Abstract
INTRODUCTION:
Genetic polymorphisms in apolipoprotein genes may be associated with alteration in lipid profile and susceptibility to gallstone disease.
AIM:
To determine the association between apolipoprotein A1 (APOA1) (−75 guanine [G] to adenine [A] and +83/84 M2+/−, MspI) and apolipoprotein C3 (APOC3) (SstI) polymorphisms with gallstone disease.
METHODS:
MspI polymorphisms of the APOA1 gene and SstI polymorphisms of APOC3 were analyzed in DNA samples of 214 gallstone patients and 322 age- and sex-matched healthy controls. All statistical analyses were performed using SPSS version 11.5 (SPSS, USA) and Arlequin version 2.0 (Arlequin, Switzerland).
RESULTS:
The APOA1 −75 G/A polymorphism was significantly associated with gallstone disease. Patients with the GG genotype (P=0.015) and G allele carriers (P=0.004) had a significantly higher risk of gallstone disease (1.087-fold and 1.561-fold, respectively), whereas patients with AA genotypes (P=0.011) and A allele carriers (P=0.004) were protected (OR 0.230 and 0.641, respectively) against gallstone disease. APOA1 +83 M2+/− and APOC3 SstI polymorphisms were not associated with gallstone disease. Case-control analysis of haplotypes showed a significant association in males only. G-M2+-S1 conferred risk for gallstone disease (P=0.036; OR 1.593, 95% CI 1.029 to 2.464), while A-M2+-S1 was protective (P=0.002; OR 0.370, 95% CI 0.197 to 0.695) against gallstone disease. In APOA1−75-APOA1+83 bilocus haplotypes, G-M2+ was associated (P=0.0001) with very high risk (OR 3.173, 95% CI 1.774 to 5.674) for gallstone disease in males only. APOA1−75-APOC3SstI haplotypes also showed significant association while APOA1+83-APOC3SstI haplotypes showed no association with gallstone disease.
CONCLUSIONS:
The APOA1 −75 G/A polymorphism is associated with gallstone disease and shows sex-specific differences. On the other hand, APOA1 M2+/− and APOC3 SstI polymorphisms may not be associated with gallstone disease. Haplotype analysis is a better predictor of risk for gallstone disease.
Keywords: APOA1-C3haplotype, APOA1 MspIpolymorphism, APOC3 SstIpolymorphism, Gallstone disease, Gene polymorphisms
Abstract
INTRODUCTION :
Les polymorphismes génétiques des gènes apolipoprotéiques peuvent s’associer à une altération du profil lipidique et à une susceptibilité aux calculs biliaires.
OBJECTIF :
Déterminer l’association entre les polymorphismes de l’apolipoprotéine A1 (APOA1) (−75 guanine [G] à adénine [A] et +83/84 M2+/−, MspI) et de l’apolipoprotéine C3 (APOC3) (SstI) et les calculs biliaires.
MÉTHODOLOGIE :
On a analysé les polymorphismes MspI du gène APOA1 et les polymorphismes SstI de l’APOC3 dans des échantillons d’ADN de 214 patients ayant des calculs biliaires et de 322 sujets témoins en santé appariés selon l’âge et le sexe. On a procédé à toutes les analyses statistiques au moyen de SPSS version 11.5 (SPSS, États-Unis) et d’Arlequin, version 2.0 (Arlequin, Suisse).
RÉSULTATS :
Le polymorphisme APOA1–75 G/A s’associait aux calculs biliaires de manière significative. Les patients présentant le génotype GG (P=0,015) et les porteurs de l’allèle G (P=0,004) étaient considérablement plus vulnérables aux calculs biliaires (1,087 et 1,561 fois plus, respectivement), tandis que les patients présentant les génotypes AA (P=0,011) et les porteurs de l’allèle A (P=0,004) étaient protégés des calculs biliaires (risque relatif rapproché [RRR] de 0,230 et 0,641, respectivement). Les polymorphismes APOA1 +83 M2+/− et APOC3 SstI n’étaient pas reliés aux calculs biliaires. Une analyse castémoin des haplotypes a révélé une association significative chez les hommes seulement. La G-M2+-SI conférait un risque de calculs biliaires (P=0,036; RRR 1,593, 95 % IC 1,029 à 2,464), tandis que l’A-M2+-SI protégeait des calculs biliaires (P=0,002; RRR 0,370, 95 % IC 0,197 à 0,695). Dans les haplotypes bilocus APOA1−75-APOA1+83, la G-M2+ s’associait (P=0,0001) à un très haut risque de calculs biliaires (RRR 3,173, 95 % IC 1,774 à 5,674), mais chez les hommes seulement. Les haplotypes APOA1−75-APOC3SstI révélaient également une association importante, tandis que les haplotypes APOA1+83-APOC3SstI n’indiquaient aucune association aux calculs biliaires.
CONCLUSIONS :
Le polymorphisme APOA1–75 G/A s’associe aux calculs biliaires et comporte des différences selon les sexes. Par contre, les polymorphismes APOA1 M2+/− et APOC3 SstI ne sont peut-être pas reliés aux calculs biliaires. Une analyse des haplotypes permet de mieux prédire le risque de calculs biliaires.
Gallstone disease is one of the common causes of abdominal pain, inflammation and infection of the gallbladder and the pancreas. However, long-standing gallstones have also been attributed to carcinoma of the gallbladder (1–3). Extensive data over the past 50 years have shown that gallstones result from complex interactions between genetic and environmental factors.
Environmental factors contribute significantly to gallstone disease. High concordance of cholelithiasis in monozygotic twins (4,5) and clustering of cases in families in whom gallstone disease is diagnosed in childhood (6) have provided evidence for a genetic basis of the disease.
