Abstract
Genomic studies in model organisms and in humans have shown that complexity in biological systems arises not from the absolute number of genes, but from the differential use of combinations of genetic programmes and the myriad ways in which these are regulated spatially and temporally during development, senescence and in disease. Nowhere is this lesson in biological complexity likely to be more apparent than in the human nervous system. Increasingly, the role of genomic non-protein coding small regulatory RNAs, in particular the microRNAs (miRNAs), in regulating cellular pathways controlling fundamental functions in the nervous system and in neurodegenerative disease is being appreciated. Not only might dysregulated expression of miRNAs serve as potential disease biomarkers but increasingly such short regulatory RNAs are being implicated directly in the pathogenesis of complex, sporadic neurodegenerative disease. Moreover, the targeting and exploitation of short RNA silencing pathways, commonly known as RNA interference, and the development of related tools, offers novel therapeutic approaches to target upstream disease components with the promise of providing future disease modifying therapies for neurodegenerative disorders.
INTRODUCTION
Progress in the development of neuroprotective and disease modifying treatments for neurodegenerative disease has been impeded by our still relatively poor knowledge of basic disease pathogenesis. Many current treatments target single cellular pathways downstream of disease initiation and which may be beyond an effective therapeutic window (1–3). Moreover, functional characterization of such diseases has often centred on the study of rare monogenic disease variants which are not necessarily informative of the commoner sporadic forms of neurodegenerative disease where combinations of multiple genetic loci and non-genetic determinants are thought to play crucial roles (4,5). Furthermore, recent genomic studies are revealing a multi-layered complexity to the organization of biological systems and gene regulatory networks and it is therefore likely that a deeper understanding of such networks will be necessary to fully appreciate the pathophysiological complexity underlying the common neurodegenerative disease phenotypes such as Alzheimer's disease (AD) and Parkinson's disease (PD). An important layer of biological complexity where new understanding is emerging relates to RNA biology and in particular to the roles of small non-coding RNAs and RNA-based gene regulatory networks. Non-coding RNAs are abundantly expressed in the central nervous system (CNS) and increasingly such RNAs, in particular the genome-encoded microRNAs (miRNAs), are being found to have important functions in nervous system development and function, as well as in neurodegenerative disease pathogenesis (6–9). miRNAs are able to negatively regulate gene targets via sequence-specific post-transcriptional gene silencing (PTGS), which is the principle mechanism behind the RNA interference (RNAi) pathway(s). With an increased understanding of such short RNA regulatory networks, the ability to target such networks or to utilize RNA-based methods will allow the development of a new class of disease modifying therapies to emerge (10,11).
GENE SILENCING BY SHORT NON-CODING RNA REGULATORY NETWORKS
In mammals, RNAi represents a set of highly conserved cellular pathways whereby double-stranded RNA (dsRNA) is processed into short RNAs of ∼20–30 nt in length. These short RNAs associate with members of the Argonaute (Ago) family of proteins to regulate gene expression at the transcriptional and post-transcriptional level (recently reviewed in 12,13). RNAi has a myriad of roles in every fundamental aspect of mammalian cellular function and its discovery has led to a widened appreciation for the role of very small regulatory RNAs in eukaryote biology. One of the most exciting developments since the discovery of RNAi in 1998 has been the application of exogenous RNAi tools as artificial regulators of gene expression; with particular emphasis on the generation of a special class of drugs that are capable of inhibiting rogue gene elements.
The therapeutic development of RNAi has been made possible by usurping elements of the endogenous mammalian miRNA biogenesis pathway for PTGS (Fig. 1A). miRNAs consist of a class of short ∼22 nt RNAs derived from longer processed dsRNA precursors. RNA Pol II transcripts with hairpin motifs or primary miRNAs (pri-miRNAs) are processed by the RNase III enzymes Drosha and Dicer into short miRNA duplexes. Single or multiple (polycistronic) pri-miRNA motifs can be found within exonic or intronic coding and non-coding mRNA, or within antisense orientation transcripts or transcripts that span intergenic regions (reviewed in 14,15). The mature miRNA associates with the Ago2-containing RNA-induced silencing complex (RISC) and RISC-loaded miRNAs are guided to the 3′-untranslated regions (UTRs) of target mRNAs by a ‘seed region’: a specific complementary region between nucleotides 2–7 of the guide strand (16). Seed region matches within the 3′-UTRs of mRNAs primarily induce translation repression by one or more of the following mechanisms: transcriptional cleavage, blocking of ribosomal function, deadenylation or shunting of mRNAs to transcriptionally inactive cytoplasmic P bodies (reviewed in 17,18). However, Ago2 is responsible for the post-transcriptional cleavage of the target RNA and is confined to miRNAs or short interfering RNAs (siRNAs) with near-perfect sequence complementarity with their cognate target. Interestingly, endogenous mammalian miRNAs are rarely found to bind their targets completely. Since seed region matches alone can suffice for translational suppression, this suggests that a single miRNA potentially regulates up to a 100 mRNA targets (19). RNAi guide sequences may additionally associate with Ago1 in a RNA-induced transcriptional silencing complex (RITS) to induce transcriptional gene silencing (TGS), which is characterized by the targeting of siRNAs or short antisense RNAs to promoter elements, resulting in transcriptional inhibition through silent-state epigenetic modifications of DNA and associated nucleosomes (20–23) (Fig. 1B). As siRNA-directed TGS elicits more permanent epigenetic modifications it suggests that, unlike PTGS, TGS may offer a more sustained inhibition of gene expression. This represents an exciting and important novel approach for the development of therapeutic gene silencing modalities.
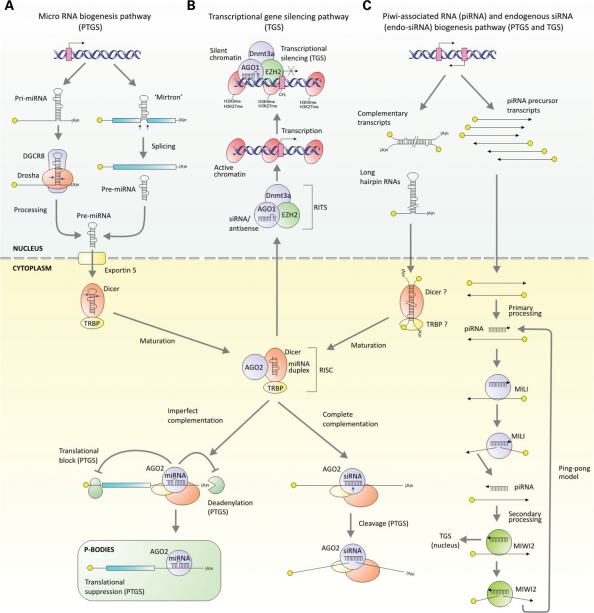
Figure 1.
