derived from MeCP2-deficient mice (58).
The cAMP response element-binding (CREB) protein is known to be a critical transcription factor regulating neuronal plasticity and activity-dependent refinement of dendritic branching, both of which are defective processes in RTT patients. Initial identification of CREB protein targets revealed an miRNA (miR-132) that was predicted to post-transcriptionally regulate MeCP2. In postnatally cultured rat neurons, miR-132 did in fact directly repress the expression of MeCP2. However, by blocking miR-132-mediated regulation of MeCP2 and thereby increasing MeCP2 levels, the expression of brain-derived neurotrophic factor (BDNF) was found to be increased (59). Since BDNF is both a known target of MeCP2 and an activator of CREB, these findings together led to the hypothesis that miR-132 functions within a feedback loop involving homeostatic regulation of MeCP2 expression via BDNF-activated CREB (Fig. 3). Homeostatic regulation of MeCP2 by miR-132 may indicate a mechanism by which MeCP2 levels are normally maintained within the narrow range required for proper neuronal development and synaptic maturation in the postnatal brain, highlighting the importance of miRNA in these processes (60).
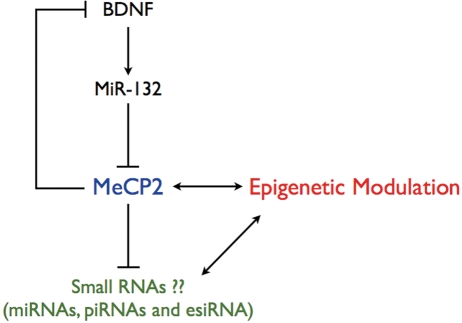
Figure 3.
Small RNAs in MeCP2-mediated epigenetic modulation. Homeostatic regulation of MeCP2 expression by miR-132 is mediated by BDNF-activated CREB. Besides directly influencing the expression of mRNA protein-coding transcripts, MeCP2 may also regulate the transcription of non-coding RNA elements, such as miRNAs, piRNAs and esiRNAs. The altered expression of small regulatory RNAs could impact epigenetic modulation as well.
Regulation of miRNA expression provides an alternative means by which MeCP2-mediated epigenetic regulation could ultimately influence protein expression and phenotype. Rather than directly influencing the expression of mRNA protein-coding transcripts, MeCP2 may also regulate the transcription of non-coding RNA elements, such as miRNA. Thus, in the absence of MeCP2, some miRNAs might display increased expression, which may result in a negative effect on the translation of mRNAs targeted by that particular miRNA (Fig. 3). So it is important now to determine whether MeCP2 can directly regulate the expression of miRNA genes and the role of miRNA(s) in the pathogenesis of RTT.
DiGeorge syndrome
DiGeorge syndrome (DGS) is a rare congenital disease that is inherited in an autosomal dominant manner. Symptoms vary greatly among individuals, but commonly include a history of recurrent infection, heart defects and characteristic facial features. Individuals with DGS have behavioral and cognitive deficits that lead to childhood pathologies, including ADHD, obsessive-compulsive disorder and autism spectrum disorder (61,62). These manifestations are the result of a common chromosomal abnormality, a large 3 Mb hemizygous deletion on chromosome 22 (22q11.2) that is produced by an error in recombination at meiosis (63). This region (called the DiGeorge critical region) comprises more than 25 genes, making this syndrome a classic contiguous gene syndrome. Despite numerous human and mouse studies implicating a small subset of these genes (e.g. Tbx1, Comt, Prodh and Gngl1) as contributors to the morphological or behavioral phenotypes of this syndrome (64–67), the genetic basis of the cognitive impairments has gone largely unexplained. Recently, the heterozygous disruption of a single gene found in the DiGeorge critical region, Dgcr8, was found to result in cognitive delay, specifically in spatial working and memory-based learning (68). DGCR8 forms a microprocessor complex along with Drosha to process the pri-miRNAs (Fig. 1) (5). Mature miRNAs were reduced in the brains of mice containing either the Dgcr8 disruption or the syntenic hemizygous deletion of the DiGeorge critical region (68). Together, these data argue that the heterozygous loss of DGCR8 causes abnormal miRNA biogenesis and leads to a deficit in cognitive performance; however, whether specific miRNAs are responsible for cognitive deficits associated with these mutants remains to be determined. The identification of the downstream targets that are misregulated in these miRNA-deficient mutants would also provide further insight into the pathogenesis of DGS, as well as a better understanding of learning and cognition more generally.