The pathophysiology of gallstone formation is complex. A number of epidemiological surveys have shown an association between altered plasma lipid levels and gallstone disease, especially decreased levels of high-density lipoprotein (HDL) cholesterol (7) and increased levels of both low-density lipoprotein cholesterol (8) and triglycerides (9). Plasma lipid and lipoprotein metabolism is controlled by activities of various enzymes and apolipoproteins (APO), which are the structural components of the lipoproteins (10). One candidate locus that has produced inconsistent linkage and association results for dyslipidemias is the APOA1-C3-A4-A5 gene complex, located on 11q23 (11). APOA1 constitutes a key component of the reverse cholesterol transport process (12). Furthermore, it is an activator of lecithin cholesterol acyltransferase, an enzyme that catalyzes the esterification of cholesterol in plasma (13).
A common variant due to guanine (G) to adenine (A) transition (G/A) has been described 75 base pairs upstream (−75 bp) from the APOA1 gene transcription start site. Several studies have reported that individuals with the A allele, which occurs at a frequency of 0.15 to 0.20 in Caucasian populations, have higher levels of HDL cholesterol and/or APOA1 than individuals that are homozygous for the most common G allele (14,15). Another polymorphism (M2+/−) is present at the +83/84 bp site in the first intron of the APOA1 gene, which is created by a cytosine (C) to thymine (T) (+83 bp) and/or a G/A (+84 bp) transition. Some studies have been performed on the association of the M2+/− polymorphism with lipid traits but the results have not been consistent (16–21).
APOC3 is a major constituent of chylomicrons and very low-density lipoprotein particles. Several DNA polymorphisms have been reported in the APOC3 promoter region (22). These mutations are in linkage disequilibrium with an SstI polymorphism in the 3′ untranslated region (C3238G) (23).
Polymorphisms of these genes may alter the lipid levels in individuals, which may predispose a person to gallstone disease. If alleles that predispose individuals to lipid alterations can be identified, screening for the presence of these alleles may identify a substantial proportion of high-risk individuals. Appropriate monitoring of these individuals, in conjunction with targeted intervention, could then influence the onset of disease.
The present study was undertaken to determine the association between APOA1 MspI polymorphisms and APOC3 SstI polymorphisms with gallstone disease.
METHODS
Subjects
The present study comprised 214 gallstone patients (mean age 44.71±13.20 years) and 322 controls (mean age 43.98±11.46 years). All subjects were from north India. The gallstone patients were recruited among inpatients undergoing cholecystectomy and outpatients attending the clinics of the Department of Gastroenterology, Sanjay Gandhi Postgraduate Institute of Medical Sciences in Lucknow, India. Initially, 350 controls were recruited from the healthy staff members of the institute and the general population of the region. Three hundred twenty-two age- and sex-matched subjects who were found to be negative for gallstone disease (by ultrasound), diabetes mellitus, obesity and other chronic debilitating diseases were included in the study. The study was approved by the local ethical committee of the institute. After informed consent was given, blood was taken in EDTA for analysis of DNA. The genomic DNA was extracted from peripheral blood leukocyte pellets using the standard salting-out method (24).
Genotyping
APOA1 and APOC3 gene fragments that encompassed polymorphisms were amplified by polymerase chain reaction (PCR) in a DNA thermal cycler (DNA Engine PTC-100, MJ Research, Inc, USA). The APOA1 gene for the MspI polymorphism was amplified using the following primers: forward 5′-AGG GAC AGA GCT GAT CCT TGA ACT CTT AAG-3′ and reverse 5′-TTA GGG GAC ACC TAC CCG TCA GGA AGA GCA-3′ (25). For APOC3 SstI polymorphism analysis, the primers used to amplify were: forward 5′-CAT GGT TCC CTA CAG AGG AGT-3′ and reverse 5′-TGA CCT TCC GCA CAA AGC TGT-3′ (26). Each amplification was performed using 100 ng to 300 ng of genomic DNA in a volume of 25 μL using 12.5 pmol of each primer, 200 μM dinucleotide triphosphate, 15 mM magnesium chloride, 100 mM Tris (pH 8.0) and two units of Taq polymerase (Fermantas Inc, USA). DNA templates were initially denatured at 95°C for 3 min; for APOA1 MspI, that procedure was followed by 30 cycles with denaturation at 95°C for 30 s, annealing at 60°C for 45 s, extension at 72°C for 60 s and final extension at 72°C for 5 min. The 435 bp PCR product was digested with five units of restriction enzyme MspI (Fermantas Inc) for 3 h at 37°C. The digested product was run on 15% polyacrylamide gel. Two sites located at −75 bp and +83/+84 bp were present in the amplified fragment. Based on restriction pattern, allelic pattern (M2+, 209 bp and 46 bp; G, 114 bp and 66 bp) was determined.
For the APOC3 SstI polymorphism, genomic DNA was initially denatured at 95°C for 5 min, and cycling conditions included denaturation at 95°C for 60 s, annealing at 59.5°C for 60 s, extension at 72°C for 60 s for 30 cycles and, finally, an extension at 72°C for 5 min. PCR product was digested with 5 U of SstI for a period of 3 h to 5 h and was run on 1.5% agarose gel for genotyping. The S1 allele gives a 596 bp band and the S2 allele gives two bands of 371 bp and 225 bp.
Statistical evaluation
To examine whether the genotype frequencies were in Hardy-Weinberg equilibrium, the χ2 goodness of fit test was used. Haplotype frequencies were determined by the maximum likelihood method, using the expectation maximization algorithm. Pair-wise linkage disequilibrium between each pair of APOA1-C3 loci was analyzed using a likelihood ratio test, whose empirical distribution was obtained by a permutation procedure. The above calculations were performed using Arlequin version 2.0 software (Arlequin, Switzerland).