Mammalian RNAi regulatory pathways. (A) miRNAs are encoded in pri-miRNAs (124,125). These ∼100 nt inverted repeat motifs are usually found embedded once or multiple times within coding or non-coding RNA Pol II-derived transcripts (126). Pri-miRNAs are first processed in the nucleus where their hairpin-like structures are recognized and cleaved by the RNase III enzyme Drosha together with DiGeorge critical region 8 protein (DGCR8), to produce shorter hairpin duplexes known as a 70–80 nt pre-miRNAs (127,128). For a small minority of miRNAs, short intronic sequences, referred to ‘mirtrons’, can be directly processed by the spliceosome into pre-miRNA-like hairpins without requiring Drosha cleavage (30,129). Spliced lariats are de-branched and likely produce functional pre-miRNAs for export. Pre-miRNAs are exported from the nucleus to the cytoplasm by the exportin-5 (130,131) followed by recognition and cleavage by a second RNase III enzyme, Dicer and its partner, TAR RNA-binding protein (TRBP), to produce a ∼22 bp, staggered miRNA/miRNA* duplex with 2 nt 3′ overhangs (132–134). Dicer/TRBP, loads one of the strands, the ‘guide strand’, into a RISC consisting in its simplest form of Ago2 (135,136) directing cleavage of translational suppression of cognate RNA. (B) The mechanism of TGS is poorly understood in mammals but is thought to include a complex consisting (RITS) of Ago1, (and possibly Ago2) a polycomb group component, enhancer of zeste 2 (EZH2) and DNA methyltransferase 3a (Dnmt3a) (21,23,137). Moreover, TGS may require the presence of low-copy promoter-derived transcripts to direct silent heterochromatin marks (H3K9 and/or H3K27 methylation) and DNA methylation at the targeted locus (138). (C) Endogenous siRNAs are derived from long hairpin sequences and complementary transcripts which are processed by Dicer into siRNAs. piRNAs are 24–31 nt short RNAs processed from single-stranded precursors derived from transposons or genomic repeat elements in the germline (13). In the ‘ping-pong model’, primary piRNAs interact with the Piwi protein MILI to cleave a transcript that generates a piRNA for incorporation into MIWI2, which in turn cycles back to produce new MILI-interacting piRNAs (24,25).
miRNAs are not the only source of short RNA duplexes and it remains to be seen what role these newer RNAi pathways will have in the development of novel therapeutics. A number of short 24–31 nt RNAs in the germline are associated with Piwi-family proteins and are referred to as a Piwi-interacting RNAs (piRNAs) (reviewed in 13) (Fig. 1C). These short RNAs, which originate from repeat-rich regions of the genome, are processed through a distinct Dicer-independent mechanism (24) and transcriptionally silence transposons by establishing de novo DNA methylation in murine fetal testes (25–27). A more recent RNAi-related pathway was discovered in studies of mouse oocytes and embryonic stem (ES) cells. An abundant class of endogenous siRNAs or endo-siRNAs was found to be derived from transcripts with long inverted repeats or from convergent and divergent transcripts of pseudogenes or transposons (28–30) (Fig. 1C). Other than blocking retrotransposition, little is known about the function of endo-siRNAs; it is unlikely that these RNAi species are limited to embryonic/oocyte cells since complementary RNA hairpins or duplexes form ubiquitously between cis- and/or trans-associating RNAs.
SHORT NON-CODING RNA REGULATORY NETWORKS AND NEURODEGENERATIVE DISEASE
The idea that dysregulation of the PTGS pathway might be a manifestation of or that specific non-coding RNAs, in particular miRNAs, might be directly causative of neurological disease is gaining ground. That specific non-coding miRNAs play fundamental roles in mammalian development, ES cell differentiation and in CNS development and function is now increasingly appreciated. A diverse repertoire of miRNAs is abundantly expressed in the CNS in tightly regulated and highly specific spatial and temporal patterns, and numerous miRNAs have now been associated with fundamental roles in neurobiology including in neuron-specific gene regulation and neuron-specific pre-mRNA splicing, neural cell lineage specification, neurogenesis and synaptogenesis (31–37). Moreover, altered miRNA expression and function has been strongly implicated in cancer and cardiovascular disease and so the notion of non-coding RNA involvement in disease pathogenesis is not new; indeed, a comprehensive resource relating miRNAs to human disease, miR2Disease, has recently been made available to the scientific community (38). A putative role for specific miRNA involvement in the control of neuronal cell number and in neurodegenerative disease (see Table 1 for summary) was first hinted at from experiments in model organisms, including in mice, in which Dicer was inactivated (39–43). For example, mice homozygous for conditionally floxed Dicer allele and expressing Cre recombinase under control of the dopamine transporter regulatory elements, resulting in conditional Dicer knockout in midbrain dopamine neurons, show specific and progressive dopamine neuron loss (41). Evidence for disruption of specific miRNAs in PD is provided by the finding of miR-133b deficiency in the midbrains of PD patients and in mouse models of dopamine neuron degeneration (41). Interestingly, Kim et al. uncover an important negative regulatory relationship in which the essential dopaminergic transcription factor Pitx3 regulates miR-133b transcription which in turn is found to suppress Pitx3 expression, providing an RNA regulatory network that controls the dopamine neurogenic gene programme. Further evidence linking regulatory miRNAs and PD comes from work identifying variation in the miRNA binding site for miR-433 in the FGF20 gene which is preferentially expressed in the midbrain. Such variation appears to confer increased susceptibility to PD as a result of increased alpha synuclein gene expression, a gene regulated by FGF20 and which is strongly linked to PD through both over-expression and point mutation mechanisms (44,45). Similarly, the recent discovery of variation in the miR-659 binding-site in the 3′-UTR of the progranulin gene as a risk factor for TDP43-positive frontotemporal dementia (46) and an earlier finding that the neurological disorder Tourette's syndrome is associated with a variation in the miR-189 binding site in mRNAs encoding the neuronal proteins Slit and Trk-like 1 (47), together implicates common genetic variation within non-coding regions of disease-related transcripts and regulatory miRNA function as factors in the origin of complex neurodegenerative disease.
Table 1.