Down syndrome
Down syndrome (DS), which affects 1 in 700 newborns, has a variable phenotype that includes congenital heart defects, craniofacial abnormalities and cognitive impairment (69). DS is caused by triplication of all or part of human chromosome 21 and is often referred to as trisomy 21. The extra chromosomal segment results in an increase in gene dosage by as much as 50% in multiple genes, which perhaps explains the DS phenotype (70,71). Genotype and phenotype correlations of partial trisomy cases allowed for the identification of a Down syndrome critical region (DSCR); duplication of this region is associated with many of the DS phenotypes, particularly mental retardation (72,73). To date, we know of more than 30 genes overexpressed in key brain regions in DS individuals; 13 of these genes reside in the DSCR (74).
Recently, the potential contribution of miRNAs to the pathogenesis of DS has been investigated. Bioinformatic analyses revealed that chromosome 21 encodes five miRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802), all of which are overexpressed in fetal brain and heart tissues from DS individuals, suggesting that they might contribute, at least in part, to the cognitive and cardiac defects seen in DS (75). Notably, none of these miRNAs are located in the DSCR; however, a role for miRNAs in DS is supported by the finding that miR-155 downregulates a human gene associated with hypertension, angiotensin II type 1 receptor (AGTR1) (76). Indeed, DS individuals do have lower blood pressure and lower AGTR1 protein levels than those without DS. Associations between an miRNA and a DS phenotype are unlikely to be rare, even for miR-155, since each miRNA has the ability to regulate a large number of protein-coding genes (77). Moreover, improved computational and experimental methods continue to reveal the location of new miRNAs, suggesting that there remain unidentified miRNAs residing on chromosome 21, and in the DSCR, which could make excellent candidates to study the molecular pathogenesis of DS further.
Other neurodevelopmental disorders linked to miRNAs
In addition to the disorders discussed above, there are several others that have a potential link to altered miRNA expression. The region associated with MRX3 and Waisman syndrome (early-onset Parkinsonism with mental retardation) harbors an miRNA, miR-175 (78). Furthermore, a microdeletion at chromosome Xp11.3 that accounts for cosegregation of retinitis pigmentosa and X-linked mental retardation in a large kindred contains two highly conserved miRNAs, miR-221 and miR-222 (79). Segmental duplications at breakpoints (BP4–BP5) of chromosome 15q13.2q13.3 result in microdeletions/duplications that are associated with a variety of neuropsychiatric abnormalities, including features of autism, ADHD, anxiety disorder, mood disorder, mental retardation, epilepsy and in some instances EEG abnormality. The ∼1.5 Mb region spanning BP4–BP5 includes six reference genes and one miRNA, miR-211, although some patients were found to have a smaller ∼500 kb deletion that includes only three reference genes and miR-211 (80). Furthermore, a recent study finds that altered miRNA expression is observed in postmortem cerebellar cortex from autistic patients (81). Nonetheless, it is important to note that whether the miRNAs associated with each disorder could contribute to disease pathogenesis remains to be determined.
In summary, miRNAs are abundant in the nervous system, where they are involved in neural development and are likely an important mediator of neuronal plasticity. Given that miRNAs play a role in the fine-tuning of protein production, they could contribute significantly to the molecular pathogenesis of neurodevelopmental disorders. Aside from the altered miRNA transcription and biogenesis, the dosage of miRNA genes associated with segmental duplication could contribute to the phenotypes, as well. Furthermore, there are significant numbers of single nucleotide polymorphisms in the human genome, which could potentially create or disrupt the putative miRNA target sites. Therefore, variations in the target mRNA sequences could also modulate the activity of specific miRNAs and contribute to phenotypic variation (82–84). It is likely that many of these variations will affect neuronal miRNAs.
piRNAs AND esiRNAs IN NEURODEVELOPMENTAL DISORDERS?
Besides miRNAs, numerous piRNAs and esiRNAs have been identified in genomes (6–8,20). piRNAs in particular have been linked to control of the mobilization of transposable elements in both mouse and Drosophila (22). Although they were initially discovered only in reproductive systems, mounting evidence from recently published work suggests that piRNAs and esiRNAs are present in both germline and somatic tissues. So the question becomes what other functions these small RNAs could play, besides modulating the activity of transposable elements.