All other analyses were performed using SPSS version 11.5 (SPSS Inc, USA) in the whole study population and in the male and female populations separately. Genotype and allele frequencies were determined by direct counting and compared by the χ2 test or by Fisher’s exact test.
Logistic regression analysis was used to determine the contribution of genetic polymorphisms to the risk of disease. Models were constructed to obtain ORs for each allele after adjustment for the effect of age, sex and body mass index (BMI).
RESULTS
Table 1 shows age, male to female ratio and BMI in gallstone patients and controls. Differences in age, male to female ratio and BMI were insignificant.
TABLE 1.
Demographic profile of gallstone patients and controls
| Demographic profile | Patients | Controls | P |
|---|---|---|---|
| Age, years (mean ± SD) | 44.71±13.20 | 43.98±11.46 | 0.500 |
| Sex, male : female | 69:145 | 116:206 | 0.368 |
| Body mass index, kg/m2 (mean ± SD) | 22.94±3.90 | 23.13±3.85 | 0.732 |
All genotype distributions were in Hardy-Weinberg equilibrium except M2+/− polymorphisms in the control population. This may be due to a low frequency of homozygote M2–/−. Genotype and allele frequencies of APOA1 −75 G/A and M2+/− MspI polymorphisms in gallstone patients and controls are given in Tables 2 and 3, respectively.
TABLE 2.
APOA1 gene −75 and +83 MspI polymorphism genotype frequencies in gallstone patients and controls
| Genotype |
Total |
Male |
Female |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (n=208), n (%) | Control (n=317), n (%) | P | OR (95% CI) | Patient (n=68), n (%) | Control (n=115), n (%) | P | OR (95% CI) | Patient (n=140), n (%) | Control (n=202), n (%) | P | OR (95% CI) | |
| APOA1 −75 guanine (G) to adenine (A) polymorphism | ||||||||||||
| GG | 135 (64.90) | 172 (54.26) | 0.015 | 1.559 (1.087–2.236) | 51 (75.00) | 61 (53.04) | 0.003 | 2.656 (1.373–5.138) | 84 (60.00) | 111 (54.95) | 0.354 | 1.230 (0.794–1.904) |
| GA | 70 (33.65) | 126 (39.75) | 0.158 | 0.769 (0.534–1.108) | 16 (23.53) | 45 (39.13) | 0.031 | 0.479 (0.244–0.939) | 54 (38.57) | 81 (40.10) | 0.776 | 0.938 (0.603–1.459) |
| AA | 3 (1.44) | 19 (5.99) | 0.011 | 0.230 (0.067–0.786) | 1 (1.47) | 9 (7.83) | 0.094 | 0.176 (0.022–1.419) | 2 (1.43) | 10 (4.95) | 0.133 | 0.278 (0.060–1.290) |
| APOA1 M2+/− | ||||||||||||
| M2+/+ | 192 (92.31) | 287 (90.54) | 0.483 | 1.254 (0.666–2.364) | 65 (95.59) | 103 (89.57) | 0.151 | 2.524 (0.686–9.287) | 127 (90.71) | 184 (91.09) | 0.905 | 0.956 (0.452–2.020) |
| M2+/− | 15 (7.21) | 26 (8.20) | 0.679 | 0.870 (0.449–1.685) | 3 (4.41) | 10 (8.70) | 0.377 | 0.485 (0.129–1.826) | 12 (8.57) | 16 (7.92) | 0.829 | 1.090 (0.499–2.381) |
| M2−/− | 1 (0.48) | 4 (1.26) | 0.653 | 0.378 (0.042–3.406) | 0 (0.00) | 2 (1.74) | 0.530 | – | 1 (0.71) | 2 (0.99) | 1.000 | 1.130 (0.505–2.528) |
−75 75 base pairs upstream; APOA1 Apolipoprotein A1;
TABLE 3.
APOA1 gene −75 and +83 MspI polymorphism allele frequencies in gallstone patients and controls
| Allele |
Total |
Male |
Female |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (n=416)*, n (%) | Control (n=634)*, n (%) | P | OR (95% CI) | Patient (n=136)*, n (%) | Control (n=230)*, n (%) | P | OR (95% CI) | Patient (n=280)*, n (%) | Control (n=404)*, n (%) | P | OR (95% CI) | |
| APOA1 −75 guanine (G) to adenine (A) polymorphism | ||||||||||||
| G | 340 (81.73) | 470 (74.13) | 0.004 | 1.561 (1.150–2.119) | 118 (86.76) | 167 (72.61) | 0.002 | 2.473 (1.392–4.392) | 222 (79.29) | 303 (75.00) | 0.192 | 1.276 (0.884–1.840) |
| A | 76 (18.27) | 164 (25.87) | 0.004 | 0.641 (0.472–0.870) | 18 (13.24) | 63 (27.39) | 0.002 | 0.404 (0.228–0.718) | 58 (20.71) | 101 (25.00) | 0.192 | 0.784 (0.543–1.131) |
| APOA1 M2+/− | ||||||||||||
| M2+ | 399 (95.91) | 600 (94.64) | 0.347 | 1.330 (0.733–2.413) | 133 (97.79) | 216 (93.91) | 0.088 | 2.873 (0.811–10.186) | 266 (95.00) | 384 (95.05) | 0.977 | 0.990 (0.491–1.994) |
| M2− | 17 (4.09) | 34 (5.36) | 0.347 | 0.752 (0.414–1.364) | 3 (2.21) | 14 (6.09) | 0.088 | 0.348 (0.098–1.234) | 14 (5.00) | 20 (4.95) | 0.977 | 1.011 (0.501–2.036) |
Total number of chromosomes. −75 75 base pairs upstream; APOA1 Apolipoprotein A1
Case-control analysis of genotype and allele
Genotype GG was found to be significantly higher (P=0.015; OR 1.559, 95% CI 1.087 to 2.236) in patients than in controls (64.90% versus 54.26%). Contrary to these results, the AA genotype was significantly lower (P=0.011) in patients than in controls (1.44% versus 5.99%) and was found to be protective (OR 0.230, 95% CI 0.067 to 0.786) against the disease (Table 2). After data were stratified based on sex, the GG genotype was found to pose a risk for the disease in males only (P=0.003; OR 2.656, 95% CI 1.373 to 5.138). In females, the GG genotype was not associated with the disease. The heterozygote GA genotype showed an association (P=0.031; OR 0.479, 95% CI 0.244 to 0.939) with the disease in the males, and the frequency of the GA genotype was significantly higher in controls than in patients (39.13% versus 23.53%). Although the frequency of the AA genotype was higher in controls than in patients, both in males and in females, the difference was not statistically significant.