Non-coding miRNAs and neurological disease
| Disease | miRNAs implicated | Putative mechanism | Reference |
|---|---|---|---|
| Parkinson's disease | Multiple | Neurodegeneration in dicer knockout mouse | (41) |
| miR-133b | Dopamine neurogenesis | (41) | |
| miR-433 | Variation in miR-433 binding site in FGF20 gene leading to increased alpha synuclein levels | (44) | |
| Alzheimer's disease | Multiple | Dysregulation multiple miRNAs in AD patient brains | (48) |
| miR-659 | Variation in miR-659 binding site in progranulin gene | (46) | |
| miR-29 | Correlate with increased BACE1/β secretase expression | (49) | |
| miR-298/328 | Direct regulation of BACE1 transcript | (50) | |
| miR-20a | Regulation of APP expression | (51) | |
| miR-107 | Regulation of APP cleaving enzyme 1 | (148) | |
| Huntington's disease | Multiple | Interaction of htt protein with Ago2 and localization to P-bodies | (52) |
| Multiple | Dysregulated miRNA expression | (55) | |
| miR-9/miR-9* | Interaction with/regulation of REST and CoREST | (56) | |
| Spinocerebellar ataxias and cerebellar degeneration | Multiple | Purkinje cell ablation of Dicer leads to neurodegeneration | (43) |
| miR-19, miR-101 and miR-130 | miRNA regulation of ataxin 1 in SCA1 | (57) | |
| Other | |||
| Prion-induced neurodegeneration | Multiple | Dysregulated miRNA expression | (58) |
| Schizophrenia | Multiple | SNPs within or near brain-expressed miRNAs | (59) |
| Fragile X syndrome | Multiple | Disordered miRNA biogenesis | (60) |
| Autism | miR-181b | Regulation of genes linked to schizophrenia | (61) |
| Tourette's syndrome | Multiple | FMRP product of FMR1 gene interacts with miRNAs which assemble into miRNPs containing FMRP which functions as a translational suppressor. Loss of FMRP function results in de-repression of multiple miRNA targets | (149) |
| Multiple | Dysregulated miRNA expression, some targeting autism-linked genes Neurexin and SHANK3 | (150) | |
| miR-189 | 3′-UTR of the SLITRK1 gene contains the binding site of miR-189, which is mutated in some patients | (47) |
Further evidence for altered miRNA expression linked to neurodegenerative disease comes from studies related to AD. Here dysregulated miRNA expression has been documented in AD patient brains relating to the control of cellular pathways involving neurogenesis, amyloid processing, insulin resistance and innate immunity (48). Specific miRNAs have now also been linked mechanistically to AD pathogenesis; for example, loss or reduced expression of the miR-29 cluster in sporadic AD has been found to correlate closely with a subgroup of patients in whom increased BACE1/β-secretase expression was observed (49). BACE1/β secretase is a rate-limiting step for β-amyloid production and its increased expression has been reported among AD patients. Additionally, Boissonneault et al. (50) have identified miR-298 and miR-328 which directly interact with the 3′-UTR of the BACE1 transcript. Further, the miR-20a family (i.e. miR-20a, miR-17-5p and miR-106b) has been shown to regulate amyloid precursor protein (APP) expression suggesting a possible role during disease (49,51), a finding corroborated specifically for miR-106b where decreased levels were found by qRT-PCR in the brains of sporadic AD patients.
There is now also accumulating evidence of dysregulated miRNA expression or involvement of regulatory miRNAs in other neurodegenerative diseases, notably hereditary diseases of polyglutamine origin. Interestingly, the huntingtin (htt) protein, which harbours an expanded polyglutamine tract in Huntington's disease (HD), has been implicated directly in the RNAi silencing pathway by virtue of its biochemical interaction with Ago2, an essential PTGS component, and co-localization with Ago2 in P-bodies (52), cytoplasmic sites of RNA metabolism, RNAi and miRNA activity (53,54). In addition, altered expression of a number of neuron-specific miRNAs has been found in both murine and human HD brains, the authors suggesting that this may be due to increased repression by the essential repressor factor REST, also known as neuron-restrictive silencing factor, which itself regulates numerous miRNA transcripts (55). Interestingly, Packer et al. (56) have observed downregulation of several miRNAs in HD patient brains and one of these, brain-enriched miR-9/miR-9*, is implicated in the regulation of components of the REST transcription factor complex: REST and CoREST, providing a possible autoregulatory link between miRNAs and neuron-specific gene transcription networks, which may be defective in HD. Similarly, in the polyglutamine expansion disorder spinocerebellar ataxia type 1 (SCA1) in which a translated CAG repeat expansion is found in the ataxin 1 gene, three miRNAs have now been found to directly co-regulate ataxin 1 expression and hence have potential roles in disease pathogenesis and as possible therapeutic targets (57). Disruption of regulatory miRNA networks has been observed in other neurological disorders including prion-induced neurodegeneration (58) and also in the neurodevelopmental disorder schizophrenia. In the schizophrenia SNPs within or near brain-expressed miRNAs (59), disordered miRNA biogenesis (60) and upregulation of a specific miRNAs shown to regulate genes linked to schizophrenia (61), have all directly implicated regulatory miRNAs mechanistically in the disease.
Thus regulatory non-coding miRNAs are increasingly being associated with the complex aetiologies of neurodegenerative and other major neurological diseases. Moreover, a range of mechanisms are now being uncovered to implicate some of these miRNAs directly in disease pathogenesis including, miRNA dysregulation leading to defects in specific cellular networks. This is seen in neurogenesis and innate immunity pathways that affect the expression of disease-linked transcripts, variations in miRNA binding domains in disease-associated genes and even defects in fundamental regulatory miRNA biogenesis pathways. While no single specific miRNA mutation has yet been associated with neurological disease, these findings suggest that as understanding of involvement of regulatory non-coding RNA networks in neurodegenerative disease accrues, non-coding RNAs will represent at the very least a new category of diagnostic markers for this group of diseases and probably an important regulatory component contributing directly to disease aetiology.