Recent studies in Drosophila suggest that piRNAs could play important roles in epigenetic regulation. Piwi protein was found to colocalize with Polycomb group (PcG) proteins to cluster PcG response sequences in the genome, as well as with HP1 protein to modulate epigenetic silencing (85–87). Conversely, Piwi protein and its associated piRNA can also promote the euchromatic character of certain heterochromatin regions and their transcriptional activity (88). Interestingly, it was found that Piwi protein interacts with dFmrp in Drosophila; however, whether dFmrp is involved in the piRNA pathway and related epigenetic regulation remains to be determined (89). More recently, maternally deposited piRNAs were found to play a significant role in mounting an effective silencing response, and a lack of maternal piRNA inheritance was revealed to be behind hybrid dysgenesis, in which crosses between different fly strains that differ in the presence of a particular transposon could produce sterile progeny (90). Thus, maternally inherited piRNAs could contribute to epigenetic control; however, it remains to be determined whether there is a similar phenomenon in mammals. These data together suggest that piRNAs could be involved in epigenetic modulation. In humans, the belief is that epigenetic modulations may serve as an intermediate process that imprints dynamic environmental experiences on the ‘fixed’ genome, resulting in the stable alteration of phenotypes (91,92). Disturbance in epigenetic regulation could lead to the inappropriate expression or silencing of genes, causing an array of multisystem disorders, particularly neurodevelopmental disorders (92). Given the role of piRNAs in epigenetic modulation, it would be interesting to explore the potential role(s) of piRNAs in neurodevelopmental disorders.
In addition to the above, in recent years, segmental changes in DNA copy number have been recognized as particularly common in mammals. A substantial fraction of genomic DNA (∼2–6%) is contained within segmental duplications, and copy number variation (CNV) is widespread among different humans and chimpanzees, as well as among inbred mouse strains (93–95). This active acquisition, duplication and dispersal of large gene-containing genomic segments are part of an ongoing evolutionary process and could contribute to the pathogenesis of neurodevelopmental disorders, including autism (96–99). However, what drives this genomic evolution remains unknown. In prokaryotes, genomic evolution is assisted by the integration of gene pools from phages and plasmids, or genomic islands. Hot regions for the integration of genomic islands are close to non-coding RNAs, such as tRNAs or small RNAs (100). In addition, RNA is known to be capable of guiding genome rearrangement in ciliates, a lower eukaryote (101). So it will be intriguing to test whether small non-coding RNAs, including both piRNAs and esiRNA, could be the components of the pathway modulating the dynamics of CNV in mammals.
SUMMARY
Recent discoveries of different small regulatory RNAs, including miRNAs, piRNAs and esiRNAs, have revealed a new layer of gene regulation. These ‘micro’ regulatory RNAs could play ‘macro’ roles in shaping diverse cellular pathways. Emerging data suggest that small regulatory RNAs, particularly miRNA, could contribute to the pathogenesis of neurodevelopmental disorders. We expect that these findings are just the tip of the iceberg, with different types of small RNAs possibly being involved in disease pathogenesis at different levels and via multiple distinct mechanisms. We therefore must take small regulatory RNAs into consideration when trying to identify disease-causing gene(s) and dissect the biological pathway(s) altered in neurodevelopmental disorders.
FUNDING
This work was supported by the National Basic Research Program of China [2007CB947502, 2006CB944000, 2007CB507400 to D.C.]; the National Natural Science Foundation of China (NSFC) [30630042, 30825026 to D.C., 30800647 to S.W.]; and the National Institutes of Health [R01 NS051630, R01 MH076090 to P.J.]. P.J. is also a recipient of the Beckman Young Investigator Award and the Basil O’Connor Scholar Research Award, as well as an Alfred P. Sloan Research Fellow in Neuroscience.