The frequency of the G allele was significantly higher in gallstone patients (P=0.004; OR 1.561, 95% CI 1.150 to 2.119) than in controls (Table 3). Alternatively, the A allele was found to be protective against the disease (OR 0.641, 95% CI 0.472 to 0.870). The G allele (P=0.002; OR 2.473, 95% CI 1.392 to 4.392) was associated with high risk in males. In females, frequency was not very different between patients and controls.
A comparison of frequencies between patients and controls indicated that the APOA1 M2+/− polymorphism was not distributed differently at the genotype level. Study subjects were then analyzed separately in the male and female populations, but no association was observed (Table 2). Analysis at the allele level also showed that frequencies were almost similar between patients and controls (Table 3). When data were further stratified according to sex, it was observed that in males, the frequency of the M2+ allele was higher (97.79% versus 93.91%) and the frequency of the M2− allele was lower (2.21% versus 6.09%) in patients than in controls, but the difference was not statistically significant (P=0.088).
Table 4 shows the APOC3 SstI polymorphism genotype and allele frequencies in patients and in the control group. The S1 and S2 alleles were defined based on the absence or presence, respectively, of the SstI restriction site. In patients, the frequency of the S1S1 genotype was lower (48.02% versus 55.38%) and the frequency of the S1S2 genotype was higher (43.07% versus 37.03%) than in controls, but the differences were not statistically significant. Genotype S2S2 showed an almost similar frequency in both groups. Sex-based stratification of the study population revealed that the APOC3 SstI polymorphism was not associated with gallstone disease in either sex (Table 4).
TABLE 4.
APOC3 SstI polymorphism genotype and allele frequencies in total subjects and stratified in male and female patients and controls
| Genotype |
Total |
Male |
Female |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (n=202), n (%) | Control (n=316), n (%) | P | OR (95% CI) | Patient (n=65), n (%) | Control (n=113), n (%) | P | OR (95% CI) | Patient (n=137), n (%) | Control (n=203), n (%) | P | OR (95% CI) | |
| S1S1 | 97 (48.02) | 175 (55.38) | 0.102 | 0.744 (0.522–1.061) | 30 (46.15) | 66 (58.41) | 0.114 | 0.610 (0.330–1.129) | 67 (48.91) | 109 (53.69) | 0.386 | 0.825 (0.535–1.274) |
| S1S2 | 87 (43.07) | 117 (37.03) | 0.170 | 1.287 (0.898–1.845) | 29 (44.62) | 37 (32.74) | 0.114 | 1.655 (0.884–3.099) | 58 (42.34) | 80 (39.41) | 0.590 | 1.129 (0.727–1.754) |
| S2S2 | 18 (8.91) | 24 (7.59) | 0.593 | 1.190 (0.629–2.254) | 6 (9.23) | 10 (8.85) | 0.932 | 1.047 (0.362–3.028) | 12 (8.76) | 14 (6.90) | 0.526 | 1.296 (0.580–2.894) |
| Allele |
Total |
Male |
Female |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (n=404*), n (%) | Control (n=632*), n (%) | P | OR (95% CI) | Patient (n=130*), n (%) | Control (n=226*), n (%) | P | OR (95% CI) | Patient (n=274*), n (%) | Control (n=406*), n (%) | P | OR (95% CI) | |
| S1 | 281 (69.55) | 467 (73.89) | 0.128 | 0.807 (0.612–1.064) | 89 (68.46) | 169 (74.78) | 0.199 | 0.732 (0.455–1.179) | 192 (70.07) | 298 (73.40) | 0.343 | 0.849 (0.604–1.192) |
| S2 | 123 (30.45) | 165 (26.11) | 0.128 | 1.239 (0.940–1.633) | 41 (31.54) | 57 (25.22) | 0.199 | 1.399 (0.848–2.199) | 82 (29.93) | 108 (26.60) | 0.343 | 1.179 (0.839–1.655) |
Total number of chromosomes. APOC3 Apolipoprotein C-3; S1 Allele with no SstI restriction site; S2 Allele with an SstI restriction site
Frequency of the S1 allele was lower and frequency of the S2 allele was higher in patients than in controls (Table 4). Stratification of the study population into male and female categories showed the same trends of frequency distribution in patients and controls, but the difference was not statistically significant.
Case-control analysis of haplotype
APOA1−75-APOA1+83-APOC3 haplotype:
Haplotypes were constructed for all three polymorphisms present at the APOA1-C3 loci and for all possible combinations of two polymorphisms. APOA1−75-APOA1+83-APOC3SstI haplotypes were constructed in 197 gallstone patients and 312 controls. Case-control analysis in the unstratified population indicated that A-M2+-S1 (P=0.005) was protective against the disease (OR 0.629, 95% CI 0.45 to 0.87). Haplotypes were also constructed separately for the male and female populations (Table 5). In males, G-M2+-S1 was associated with a high risk (P=0.036; OR 1.593, 95% CI 1.029 to 2.464) for developing gallstone disease. A-M2+-S1 was also significantly (P=0.002; OR 0.370, 95% CI 0.197 to 0.695) associated with the disease. Haplotypes A-M2−-S1 and A-M2+-S2 were present only in controls, and haplotype A-M2−-S2 was absent in males and in female controls also. In females, G-M2−-S2 was absent in both patients and controls, and no other haplotype frequency showed a significant difference between patients and controls.