THERAPEUTIC RNAi SILENCING OF DISEASE GENES
Specifically targeted exogenous post-transcriptional degradation of mRNAs in the cytoplasm has been used as a technology to suppress gene expression by introducing dsRNA triggers at different levels of the RNAi pathway (Fig. 2). The most common form of silencing is typically achieved by administering chemically synthesized siRNAs. Most synthetic strategies have focused on producing siRNA mimics by standard phosphoramadite chemistry. While synthetic 19–21 bp siRNAs are loaded directly into RISC, a number of groups are using larger synthetic duplexes (>21 bp) or synthetic precursor-miRNAs (pre-miRNAs), which are first cleaved by Dicer/TRBP, enhancing strand selection upon RISC entry (62–64). For synthetic effector sequences, structures are not confined by canonical symmetrical 19–23 bp natural siRNA parameters. A number of different asymmetric RNA duplexes of between 15–21 bp have been shown to be powerful RNAi inhibitors (65,66). All synthetic siRNAs have the potential to be chemically modified such that they have desirable in vivo properties that are not found in their natural counterparts. The most important property derived by chemical modification is the prevention of degradation by serum nucleases. Additional benefits from chemical modifications may result in enhanced guide strand function, decreased immune activation and improved pharmacokinetic properties (reviewed in 67).
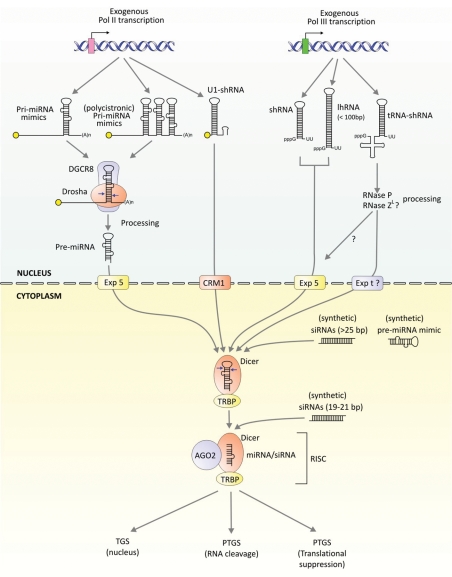
Figure 2.
Exogenous RNAi-mediated gene silencing. RNA Pol II-derived transcripts introduce hairpin duplexes which structurally mimic mono or polycistronic pri-miRNAs which are recognized and processed by Drosha/DGCR8 into pre-miRNA-like hairpins. These hairpins are cleaved by Dicer/TRBP following export via exportin-5. The Pol II-generated U1 shRNA transcripts, which contain a 3′ terminal B-box, structurally mimic pre-miRNAs but are likely exported by the CRM1 pathway prior to Dicer/TRBP cleavage (139). RNA Pol III promoters express shRNAs and long hairpin RNAs (lhRNAs) with defined 5′ and 3′ termini. U6 or H1-derived shRNAs and lhRNAs, like pre-miRNAs, consist of 2 nt 3′ overhangs and exit the nucleus via exportin-5. lhRNAs, are processed sequentially by Dicer to produce up to three siRNAs (140,141). tRNALys3 and tRNAVal Pol III promoters can also be used to produced tRNA–shRNAs for processing in the nucleus by 5′ and 3′ tRNA processing enzymes prior to export (69,142). If unprocessed by RNase ZL, tRNA–shRNAs may exit the nucleus via exportin-t (143). Synthetic siRNAs or pre-miRNA mimics can be introduced as Dicer substrates (typically >25 bp duplexes) or as 19 bp duplexes for direct loading into Ago2-RISC.
Gene-based approaches allow for the expression of hairpin duplexes that structurally mimic the miRNA precursors, pri-miRNAs and pre-miRNAs, to generate exogenous guide strands (Fig. 2). Expressed RNAi effectors have the advantage of being constantly renewed from an expression cassette, thereby providing a more sustained suppression of the target RNA. Moreover, encoded RNAi cassettes can be packaged and delivered by different viral vectors, thereby harnessing the cell-specific targeting and expression characteristics of viruses. The most commonly used expression cassette is a short hairpin RNA (shRNA)-encoding sequence that is inserted downstream of a RNA Pol III promoter. shRNAs are typically expressed from Pol III promoters such as H1 and U6, since these promoters produce defined 5′ and 3′ termini which mimic typical endogenous pre-miRNA characteristics. U1 Pol II promoters and tRNA Pol III promoters have the advantage of producing effective Dicer susbtrates that likely do not use exportin-5 for nuclear export (68–70). Most Pol III and some Pol II promoters cannot be tightly regulated and are therefore constitutively active, which may have undesirable effects (see later). More recent attention has been placed on using pri-miRNA mimics, which follow a more natural maturation pathway (71–73). Pri-miRNAs have the advantage of tissue-controlled expression of RNAi-precursors and can be organized in polycistronic clusters, allowing for the simultaneous inhibition of multiple targets (74,75).
RNAi-BASED THERAPEUTICS FOR NEURODEGENERATIVE DISEASE
While regulatory non-coding RNA networks are increasingly implicated in the complex pathogenesis of specific neurodegenerative diseases, the ability to exploit in parallel the PTGS pathway in a therapeutic context to specifically modify the expression of neurodegenerative disease-associated transcripts will offer novel approaches for developing disease modifying therapies for both familial and sporadic disease (10,11). The major challenges confronting approaches to develop RNA-based therapies are: (i) the relatively poor understanding of complex disease pathways and of appropriate target transcripts in the case of sporadic neurodegenerative disease and (ii) the optimal design and delivery of therapeutic RNAi trigger molecules to the nervous system. However, that such approaches are likely to be of therapeutic benefit for neurodegenerative disease, i.e. that such disease processes might be halted or reversed by targeted gene inhibition, is suggested from studies in conditional transgenic model systems harbouring mutant (expanded) htt transgene (76), a mutant SCA1[82Q] transgene (77) or an over-expressed alpha synuclein allele (78), where in each case switching off the disease-causing transgene resulted in concomitant improvement in disease phenotypes.
Given the difficulty in defining appropriate therapeutic targets in sporadic neurodegenerative disease, it is unsurprising that most progress in the development of such RNA-based therapies has been made in hereditary neurodegenerative disease, most notably HD. Harper et al. (79) provided the first demonstration that targeting mutant human htt in the mouse brain using an adeno-associated virus serotype 1 (AAV1) vector delivered U6 promoter-driven shRNA effector system could yield improvements in HD-associated neuropathology and behaviour. A series of subsequent studies by this and other groups provide further evidence in support of this approach (80–85), however, certain important issues in the development of this therapeutic approach are highlighted. The first is the appropriateness of different animal models, and in particular the use of relatively rapid disease-onset animal models by some groups (81,82) seems simplistic and questionable in the context of complex, chronic neurodegenerative disease. A second important issue concerns the optimal delivery method for therapeutic RNAi effectors. Most investigators have utilized vector expression systems and Machida et al. (84) recently reported improved delivery in an HD model using an AAV5 delivery system. In contrast, DiFiglia et al. (81) have shown encouraging data utilizing cholesterol-conjugated synthetic siRNAs. However, questions relating to efficiency of synthetic siRNA delivery to the CNS remain, and in addition whether or not such conjugated siRNAs are likely to prove toxic in human subjects or indeed, how such synthetic compounds might be repeatedly administered to patients over time are major unresolved issues. A further unresolved question relates to whether or not specific silencing of the mutant disease transcript will be required for therapeutic benefit in HD; current work involves indiscriminate targeting of both wild-type and mutant alleles. While several groups have developed successful allele-specific silencing of neurological disease transcripts (86–89) at present there remains no motif for the targeted discrimination of mutant from wild-type HD transcripts, although work to identify possible disease-linked and CAG expansion-linked SNPs is in progress (90). Finally, there is the crucial question of safety and the potential side- and off-target effects of such RNA-based therapies which have been identified in a number of studies (80,85) and are discussed in detail below.