ACKNOWLEDGEMENTS
We would like to thank C. Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Amaral P.P., Dinger M.E., Mercer T.R., Mattick J.S. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick J.S. The functional genomics of noncoding RNA. Science. 2005;309:1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 5.Plasterk R.H. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 6.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T.N., Siomi M.C., Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 8.Okamura K., Chung W.J., Ruby J.G., Guo H., Bartel D.P., Lai E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehninger D., Li W., Fox K., Stryker M.P., Silva A.J. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60:950–960. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krichevsky A.M., Sonntag K.C., Isacson O., Kosik K.S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosik K.S., Krichevsky A.M. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Dykxhoorn D.M., Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 13.Du T., Zamore P.D. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan S., Tong Y., Steitz J.A. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasudevan S., Steitz J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 19.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siomi H., Siomi M.C. Interactions between transposable elements and Argonautes have (probably) been shaping the Drosophila genome throughout evolution. Curr. Opin. Genet. Dev. 2008;18:181–187. doi: 10.1016/j.gde.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- 22.Aravin A.A., Hannon G.J., Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 23.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 24.Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 25.Kim V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 26.Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 28.Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z., et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura K., Lai E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caudy A.A., Myers M., Hannon G.J., Hammond S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishizuka A., Siomi M.C., Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin P., Zarnescu D.C., Ceman S., Nakamoto M., Mowrey J., Jongens T.A., Nelson D.L., Moses K., Warren S.T. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 33.Warren S.T., Sherman S.L. The fragile X syndrome. In: Scriver C.R., Beaudet A.L., Valle D., Childs B., Kinzler K.W., Vogelstein B., editors. The Metabolic & Molecular Bases of Inherited Disease. Vol. I. New York: McGraw-Hill Companies; 2001. pp. 1257–1290. [Google Scholar]
- 34.Oberle I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boue J., Bertheas M.F., Mandel J.L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 35.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 36.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S.T., Schlessinger D., Sutherland G.R., Richards R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 37.Ashley C.T., Jr, Wilkinson K.D., Reines D., Warren S.T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 39.Laggerbauer B., Ostareck D., Keidel E.M., Ostareck-Lederer A., Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 40.Li Z., Zhang Y., Ku L., Wilkinson K.D., Warren S.T., Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y., Gutekunst C.A., Eberhart D.E., Yi H., Warren S.T., Hersch S.M. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 45.Stefani G., Fraser C.E., Darnell J.C., Darnell R.B. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:9272–9276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K., Bogert B.A., Li W., Su K., Lee A., Gao F.B. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr. Biol. 2004;14:1025–1034. doi: 10.1016/j.cub.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 47.Barbee S.A., Estes P.S., Cziko A.M., Hillebrand J., Luedeman R.A., Coller J.M., Johnson N., Howlett I.C., Geng C., Ueda R., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli G., Larson J., Martone M.E., Jones Y., Smalheiser N.R. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 49.Jin P., Alisch R.S., Warren S.T. RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 50.Xu X.L., Li Y., Wang F., Gao F.B. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J. Neurosci. 2008;28:11883–11889. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang L., Duan R., Chen D., Wang J., Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum. Mol. Genet. 2007;16:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y., Xu S., Xia L., Wang J., Wen S., Jin P., Chen D. The bantam microRNA is associated with Drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. doi: 10.1371/journal.pgen.1000444. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 54.Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 55.Chahrour M., Zoghbi H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Nan X., Cross S., Bird A. Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp. 1998;214:6–16. doi: 10.1002/9780470515501.ch2. discussion 16–21, 46–50. [DOI] [PubMed] [Google Scholar]
- 57.Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nomura T., Kimura M., Horii T., Morita S., Soejima H., Kudo S., Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum. Mol. Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- 59.Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl Acad. Sci. USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein M.E., Lioy D.T., Ma L., Impey S., Mandel G., Goodman R.H. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 61.Gothelf D., Presburger G., Levy D., Nahmani A., Burg M., Berant M., Blieden L.C., Finkelstein Y., Frisch A., Apter A., et al. Genetic, developmental, and physical factors associated with attention deficit hyperactivity disorder in patients with velocardiofacial syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;126B:116–121. doi: 10.1002/ajmg.b.20144. [DOI] [PubMed] [Google Scholar]