TABLE 5.
APOA1-C3 haplotype frequencies in gallstone patients and controls
| Haplotype, males | Patients (frequency), n=128 | Controls (frequency), n=226 | P | OR |
95% CI |
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| G-M2+-S1 | 0.56 | 0.44 | 0.04 | 1.59 | 1.03 | 2.46 |
| G-M2+-S2 | 0.31 | 0.23 | 0.11 | 1.49 | 0.92 | 2.42 |
| G-M2−-S1 | 0.02 | 0.04 | 0.37 | 0.55 | 0.15 | 2.05 |
| G-M2−-S2 | 0.00 | 0.01 | 0.35 | – | – | – |
| A-M2+-S1 | 0.11 | 0.25 | 0.00 | 0.37 | 0.20 | 0.69 |
| A-M2+-S2 | 0.00 | 0.01 | 0.15 | – | – | – |
| A-M2−-S1 | 0.00 | 0.02 | 0.19 | – | – | – |
| A-M2−-S2 | 0.00 | 0.00 | – | – | – | – |
| Haplotype, females | Patients (frequency), n=266 | Controls (frequency), n=398 | P | OR |
95% CI |
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| G-M2+-S1 | 0.476 | 0.466 | 0.803 | 1.040 | 0.762 | 1.420 |
| G-M2+-S2 | 0.287 | 0.249 | 0.274 | 1.215 | 0.857 | 1.724 |
| G-M2−-S1 | 0.039 | 0.037 | 0.923 | 1.041 | 0.462 | 2.346 |
| G-M2−-S2 | 0.000 | 0.000 | – | – | – | – |
| A-M2+-S1 | 0.185 | 0.220 | 0.274 | 0.805 | 0.545 | 1.188 |
| A-M2+-S2 | 0.011 | 0.018 | 0.492 | 0.622 | 0.159 | 2.438 |
| A-M2−-S1 | 0.000 | 0.011 | 0.091 | – | – | – |
| A-M2−-S2 | 0.003 | 0.000 | 0.286 | – | – | – |
A Adenine; APOA1-C3 Apolipoprotein A1-C3; G Guanine; S1 Allele with no SstI restriction site; S2 Allele with an SstI restriction site
APOA1−75-APOA1+83 haplotype:
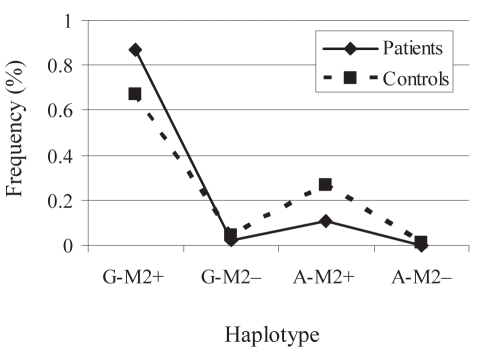
In the APOA1−75-APOA1+83 bilocus haplotype, G-M2+ and A-M2+ showed a significant (P=0.001 and P=0.003, respectively) difference in the frequency between patients and controls. G-M2+ was found to impose risk (OR 1.665, 95% CI 1.236 to 2.244), whereas A-M2+ was protective (OR 0.620, 95% CI 0.451 to 0.854) against the disease. Analysis of the male population revealed that G-M2+ was associated (P=0.0001) with a very high risk (OR 3.173, 95% CI 1.774 to 5.674) for gallstone disease (Figure 1). Haplotype A-M2+ was protective (P=0.001; OR 0.340, 95% CI 0.182 to 0.639) against the disease. In females, no haplotype was significantly associated with gallstone disease.
Figure 1).
Apolipoprotein A1 (APOA1) guanine (G) to adenine (A)-APOC3 SstI haplotype frequency in gallstone patients and controls in males. Continuous line represents the frequency in patients and the dotted line shows the frequency in controls. G-M2+ haplotype showed a remarkably higher frequency in patients (0.867 versus 0.673) than in controls and conferred very high risk (P=0.0001, OR 3.173, 95% CI 1.774 to 5.674)
APOA1−75-APOC3SstI haplotype:
Analysis of the APOA1−75-APOC3SstI loci in the total population showed that A-S1 (P=0.002) was protective (OR 0.601, 95% CI 0.435 to 0.832) against gallstone disease. When analyzed separately for males and females, this haplotype was protective (P=0.001; OR 0.345, 95% CI 0.184 to 0.647) against the disease in males only. In females, no haplotype showed a significantly different distribution of frequency between patients and controls.
APOA1+83-APOC3SstI haplotype: In the total population, all APOA1+83-APOC3SstI haplotypes showed almost similar frequencies in patients and controls. Separate analyses in male and female populations also did not reveal any difference (data not shown).
Risk assessment
To determine the contribution of different alleles to the risk of gallstone disease, logistic regression analysis was used (Table 6). Different models were constructed to determine the contribution of risk alleles alone and in association with environmental factors for all three polymorphisms. The APOA1 G allele gave 1.4% variance when adjusted for age and sex, and with inclusion of BMI in the model, it predicted 1.6% variance for gallstone disease. In the APOA1 M2+/− polymorphism, the M2+ allele, and in the APOC3 SstI polymorphism, the S2 allele did not show any significant risk, even after adjustment for the covariates of age, sex and BMI.