The related spinocerebellar ataxias (SCAs), several of which also have polyglutamine expansions as the causative mutation, have been investigated by a number of groups. Xia et al. (91) first demonstrated proof of principle for silencing the human SCA1 disease transcript in cerebellum with concomitant phenotypic benefit. Interestingly, discriminatory SNPs have been identified for SCA3 and SCA7 (92,93), and in the case of the former, allele-specific silencing of mutant ataxin-3 has been demonstrated in vivo, albeit via the targeting of a virally expressed mutant transcript (92). Schwarz et al. (89) have attempted to understand and define the biochemical parameters for allele-specific siRNA silencing, while in the case of SCA7, Scholefield et al. (unpublished data) have similarly defined parameters for shRNA- and miRNA-based allele-specific silencing of a disease-associated SNP in the mutant ataxin-7 transcript, highlighting the importance of structural position 16 mismatches in construct design to achieve optimal allele-specific discrimination and phenotype correction in this particular case. Similar structural constraints at this or other single nucleotide mismatch positions within the effector guide sequence may well apply in other cases. A further strategic option as an alternative to allele-specific silencing is illustrated in the case of SCA6, where Kubodera et al. (94) have developed a gene knockdown and replacement strategy whereby both mutant and wild-type transcripts are suppressed and the essential wild-type protein is replaced by co-expression of an siRNA-resistant wild-type mRNA.
Limited progress has been made to date in the RNA-based silencing of targets linked to common sporadic forms and less common familial forms of PD and AD. Sapru et al. (95) have successfully targeted the alpha synuclein pathway in the context of PD. While in the case of AD, the amyloid (96,97) and BACE1/β secretase (98) pathways have both been targeted, the latter yielding a striking improvement in disease phenotype.
There are considerable challenges facing the clinical application of exogenous RNAi effector sequences. A number of toxicities are associated with siRNAs and exogenous RNAi precursors. High levels of expressed shRNAs in the liver are known to cause fatalities in mice due in part to the saturation of the endogenous RNAi machinery (99) and McBride and colleagues have observed toxicities arising from shRNA-based vectors in brain (71,85). Expressed shRNAs likely abrogate the function of natural miRNAs (73,99,100). Therefore, careful consideration should be given to the dosage used when applying ectopically introduced RNAi effectors. Pol II promoters, which can be regulated more easily by cell-specific environmental factors, have since become more popular for the expression of pri-miRNA mimics as effective guide strand ‘shuttles’. At present there is no evidence to suggest that pri-miRNA shuttles, even if highly expressed, can saturate the endogenous RNAi pathway (71–73). Another area of concern is the possibility of off-target inhibition of unintended mRNAs through interactions between the 6–7 nt seed region of exogenous siRNA guide sequences and target mRNAs. Many off-target effects of this nature have been observed when introducing exogenous siRNAs (101–103). Off-target effects can in some instances be mitigated by introducing specific base modifications within the siRNA duplex (104). However, to determine the full extent of any off-target inhibition, prior screening using candidate RNA and protein expression array analyses may be required (105). Lastly, dsRNA duplexes can potentially activate the innate cellular immune system through the unwanted release of inflammatory cytokines and induction of an interferon response. dsRNAs activate cytoplasmic pattern recognition receptors such as dsRNA-dependent protein kinase receptor and membrane-associated toll-like receptors (reviewed in 106). Careful consideration should be given to the innate immune response, which can mask the effects of RNAi-mediated gene silencing. Recent evaluation of a high-profile study using siRNAs targeted to vascular endothelial growth factor (VEGF) showed that an off-target immune response was responsible for the observed suppression of VEGF (107). Interestingly, ectopic expression of RNAi effectors from vectors, however, is less likely to induce a non-specific immunostimulatory response (108).
FUTURE POSSIBILITIES FOR THE THERAPEUTIC REGULATION OF NEURODEGENERATIVE DISEASE-LINKED MIRNAS
An increased understanding of the role of regulatory non-coding RNAs, and especially miRNAs, in the pathogenesis of sporadic complex neurodegenerative disease will lead to therapeutic possibilities aimed at modulating the activity of these non-coding RNA networks in diseased cells (Fig. 3). miRNA biogenesis and target gene silencing are governed by general and miRNA-specific feedback circuits (reviewed in 13), which are regulated transcriptionally and post-transcriptionally. Ablation of normal miRNA regulation is often associated with disease, as is increasingly being seen for cancer, including nervous system tumours (109,110), and cardiovascular disease (111). While miRNAs are regulated at the transcriptional level, post-transcriptional miRNA regulation during biogenesis suggests a fine-tuning of spatio-temporal miRNA activity and provides the possibility of exogenously controlling specific miRNAs linked to neurodegenerative disease. Currently, factors known to modulate miRNA biogenesis include the RNA-binding protein hnRNAP A1, which facilitates processing of pri-miR-18a by binding to conserved loop sequences (112); and SMAD proteins, which associate with Drosha to process pri-miR-21 in smooth vascular muscle cells (113). Many more factors are likely to be discovered in future which control specific miRNAs. Interestingly, in human gliomas where miR-21 levels have been reported to be elevated, the suppression of miR-21 in neural precursor cells was shown, together with tumour necrosis factor-related apoptosis inducing ligand (S-TRAIL), to sensitize gliomas for increased apoptotic activity (114). Post-transcriptional control was also shown for precursors of miR-138, which is a miRNA restricted to neuronal cells. The precursor, pre-miR-138-2, is ubiquitously expressed throughout all tissues, indicating cell- or tissue-selective Dicer cleavage of pre-miRNAs in neurons (115). Lee et al. (116) showed in a recent study using normal tissues, tumours and cell lines, that a large number of pri-miRNAs are transcribed but are not processed to the mature miRNA, especially in cancer cells, implying that post-transcriptional dysregulation of miRNAs may be at the root of many disease aetiologies.