- 62.Gothelf D., Presburger G., Zohar A.H., Burg M., Nahmani A., Frydman M., Shohat M., Inbar D., Aviram-Goldring A., Yeshaya J., et al. Obsessive-compulsive disorder in patients with velocardiofacial (22q11 deletion) syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;126B:99–105. doi: 10.1002/ajmg.b.20124. [DOI] [PubMed] [Google Scholar]
- 63.Edelmann L., Pandita R.K., Morrow B.E. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paylor R., Glaser B., Mupo A., Ataliotis P., Spencer C., Sobotka A., Sparks C., Choi C.H., Oghalai J., Curran S., et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl Acad. Sci. USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paterlini M., Zakharenko S.S., Lai W.S., Qin J., Zhang H., Mukai J., Westphal K.G., Olivier B., Sulzer D., Pavlidis P., et al. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat. Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- 66.Torres-Juan L., Rosell J., Morla M., Vidal-Pou C., Garcia-Algas F., de la Fuente M.A., Juan M., Tubau A., Bachiller D., Bernues M., et al. Mutations in TBX1 genocopy the 22q11.2 deletion and duplication syndromes: a new susceptibility factor for mental retardation. Eur. J. Hum. Genet. 2007;15:658–663. doi: 10.1038/sj.ejhg.5201819. [DOI] [PubMed] [Google Scholar]
- 67.Gothelf D., Eliez S., Thompson T., Hinard C., Penniman L., Feinstein C., Kwon H., Jin S., Jo B., Antonarakis S.E., et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat. Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 68.Stark K.L., Xu B., Bagchi A., Lai W.S., Liu H., Hsu R., Wan X., Pavlidis P., Mills A.A., Karayiorgou M., et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 69.Epstein C.J. Down syndrome (trisomy 21) In: Scriver C.R., Beaudet A.L., Valle D., Childs B., Kinzler K.W., Vogelstein B., editors. The Metabolic & Molecular Bases of Inherited Disease. Vol. 1. New York: McGraw-Hill Companies; 2001. pp. 1223–1249. [Google Scholar]
- 70.Gardiner K., Costa A.C. The proteins of human chromosome 21. Am. J. Med. Genet. C Semin. Med. Genet. 2006;142C:196–205. doi: 10.1002/ajmg.c.30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mao R., Zielke C.L., Zielke H.R., Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 72.Korenberg J.R., Chen X.N., Schipper R., Sun Z., Gonsky R., Gerwehr S., Carpenter N., Daumer C., Dignan P., Disteche C., et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl Acad. Sci. USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delabar J.M., Theophile D., Rahmani Z., Chettouh Z., Blouin J.L., Prieur M., Noel B., Sinet P.M. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 74.Rachidi M., Lopes C. Mental retardation in Down syndrome: from gene dosage imbalance to molecular and cellular mechanisms. Neurosci. Res. 2007;59:349–369. doi: 10.1016/j.neures.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 75.Kuhn D.E., Nuovo G.J., Martin M.M., Malana G.E., Pleister A.P., Jiang J., Schmittgen T.D., Terry A.V., Jr, Gardiner K., Head E., et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem. Biophys. Res. Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Sethupathy P., Borel C., Gagnebin M., Grant G.R., Deutsch S., Elton T.S., Hatzigeorgiou A.G., Antonarakis S.E. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am. J. Hum. Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bushati N., Cohen S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 78.Dostie J., Mourelatos Z., Yang M., Sharma A., Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L., Wang T., Wright A.F., Suri M., Schwartz C.E., Stevenson R.E., Valle D. A microdeletion in Xp11.3 accounts for co-segregation of retinitis pigmentosa and mental retardation in a large kindred. Am. J. Med. Genet. A. 2006;140:349–357. doi: 10.1002/ajmg.a.31080. [DOI] [PubMed] [Google Scholar]
- 80.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J., et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J. Med. Genet. 2008 doi: 10.1136/jmg.2008.059907. November 26 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abu-Elneel K., Liu T., Gazzaniga F.S., Nishimura Y., Wall D.P., Geschwind D.H., Lao K., Kosik K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 82.Abelson J.F., Kwan K.Y., O’Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 83.Georges M., Clop A., Marcq F., Takeda H., Pirottin D., Hiard S., Tordoir X., Caiment F., Meish F., Bibe B., et al. Polymorphic microRNA-target interactions: a novel source of phenotypic variation. Cold Spring Harb. Symp. Quant. Biol. 2006;71:343–350. doi: 10.1101/sqb.2006.71.056. [DOI] [PubMed] [Google Scholar]
- 84.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibe B., Bouix J., Caiment F., Elsen J.M., Eychenne F., et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 85.Pal-Bhadra M., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 86.Brower-Toland B., Findley S.D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S.C., Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grimaud C., Bantignies F., Pal-Bhadra M., Ghana P., Bhadra U., Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 88.Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 89.Megosh H.B., Cox D.N., Campbell C., Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 90.Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 92.Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 93.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 94.Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl Acad. Sci. USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Egan C.M., Sridhar S., Wigler M., Hall I.M. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 2007;39:1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- 96.Bailey J.A., Eichler E.E. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 97.Abrahams B.S., Geschwind D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geschwind D.H. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geschwind D.H. Autism: family connections. Nature. 2008;454:838–839. doi: 10.1038/454838a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sridhar J., Rafi Z.A. Identification of novel genomic islands associated with small RNAs. In Silico Biol. 2007;7:601–611. [PubMed] [Google Scholar]
- 101.Nowacki M., Vijayan V., Zhou Y., Schotanus K., Doak T.G., Landweber L.F. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]