TABLE 6.
Adjusted ORs for gallstone disease according to alleles
| Polymorphism | Model | P | OR | 95% CI | R2×100 |
|---|---|---|---|---|---|
| APOA1 −75 G/A | G allele + age + sex | 0.004 | 1.562 | 1.150–2.122 | 1.4 |
| G allele + age + sex + BMI | 0.004 | 1.559 | 1.148–2.118 | 1.6 | |
| APOA1 M2+/− | M2+ allele + age + sex | 0.371 | 1.314 | 0.723–2.386 | 0.5 |
| M2+ allele + age + sex + BMI | 0.335 | 1.343 | 0.738–2.445 | 0.6 | |
| APOC3 SstI | S2 allele + age + sex | 0.123 | 1.243 | 0.943–1.640 | 0.7 |
| S2 allele + age + sex + BMI | 0.116 | 1.249 | 0.947–1.649 | 0.7 |
−75 75 base pairs upstream; APOA1 Apolipoprotein A1; APOC3 Apolipoprotein C-3; BMI Body mass index; G/A Guanine (G) to adenine (A) polymorphism; R2×100 Part of variance explained by predictors
DISCUSSION
Gallstone disease is a major problem in India and, with privileged circumstances and urbanization in rural villages, its rate may increase in the future. Alterations in lipids have been associated with disease but the mechanism is unknown. Genetic association studies in humans have the statistical power to reveal the contribution of risk alleles to a polygenic disease like gallstone formation. The low HDL cholesterol and high triglyceride concentrations reported in gallstone patients led us to search for a possible association between genetic polymorphisms present on APOA1-C3 loci and gallstone disease. Juvonen et al (27) studied the association between APOA1 MspI and gallstone disease in Finland, but no significant association was reported.
The present study is, to the authors’ knowledge, the first to report on the association of APOA1-C3 MspI and SstI polymorphisms with gallstone disease in the Indian population. The most important finding of this study was that a −75 G/A polymorphism is independently associated with gallstone disease. A model that included only genetic polymorphisms revealed that an APOA1 −75 G/A polymorphism alone could explain 1.1% of cases, suggesting a significant contribution of this polymorphism to genetic risk. This polymorphism was definitely a risk factor for gallstone disease because an adjusted OR of the G allele for the environmental factors of age, sex and BMI also indicated a risk for the disease.
Several studies have reported that individuals with the A allele have higher levels of HDL cholesterol and/or APOA1 (14,15,21,28,29). Therefore, APOA1 −75 G allele carriers may have low levels of HDL cholesterol or high triglyceride levels. The elevated triglycerides and reduced HDL cholesterol have been associated with gallstone disease (7,9). To determine whether an APOA1 −75 G/A polymorphism determines lipid levels, plasma levels of total cholesterol, HDL cholesterol, triglycerides and low-density lipoprotein cholesterol were analyzed in 156 controls. However, in various genotypes of the APOA1 −75 G allele polymorphism, lipid levels were not significantly different (data not shown). Therefore, the APOA1 −75 G/A polymorphism may not affect gallstone development, due to altered levels of circulating lipids. Another mechanism might be associated with the high risk for gallstone disease in G allele carriers or protection in A allele carriers. It has been reported that APOA1 and APOA2 retard the nucleation of cholesterol monohydrate crystals and are potential antinucleating factors (30). This G to A substitution is between the CACAT sequence and the TAAATA box of the transcription start site of APOA1 and creates a 6 bp perfect repeat (CAGGGC) which has homology to known nuclear-protein binding sites (31,32). On the other hand, studies surrounding transcription rates have associated the APOA1 A allele with a four- to sevenfold increase in the transcription rate of APOA1 in vitro (28,33). Therefore, we hypothesize that in APOA1 A allele carriers, the level of APOA1 protein is increased, which acts as an antinucleating agent and provides protection against gallstone formation.
Sex-specific differences were clearly seen in the distribution of the −75 G/A polymorphism between gallstone patients and controls, and the association was observed only in males. A meta-analysis has also shown a more apparent effect of this polymorphism on lipids in male subjects (14). This sex-specific difference can be attributed to the interaction between sex-specific hormones and genetic variants. To date, there is no study showing the molecular mechanism of gene-hormone interactions in gallstone disease.
The present study also considered the APOA1 M2+/− polymorphism in gallstone patients for the first time. Though this polymorphism was not associated with gallstone disease in the present study, trends toward an association were observed. A larger sample size may reveal a significant association with gallstone disease.
Another widely studied polymorphism of the APOA1-C3 cluster is the SstI polymorphism in the 3′ untranslated region of APOC3. The S2 allele has also been associated with elevated triacylglycerol, cholesterol and APOC3 concentration, and increased coronary artery disease risk (34–36). In our study, the SstI polymorphism in APOC3 was not found to be associated with gallstone disease. Regression analysis for adjustment of environmental factors only revealed trends toward an association (Table 6). A recent study using twins also reported a significant contribution of environmental factors to the development of gallstone disease (5).
In gallstone disease, multiple candidate genes are proposed (37) and each gene may have multiple genetic variants. Some alleles may provide protection and others may be associated with a higher risk for gallstone disease. To estimate the overall risk for gallstone disease in an individual, it is pertinent to analyze all genetic polymorphisms simultaneously. Therefore, the haplotype analysis is more accurate than the genotype or allele analyses. It was found that haplotype frequencies of APOA1-C3 in males were significantly different between patients and controls (Table 5). The combined effect of all three polymorphisms of APOA1-C3 suggests that the major contribution is from the APOA1 −75 G/A polymorphism. However, a significant association of APOA1 haplotype (G-M2+) with gallstone disease was observed, where measures of association (P=0.0001, OR 3.173) were remarkably higher (Table 5) than the APOA1 −75 G/A polymorphism alone. Although APOA1 M2+/− was not significantly associated with disease it did show a weak association (P=0.088). This indicates that the APOA1 M2+/− polymorphism also contributes toward risk, and studies in large populations might reveal this.