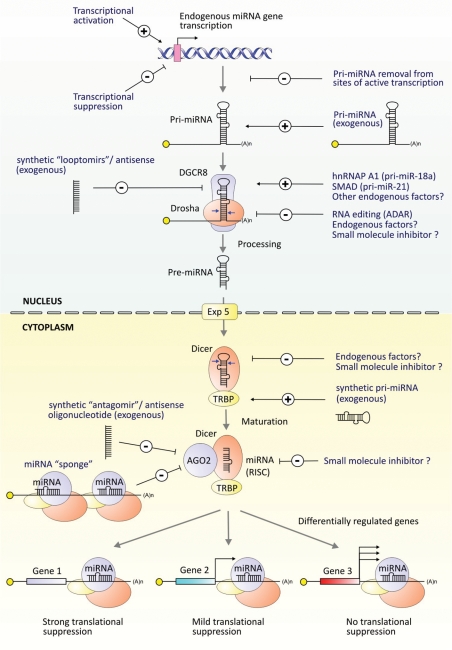
Figure 3.
Regulation and modulation of miRNA biogenesis and function. A number of endogenous and exogenous factors can control miRNA biogenesis, and these have the potential to be exploited as novel therapies. Transcription factors act on promoters to activate or suppress endogenous miRNA gene expression. Post-transcriptional control can occur at the level or pri-miRNA processing in the nucleus, or pre-miRNA cleavage in the cytoplasm. In the nucleus, Drosha/DGCR8 preferentially processes pri-miRNAs that are retained at the site of transcription (144), regulating pri-miRNAs co-transcriptionally (145). To stimulate the activity of a specific miRNA, exogenous pri-miRNAs can be introduced as gene-based cassettes or as synthetic pre-miRNAs. Factors which positively regulate specific pri-miRNAs include the SMAD proteins and hnRNAP A1 (112,113), with other positive or negative regulatory factors likely to be discovered in the future. Pri-miRNAs are additionally subject to RNA editing (adenosine to inosine RNA editing), resulting in an unprocessed pri-miRNA or in miRNAs with altered seed regions (146,147). miRNAs can be blocked by ‘looptomirs’ or short ASOs targeted to the loop of pri-miRNAs (112,113). In the cytoplasm, miRNAs can be blocked by ‘antagomirs’ or ASOs (120,121). Moreover, expressed sequences that contain multiple miRNA-targets are referred to as ‘sponges’, blocking translational suppression of endogenous mRNA targets (122). Small molecule inhibitors may be found to block miRNA: mRNA interactions, pre-miRNA maturation or earlier steps in the nucleus. miRNA dysregulation has the potential to affect multiple genes, whose products in turn may positively or negatively feedback to regulate specific miRNA gene transcription or biogenesis.
As miRNA dysfunction in neurodegenerative disease is understood in greater detail, such therapeutic methods to modulate and correct the activity of aberrant nervous system miRNAs will be developed and evaluated. However, validating or blocking direct miRNA–mRNA interactions remains a challenge and tools aimed at inhibiting or augmenting the function of a specific miRNA are useful additions to research and therapeutics (Fig. 3). Mature miRNAs can be inactivated by administering short complementary synthetic antisense oligonucleotides (ASOs) (117). ASOs are usually chemically modified at the 2’ sugar moiety or have base changes such as locked nucleic acids, peptide nucleic acid or morpholino bases to improve specificity and serum half-life in vivo (114,118,119). Antagomirs are usually cholesterol-conjugated ASOs that are capable of improved degradation of specific miRNAs when administered in vivo (120). Recently, ASOs have been shown to block miRNA precursors (112) and miRNAs without degrading the target RNA (121). While a complete mechanism behind ASO-mediated inhibition of miRNA activity remains to be determined, anti-miRNA ASOs are likely to feature as important future therapeutic agents. Expressed sequences consisting of multiple miRNA seed targets, referred to as miRNA sponges, are also effective miRNA inhibitors and likely function by competing with endogenous miRNA targets (122). Perhaps the future lies in finding small-molecule inhibitors of specific miRNAs. A recent chemical screen of 1000 small molecules, identified a potent inhibitor of miR-21, thus paving the way for screening platforms to identify novel inhibitors of other miRNAs (123).
CONCLUSION AND FUTURE DIRECTIONS
Of crucial future importance will be efforts to further understand basic cellular disease mechanisms and the role of non-coding RNA networks in the complex phenotypes of sporadic neurodegenerative disease. Such knowledge will lead increasingly to opportunities to modulate non-coding RNA activity and function in neurodegenerative disease models. It will also to lead to the discovery of novel disease targets for RNA-based silencing methods. Therapies for complex multigenic diseases will likely require modulation of multiple targets or cellular disease pathways. The advent and development of experimental RNA-based therapeutics could lead to opportunities for tackling early causative factors in neurodegenerative disease pathogenesis and the development of a new generation of disease modifying therapies.
FUNDING
M.S.W. acknowledges grant support from the South African National Research Foundation (NRF), Medical Research Council (MRC) and Poliomyelitis Research Foundation (PRF), and M.J.A.W. from the UK MRC and Biotechnology and Biological Sciences Research Council (BBSRC), The Wellcome Trust, the UK Parkinson's Disease Society, the Muscular Dystrophy Campaign and Action Duchenne.