By virtue of the APOA1 −75 G/A polymorphism, which was significantly associated with gallstone disease, the maximum variance observed in the present study was 1.6%, which indicates that other genetic factors also contribute to the risk of developing gallstone disease. A recent study involving twins detected a 25% variance for heritability (5). The genes involved in gallbladder motility (cholecystokinin A receptor and cholecystokinin), cholesterol synthesis (3-hydroxy-3-methylglutaryl-coenzyme A reductase), bile acid synthesis (cholesterol 7-α hydroxylase) and mucins are important candidates for genetic studies in gallstone disease. Identification of all risk alleles will provide a definite contribution of risk due to genetic factors. Because this is the first study of its kind, additional studies in different populations with a large sample size are required for confirmation of the present study’s results.
CONCLUSIONS
The APOA1 −75 G/A polymorphism is associated with gallstone disease and shows sex-specific differences. APOA1 M2+/− and APOC3 SstI polymorphisms may not be associated with gallstone disease. Haplotype analysis is a better predictor of risk for gallstone disease than the genotype or allele analyses.
Acknowledgments
The authors acknowledge the Indian Council of Medical Research, New Delhi, for financially supporting this study.
REFERENCES
- 1.Lowenfels AB, Lindstrom CG, Conway MJ, Hastings PR. Gallstones and risk of gallbladder cancer. J Natl Cancer Inst. 1985;75:77–80. [PubMed] [Google Scholar]
- 2.Lowenfels AB, Walker AM, Althaus DP, Townsend G, Domellof L. Gallstone growth, size, and risk of gallbladder cancer: An interracial study. Int J Epidemiol. 1989;18:50–4. doi: 10.1093/ije/18.1.50. [DOI] [PubMed] [Google Scholar]
- 3.Maringhini A, Moreau JA, Melton LJ, III, Hench VS, Zinsmeister AR, DiMagno EP. Gallstones, gallbladder cancer, and other gastrointestinal malignancies: An epidemiologic study in Rochester, Minnesota. Ann Intern Med. 1987;107:30–5. doi: 10.7326/0003-4819-107-1-30. [DOI] [PubMed] [Google Scholar]
- 4.Harvald B, Hauge M. A catamnestic investigation of Danish twins; a preliminary report. Dan Med Bull. 1956;3:150–8. [PubMed] [Google Scholar]
- 5.Katsika D, Grjibovski A, Einarsson C, Lammert F, Lichtenstein P, Marschall HU. Genetic and environmental influences on symptomatic gallstone disease: A Swedish study of 43,141 twin pairs. Hepatology. 2005;41:1138–43. doi: 10.1002/hep.20654. [DOI] [PubMed] [Google Scholar]
- 6.Hagberg B, Svennerholm L, Thoren L. Cholelithiasis in childhood: A follow-up study with special reference to hereditary, constitutional factors and serum lipids. Acta Chir Scand. 1962;123:307–15. [PubMed] [Google Scholar]
- 7.Scragg RK, Calvert GD, Oliver JR. Plasma lipids and insulin in gallstone disease: A case-control study. Br Med J (Clin Res Ed) 1984;289:521–5. doi: 10.1136/bmj.289.6444.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr GC, Kritz-Silverstein D, Barrette-Connor E. Plasma lipids and gallbladder disease. Am J Epidemiol. 1991;134:78–85. doi: 10.1093/oxfordjournals.aje.a115995. [DOI] [PubMed] [Google Scholar]
- 9.Thijs C, Knipschild P, Brombacher P. Serum lipid and gallstones: A case-control study. Gastroenterology. 1990;99:843–9. doi: 10.1016/0016-5085(90)90978-a. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. Lipoprotein receptors in the liver. Control signals for plasma cholesterol traffic. J Clin Invest. 1983;72:743–7. doi: 10.1172/JCI111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnon F, Jarvik GP, Motulsky AG, Deeb SS, Brunzell JD, Wijsman EM. Evidence of linkage of HDL level variation to APOC3 in two samples with different ascertainment. Hum Genet. 2003;113:522–33. doi: 10.1007/s00439-003-1006-5. [DOI] [PubMed] [Google Scholar]
- 12.Reichl D, Miller NE. Pathophysiology of reverse cholesterol transport. Insights from inherited disorders of lipoprotein metabolism. Arteriosclerosis. 1989;9:785–97. doi: 10.1161/01.atv.9.6.785. [DOI] [PubMed] [Google Scholar]
- 13.Fielding CJ, Shore VG, Fielding PE. A protein cofactor of lecithin: Cholesterol acyltransferase. Biochem Biophys Res Commun. 1972;46:1493–8. doi: 10.1016/0006-291x(72)90776-0. [DOI] [PubMed] [Google Scholar]
- 14.Juo SH, Wyszynski DF, Beaty TH, Huang HY, Bailey-Wilson JE. Mild association between the A/G polymorphism in the promoter of the apolipoprotein A-I gene and apolipoprotein A-I levels: A meta-analysis. Am J Med Genet. 1999;82:235–41. [PubMed] [Google Scholar]
- 15.Ordovas JM. The genetics of serum lipid responsiveness to dietary interventions. Proc Nutr Soc. 1999;58:171–87. doi: 10.1079/pns19990023. [DOI] [PubMed] [Google Scholar]