ACKNOWLEDGEMENTS
We thank Mishka Blondeel, Victoria Green, Graham McClorey and Janine Scholefield for comments and critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schapira A.H. Neurobiology and treatment of Parkinson's disease. Trends Pharmacol. Sci. 2009;30:41–47. doi: 10.1016/j.tips.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Talbot K. The study of rare diseases: butterfly collecting or an entree to understanding common conditions? Pract. Neurol. 2007;7:210–211. doi: 10.1136/jnnp.2007.124396. [DOI] [PubMed] [Google Scholar]
- 3.Koller W.C., Tse W. Unmet medical needs in Parkinson's disease. Neurology. 2004;62:S1–S8. doi: 10.1212/wnl.62.1_suppl_1.s1. [DOI] [PubMed] [Google Scholar]
- 4.Oksenberg J.R., Hauser S.L. Neurogenetics in the Annals: dealing with complexity. Ann. Neurol. 2008;63:A11–A14. doi: 10.1002/ana.21248. [DOI] [PubMed] [Google Scholar]
- 5.Rubinsztein D.C. Functional genomics approaches to neurodegenerative diseases. Mamm. Genome. 2008 doi: 10.1007/s00335-008-9130-0. [Epub ahead of print July 30, 2008] [DOI] [PubMed] [Google Scholar]
- 6.Mourelatos Z. MicroRNAs: biology and roles in neurodegeneration and brain tumours. Introduction and historical background. Brain Pathol. 2008;18:110–112. doi: 10.1111/j.1750-3639.2007.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson P.T., Wang W.X., Rajeev B.W. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogelj B., Giese K.P. Expression and function of brain specific small RNAs. Rev. Neurosci. 2004;15:185–198. doi: 10.1515/revneuro.2004.15.3.185. [DOI] [PubMed] [Google Scholar]
- 9.St Laurent G., III, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Alegre P. Therapeutic RNA interference for neurodegenerative diseases: from promise to progress. Pharmacol. Ther. 2007;114:34–55. doi: 10.1016/j.pharmthera.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Jagannath A., Wood M. RNA interference based gene therapy for neurological disease. Brief. Funct. Genomic. Proteomic. 2007;6:40–49. doi: 10.1093/bfgp/elm005. [DOI] [PubMed] [Google Scholar]
- 12.Okamura K., Lai E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 14.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 16.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Flynt A.S., Lai E.C. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D.H., Saetrom P., Snove O., Jr, Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 22.Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 23.Weinberg M.S., Villeneuve L.M., Ehsani A., Amarzguioui M., Aagaard L., Chen Z.X., Riggs A.D., Rossi J.J., Morris K.V. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 25.Aravin A.A., Hannon G.J., Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 26.Aravin A.A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T.W., et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 30.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blakaj A., Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J. Biol. Chem. 2008;283:9505–9508. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosik K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 33.Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 34.Smirnova L., Grafe A., Seiler A., Schumacher S., Nitsch R., Wulczyn F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 35.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilen J., Liu N., Burnett B.G., Pittman R.N., Bonini N.M. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol. Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 40.Hebert S.S., De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 41.Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis T.H., Cuellar T.L., Koch S.M., Barker A.J., Harfe B.D., McManus M.T., Ullian E.M. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer A., O'Carroll D., Tan C.L., Hillman D., Sugimori M., Llinas R., Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G., van der Walt J.M., Mayhew G., Li Y.J., Zuchner S., Scott W.K., Martin E.R., Vance J.M. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood-Kaczmar A., Gandhi S., Wood N.W. Understanding the molecular causes of Parkinson's disease. Trends Mol. Med. 2006;12:521–528. doi: 10.1016/j.molmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Rademakers R., Eriksen J.L., Baker M., Robinson T., Ahmed Z., Lincoln S.J., Finch N., Rutherford N.J., Crook R.J., Josephs K.A., et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum. Mol. Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abelson J.F., Kwan K.Y., O'Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 48.Cogswell J.P., Ward J., Taylor I.A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C., et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 49.Hebert S.S., Horre K., Nicolai L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boissonneault V., Plante I., Rivest S., Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse }beta{-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert S.S., Horre K., Nicolai L., Bergmans B., Papadopoulou A.S., Delacourte A., De Strooper B. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol. Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Savas J.N., Makusky A., Ottosen S., Baillat D., Then F., Krainc D., Shiekhattar R., Markey S.P., Tanese N. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jagannath A., Wood M.J. Localization of double-stranded small interfering RNA to cytoplasmic processing bodies is Ago2 dependent and results in up-regulation of GW182 and Argonaute-2. Mol. Biol. Cell. 2009;20:521–529. doi: 10.1091/mbc.E08-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 55.Johnson R., Teh C.H., Kunarso G., Wong K.Y., Srinivasan G., Cooper M.L., Volta M., Chan S.S., Lipovich L., Pollard S.M., et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Packer A.N., Xing Y., Harper S.Q., Jones L., Davidson B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y., Samaco R.C., Gatchel J.R., Thaller C., Orr H.T., Zoghbi H.Y. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat. Neurosci. 2008;11:1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saba R., Goodman C.D., Huzarewich R.L., Robertson C., Booth S.A. A miRNA signature of prion induced neurodegeneration. PLoS ONE. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen T., Olsen L., Lindow M., Jakobsen K.D., Ullum H., Jonsson E., Andreassen O.A., Djurovic S., Melle I., Agartz I., et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark K.L., Xu B., Bagchi A., Lai W.S., Liu H., Hsu R., Wan X., Pavlidis P., Mills A.A., Karayiorgou M., et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 61.Beveridge N.J., Tooney P.A., Carroll A.P., Gardiner E., Bowden N., Scott R.J., Tran N., Dedova I., Cairns M.J. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 62.Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 63.Rose S.D., Kim D.H., Amarzguioui M., Heidel J.D., Collingwood M.A., Davis M.E., Rossi J.J., Behlke M.A. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiba Y., Masuda H., Watanabe N., Ego T., Takagaki K., Ishiyama K., Ohgi T., Yano J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2’-O-protecting group: structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007;35:3287–3296. doi: 10.1093/nar/gkm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X., Rogoff H.A., Li C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 66.Sano M., Sierant M., Miyagishi M., Nakanishi M., Takagi Y., Sutou S. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 68.Kato Y., Taira K. Expression of siRNA from a single transcript that includes multiple ribozymes in mammalian cells. Oligonucleotides. 2003;13:335–343. doi: 10.1089/154545703322617014. [DOI] [PubMed] [Google Scholar]
- 69.Scherer L.J., Frank R., Rossi J.J. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denti M.A., Rosa A., Sthandier O., De Angelis F.G., Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol. Ther. 2004;10:191–199. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 71.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ely A., Naidoo T., Mufamadi S., Crowther C., Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol. Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 73.Keck K., Volper E.M., Spengler R.M., Long D.D., Chan C.Y., Ding Y., McCaffrey A.P. Rational Design Leads to More Potent RNA Interference Against Hepatitis B Virus: Factors Effecting Silencing Efficiency. Mol. Ther. 2008 doi: 10.1038/mt.2008.273. [Epub ahead of print December 16, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aagaard L.A., Zhang J., von Eije K.J., Li H., Saetrom P., Amarzguioui M., Rossi J.J. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y.P., Haasnoot J., ter Brake O., Berkhout B., Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 77.Zu T., Duvick L.A., Kaytor M.D., Berlinger M.S., Zoghbi H.Y., Clark H.B., Orr H.T. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nuber S., Petrasch-Parwez E., Winner B., Winkler J., von Horsten S., Schmidt T., Boy J., Kuhn M., Nguyen H.P., Teismann P., et al. Neurodegeneration and motor dysfunction in a conditional model of Parkinson's disease. J. Neurosci. 2008;28:2471–2484. doi: 10.1523/JNEUROSCI.3040-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denovan-Wright E.M., Rodriguez-Lebron E., Lewin A.S., Mandel R.J. Unexpected off-targeting effects of anti-huntingtin ribozymes and siRNA in vivo. Neurobiol. Dis. 2008;29:446–455. doi: 10.1016/j.nbd.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G., et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franich N.R., Fitzsimons H.L., Fong D.M., Klugmann M., During M.J., Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington's disease. Mol. Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gary D.S., Davidson A., Milhavet O., Slunt H., Borchelt D.R. Investigation of RNA interference to suppress expression of full-length and fragment human huntingtin. Neuromolecular Med. 2007;9:145–155. doi: 10.1007/BF02685888. [DOI] [PubMed] [Google Scholar]
- 84.Machida Y., Okada T., Kurosawa M., Oyama F., Ozawa K., Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 85.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abdelgany A., Wood M., Beeson D. Allele-specific silencing of a pathogenic mutant acetylcholine receptor subunit by RNA interference. Hum. Mol. Genet. 2003;12:2637–2644. doi: 10.1093/hmg/ddg280. [DOI] [PubMed] [Google Scholar]
- 87.Gonzalez-Alegre P., Miller V.M., Davidson B.L., Paulson H.L. Toward therapy for DYT1 dystonia: allele-specific silencing of mutant TorsinA. Ann. Neurol. 2003;53:781–787. doi: 10.1002/ana.10548. [DOI] [PubMed] [Google Scholar]
- 88.Miller V.M., Xia H., Marrs G.L., Gouvion C.M., Lee G., Davidson B.L., Paulson H.L. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwarz D.S., Ding H., Kennington L., Moore J.T., Schelter J., Burchard J., Linsley P.S., Aronin N., Xu Z., Zamore P.D. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W., Kennington L.A., Rosas H.D., Hersch S., Cha J.H., Zamore P.D., Aronin N. Linking SNPs to CAG repeat length in Huntington's disease patients. Nat. Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 92.Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenberg J., Solomon G.A., Vorster A.A., Heckmann J., Bryer A. Origin of the SCA7 gene mutation in South Africa: implications for molecular diagnostics. Clin. Genet. 2006;70:415–417. doi: 10.1111/j.1399-0004.2006.00680.x. [DOI] [PubMed] [Google Scholar]
- 94.Kubodera T., Yokota T., Ishikawa K., Mizusawa H. New RNAi strategy for selective suppression of a mutant allele in polyglutamine disease. Oligonucleotides. 2005;15:298–302. doi: 10.1089/oli.2005.15.298. [DOI] [PubMed] [Google Scholar]
- 95.Sapru M.K., Yates J.W., Hogan S., Jiang L., Halter J., Bohn M.C. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp. Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 96.Miller V.M., Gouvion C.M., Davidson B.L., Paulson H.L. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie Z., Dong Y., Maeda U., Xia W., Tanzi R.E. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation. J. Biol. Chem. 2007;282:4318–4325. doi: 10.1074/jbc.M609293200. [DOI] [PubMed] [Google Scholar]
- 98.Singer O., Marr R.A., Rockenstein E., Crews L., Coufal N.G., Gage F.H., Verma I.M., Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 99.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 100.An D.S., Qin F.X., Auyeung V.C., Mao S.H., Kung S.K., Baltimore D., Chen I.S. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C., Hughes C.M., Shanmugam K.S., Bhattacharjee A., Meyerson M., et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burchard J., Jackson A.L., Malkov V., Needham R.H., Tan Y., Bartz S.R., Dai H., Sachs A.B., Linsley P.S. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. Position-specific chemical modification of siRNAs reduces ‘off-target’ transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 106.Sioud M. Does the understanding of immune activation by RNA predict the design of safe siRNAs? Front. Biosci. 2008;13:4379–4392. doi: 10.2741/3011. [DOI] [PubMed] [Google Scholar]
- 107.Kleinman M.E., Yamada K., Takeda A., Chandrasekaran V., Nozaki M., Baffi J.Z., Albuquerque R.J., Yamasaki S., Itaya M., Pan Y., et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robbins M.A., Li M., Leung I., Li H., Boyer D.V., Song Y., Behlke M.A., Rossi J.J. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat. Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
- 109.He X., He L., Hannon G.J. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 110.Papagiannakopoulos T., Kosik K.S. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Callis T.E., Wang D.Z. Taking microRNAs to heart. Trends Mol. Med. 2008;14:254–260. doi: 10.1016/j.molmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 112.Michlewski G., Guil S., Semple C.A., Caceres J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Corsten M.F., Miranda R., Kasmieh R., Krichevsky A.M., Weissleder R., Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 115.Obernosterer G., Leuschner P.J., Alenius M., Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee E.J., Baek M., Gusev Y., Brackett D.J., Nuovo G.J., Schmittgen T.D. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hutvagner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fabani M.M., Gait M.J. miR-122 targeting with LNA/2’-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 121.Davis S., Propp S., Freier S.M., Jones L.E., Serra M.J., Kinberger G., Bhat B., Swayze E.E., Bennett C.F., Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed. Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 125.Zeng Y., Yi R., Cullen B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 129.Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 132.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 133.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 134.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 136.MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U.S.A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Nordsell R., Shames D.S., Minna J.D., Corey D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 138.Han J., Kim D., Morris K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 140.Saayman S., Barichievy S., Capovilla A., Morris K.V., Arbuthnot P., Weinberg M.S. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weinberg M.S., Ely A., Barichievy S., Crowther C., Mufamadi S., Carmona S., Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol. Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 142.Kawasaki H., Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rodriguez M.S., Dargemont C., Stutz F. Nuclear export of RNA. Biol. Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 144.Pawlicki J.M., Steitz J.A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Blow M.J., Grocock R.J., van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang W.X., Rajeev B.W., Stromberg A.J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P.T. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y., Lin L., Jin P. The microRNA pathway and fragile X mental retardation protein. Biochim. Biophys. Acta. 2008;1779:702–705. doi: 10.1016/j.bbagrm.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Abu-Elneel K., Liu T., Gazzaniga F.S., Nishimura Y., Wall D.P., Geschwind D.H., Lao K., Kosik K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]