- 16.Wang XL, Badenhop R, Humphrey KE, Wilcken DE. New MspI polymorphism at +83 bp of the human apolipoprotein AI gene: Association with increased circulating high density lipoprotein cholesterol levels. Genet Epidemiol. 1996;13:1–10. doi: 10.1002/(SICI)1098-2272(1996)13:1<1::AID-GEPI1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17.Wang XL, Liu SX, McCredie RM, Wilcken DE. Polymorphisms at the 5′-end of the apolipoprotein AI gene and severity of coronary artery disease. J Clin Invest. 1996;98:372–7. doi: 10.1172/JCI118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmena-Ramon RF, Ordovas JM, Ascaso JF, Real J, Priego MA, Carmena R. Influence of genetic variation at the apo A-I gene locus on lipid levels and response to diet in familial hypercholesterolemia. Atherosclerosis. 1998;139:107–13. doi: 10.1016/s0021-9150(98)00064-1. [DOI] [PubMed] [Google Scholar]
- 19.Reguero JR, Cubero GI, Batalla A, et al. Apolipoprotein AI gene polymorphisms and risk of early coronary heart disease. Cardiology. 1998;90:231–5. doi: 10.1159/000006849. [DOI] [PubMed] [Google Scholar]
- 20.Pulkkinen A, Viitanen L, Kareinen A, Lehto S, Laakso M. MspI polymorphism at +83 bp in intron 1 of the human apolipoprotein AI gene is associated with elevated levels of HDL cholesterol and apolipoprotein AI in nondiabetic subjects but not in type 2 diabetic patients with coronary heart disease. Diabetes Care. 2000;23:791–5. doi: 10.2337/diacare.23.6.791. [DOI] [PubMed] [Google Scholar]
- 21.Zou Y, Hu D, Yang X, et al. Relationships among apolipoprotein A1 gene polymorphisms, lipid levels and coronary atherosclerosis disease. Chin Med J (Engl) 2003;116:665–8. [PubMed] [Google Scholar]
- 22.Moll PP, Michel VV, Weidman WH, Kottke BA. Genetic determination of plasma apolipoprotein AI in a population-based sample. Am J Hum Genet. 1989;44:124–39. [PMC free article] [PubMed] [Google Scholar]
- 23.Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3′ untranslated region polymorphisms. Proc Natl Acad Sci USA. 1993;90:4562–6. doi: 10.1073/pnas.90.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson IA, Ordovas JM, Barnard JR, et al. Effect of apolipoprotein A-I genetic variations on plasma apolipoprotein, serum lipoprotein and glucose levels. Clin Genet. 2002;61:176–84. doi: 10.1034/j.1399-0004.2002.610302.x. [DOI] [PubMed] [Google Scholar]
- 26.Waterworth DM, Ribalta J, Nicaud V, Dallongeville J, Humphries SE, Talmud P. ApoCIII gene variants modulate postprandial response to both glucose and fat tolerance tests. Circulation. 1999;99:1872–7. doi: 10.1161/01.cir.99.14.1872. [DOI] [PubMed] [Google Scholar]
- 27.Juvonen T, Savolainen MJ, Kairaluoma MI, Lajunen LH, Humphries SE, Kesaniemi YA. Polymorphisms at the apoB, apoA-I, and cholesteryl ester transfer protein gene loci in patients with gallbladder disease. J Lipid Res. 1995;36:804–12. [PubMed] [Google Scholar]
- 28.Jeenah M, Kessling A, Miller N, Humphries S. G to A substitution in the promoter region of the apolipoprotein AI gene is associated with elevated serum apolipoprotein AI and high density lipoprotein cholesterol concentrations. Mol Biol Med. 1990;7:233–41. [PubMed] [Google Scholar]
- 29.Pagani F, Sidoli A, Giudici GA, Barenghi L, Vergani C, Baralle FE. Human apolipoprotein A-I gene promoter polymorphism: Association with hyperalphalipoproteinemia. J Lipid Res. 1990;31:1371–7. [PubMed] [Google Scholar]
- 30.Kibe A, Holzbach RT, LaRusso NF, Mao SJ. Inhibition of cholesterol crystal formation by apolipoproteins in supersaturated model bile. Science. 1984;225:514–6. doi: 10.1126/science.6429856. [DOI] [PubMed] [Google Scholar]
- 31.Jones KA, Yamamoto KR, Tjian R. Two distinct transcription factors bide to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–72. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 32.Kadonaga JT, Jones KA, Tjian R. Promoter-specific activation of RNA polymerase-II transcription by SP1. Trends Biochem Sci. 1986;11:20–3. [Google Scholar]
- 33.Angotti E, Mele E, Costanzo F, Avvedimento EV. A polymorphism (G→A transition) in −78 position of apolipoprotein A-I promoter increases transcription efficiency. J Biol Chem. 1994;269:17371–4. [PubMed] [Google Scholar]
- 34.Shoulders CC, Harry PJ, Lagrost L, et al. Variation at the apo AI/CIII/AIV gene complex is associated with elevated plasma levels of apo CIII. Atherosclerosis. 1991;87:239–47. doi: 10.1016/0021-9150(91)90026-y. [DOI] [PubMed] [Google Scholar]
- 35.Talmud PJ, Humphries SE. Apolipoprotein C-III gene variation and dyslipidaemia. Curr Opin Lipidol. 1997;8:154–8. doi: 10.1097/00041433-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Surguchov AP, Page GP, Smith L, Patsch W, Boerwinkle E. Polymorphic markers in apolipoprotein C-III gene flanking regions and hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 1996;16:941–7. doi: 10.1161/01.atv.16.8.941. [DOI] [PubMed] [Google Scholar]
- 37.Lammert F, Carey MC, Paigen B. Chromosomal organization of candidate genes involved in cholesterol gallstone formation: A murine gallstone map. Gastroenterology. 2001;120:221–38. doi: 10.1053/gast.2001.20878. [DOI] [PubMed] [Google Scholar]